Abstract
The genetic and physiological mechanisms of aluminum (Al) tolerance have been well studied in certain cereal crops, and Al tolerance genes have been identified in sorghum (Sorghum bicolor) and wheat (Triticum aestivum). Rice (Oryza sativa) has been reported to be highly Al tolerant; however, a direct comparison of rice and other cereals has not been reported, and the mechanisms of rice Al tolerance are poorly understood. To facilitate Al tolerance phenotyping in rice, a high-throughput imaging system and root quantification computer program was developed, permitting quantification of the entire root system, rather than just the longest root. Additionally, a novel hydroponic solution was developed and optimized for Al tolerance screening in rice and compared with the Yoshida's rice solution commonly used for rice Al tolerance studies. To gain a better understanding of Al tolerance in cereals, comparisons of Al tolerance across cereal species were conducted at four Al concentrations using seven to nine genetically diverse genotypes of wheat, maize (Zea mays), sorghum, and rice. Rice was significantly more tolerant than maize, wheat, and sorghum at all Al concentrations, with the mean Al tolerance level for rice found to be 2- to 6-fold greater than that in maize, wheat, and sorghum. Physiological experiments were conducted on a genetically diverse panel of more than 20 rice genotypes spanning the range of rice Al tolerance and compared with two maize genotypes to determine if rice utilizes the well-described Al tolerance mechanism of root tip Al exclusion mediated by organic acid exudation. These results clearly demonstrate that the extremely high levels of rice Al tolerance are mediated by a novel mechanism, which is independent of root tip Al exclusion.
Aluminum (Al) is the most abundant metal in the earth's crust, constituting approximately 7% of the soil (Wolt, 1994). Al is predominately found as a key component of soil clays; however, under highly acidic soil conditions (pH < 5.0), Al3+ is solubilized into the soil solution and is highly phytotoxic. Al3+ causes a rapid inhibition of root growth that leads to a reduced and stunted root system, thus having a direct effect on the ability of a plant to acquire both water and nutrients. Approximately 30% of the world's total land area and over 50% of potentially arable lands are acidic, with the majority (60%) found in the tropics and subtropics (von Uexkull and Mutert, 1995). Thus, acidic soils are a major limitation to crop production, particularly in the developing world.
As a whole, cereal crops (Poaceae) provide an excellent model for studying Al tolerance because of their abundant genetic resources, large, active research communities, and importance to agriculture. In addition, work in one cereal species can rapidly translate into impact throughout the family. Previous research has focused on understanding the genetic and physiological mechanisms of Al tolerance in maize (Zea mays), sorghum (Sorghum bicolor), and wheat (Triticum aestivum). The most recognized physiological mechanism conferring Al tolerance in plants involves exclusion of Al from the root tip (Miyasaka et al., 1991; Delhaize and Ryan, 1995; Kochian, 1995; Kochian et al., 2004a, 2004b). The exclusion mechanism is primarily mediated by Al-activated exudation of organic acids such as malate, citrate, or oxalate from the root apex, the site of Al toxicity (Ryan et al., 1993, 2001; Ma et al., 2001). These organic acids chelate Al in the rhizosphere, reducing the concentration and toxicity of Al at the growing root tip (Ma et al., 2001). Phosphate has also been identified as a class of root exudates involved in cation chelation and therefore can be considered a potential exudate involved in Al exclusion from the root tip (Pellet et al., 1996).
Al-activated malate and citrate anion efflux transporters have been cloned from wheat (ALMT1; Sasaki et al., 2004) and sorghum (SbMATE; Magalhaes et al., 2007), and root citrate efflux transporters have been implicated in Al tolerance in maize (Piñeros and Kochian, 2001; Zhang et al., 2001). Recently, a maize homolog of sorghum SbMATE was shown to be the root citrate efflux transporter that plays a role in maize Al tolerance (Maron et al., 2010). Although organic acids have been shown to play a major role in Al tolerance in these species, another exclusion mechanism has been identified in an Arabidopsis (Arabidopsis thaliana) mutant, where a root-mediated increase in rhizosphere pH lowers the Al3+ activity and thus participates in Al exclusion from the root apex (Degenhardt et al., 1998). Furthermore, there is clear evidence that tolerance in maize cannot be fully explained by organic acid release (Piñeros et al., 2005). These types of findings strongly suggest that multiple Al tolerance mechanisms exist in plants.
Rice (Oryza sativa) has been reported to be the most Al-tolerant cereal crop under field conditions, capable of withstanding significantly higher concentrations of Al than other major cereals (Foy, 1988). Despite this fact, very little is known about the physiological mechanisms of Al tolerance in rice. Two independent studies have identified increased Al accumulation in the root apex in susceptible compared with Al-tolerant rice varieties, but no differences were observed in organic acid exudation or rhizosphere pH (Ma et al., 2002; Yang et al., 2008). These studies suggest that rice may contain novel physiological and/or genetic mechanisms that confer significantly higher levels of Al tolerance than those found in other cereals. A more thorough analysis is required to clarify the mechanism of Al tolerance in rice.
Cultivated rice is characterized by deep genetic divergence between the two major varietal groups: Indica and Japonica (Dally and Second, 1990; Garris et al., 2005; Hu et al., 2006; Londo et al., 2006). Extensive selection pressure over the last 10,000 years has resulted in the formation of five genetically distinct subpopulations: indica and aus within the Indica varietal group, and temperate japonica, tropical japonica, and aromatic/groupV within the Japonica varietal group (Garris et al., 2005; Caicedo et al., 2007; K. Zhao and S. McCouch, personal communication). (Note: When referring to varietal groups, the first letter will be capitalized, while lowercase letters will be used to refer to the subpopulation groups.) Subpopulation differences in trait performance are often significant, particularly with respect to biotic and abiotic stress (Champoux et al., 1995; Lilley et al., 1996; Garris et al. 2003; Xu et al., 2009). This can lead to confusion because trait or performance differences may be confounded with subpopulation structure, leading to false positives (type 1 error; Devlin and Roeder, 1999; Pritchard and Donnelly, 2001; Yu et al., 2006; Zhao et al., 2007). Therefore, it is important to consider the subpopulation origin of genotypes being compared when studying the genetics and physiology of Al tolerance in rice.
Al tolerance screening is typically conducted by comparing root growth of seedlings grown in hydroponic solutions, with and without Al (Piñeros and Kochian, 2001; Magalhaes et al., 2004; Sasaki et al., 2004). Sorghum and maize are often screened for Al tolerance in Magnavaca's nutrient solution (Piñeros and Kochian, 2001; Magalhaes et al., 2004; Piñeros et al., 2005), while rice seedlings are typically grown in Yoshida's solution (Yoshida et al., 1976). Furthermore, Al concentrations used to screen for Al tolerance in maize (222 μm), sorghum (148 μm), and wheat (100 μm) are significantly lower than those used for screening Al tolerance in rice (1,112–1,482 μm; Wu et al., 2000; Nguyen et al., 2001, 2002, 2003). These differences in chemical composition of the nutrient solutions make it difficult to directly compare plant response to Al across these cereals. In rice, the high Al concentrations required to observe significant differences in root growth between susceptible and resistant varieties also complicate Al tolerance screening due to the precipitation of Al along with other elements. The result is that control (−Al) and treatment (+Al) solutions may differ with regard to essential mineral nutrients that react with Al, leading to differences in growth not directly attributable to Al. Additionally, because the active form of Al that is toxic to root growth is Al3+, any Al that precipitates out of solution has no effect on root growth (Kochian et al., 2004a). In a hydroponic solution, Al may be found in one of four forms: (1) as free Al3+, where it actively inhibits root growth; (2) precipitated with other elements and essentially unavailable to inhibit plant growth; (3) different hydroxyl monomers of Al, which are not believed to be toxic to roots (Parker et al., 1988); or (4) complexed with other elements in an equilibrium between its active and inactive states. The degree to which Al inhibits root growth is primarily dependent upon the activity of free Al3+ in solution (Kochian et al., 2004a).
The objectives of this study were to (1) develop and optimize a suitable nutrient solution and high-throughput Al tolerance screening method for rice; (2) quantify and compare differences in Al tolerance between maize, sorghum, wheat, and rice; and (3) use the developed screening methods to determine if rice utilizes the organic acid-mediated Al exclusion mechanism that is observed in maize, sorghum, and wheat.
RESULTS
Optimization of Nutrient Solution Composition for Al Tolerance Screening in Rice
To establish a hydroponic solution for screening Al tolerance in rice seedlings that would allow us to compare levels of tolerance between rice and other cereals, we modified the Magnavaca's nutrient solution (Magnavaca et al., 1987) that has been previously used for maize and sorghum Al tolerance research (Piñeros and Kochian, 2001; Magalhaes et al., 2004; Piñeros et al., 2005). Modifications were made to ensure a sufficient supply of essential nutrients and to minimize the chemical interactions between Al and other mineral species in the nutrient solution at the high Al concentrations needed for rice. Using the chemical speciation program Geochem-EZ (Shaff et al., 2010; http://www.plantmineralnutrition.net/Geochem/geochem%20home.htm) and inductively coupled plasma emission spectrometry (ICP-ES) analysis, we first analyzed the chemical composition of the Yoshida's solution to understand what was causing the visible precipitate that was always observed when Al was supplied at necessarily high concentrations above 1 mm AlCl3. These concentrations have been shown to cause a measurable inhibition of rice root growth (Wu et al., 2000; Nguyen et al., 2001, 2002, 2003). Geochem-EZ predicted that when 1,297 μm AlCl3 was added to Yoshida's solution, it would only result in a free Al3+ activity of 116 μm, with the significant reduction in Al3+ activity due to Al interaction with HPO42−SO42−and citrate (citrate was used as the iron [Fe] chelate). The observed precipitation in the Yoshida's solution was predicted by Geochem-EZ to be due to Al precipitating with the high concentrations of SO42−HPO42− in the solution and Fe precipitating with HPO42−. ICP-ES analysis of Yoshida's control and Al-containing solutions confirmed that, in addition to the differences in Al concentrations, there were significant differences in phosphorus (P) and Fe availability. Chemical analysis of the nutrient solution to determine soluble and precipitated minerals identified that available P was reduced 85.8% (from 321 to 45.6 μm) and available Fe was reduced 85.2% (from 35.8 to 5.3 μm; Table I). Furthermore, 40% of the total Al added to the solution was lost as a precipitate.
Table I. Comparison of total (supplied) and soluble P, Fe, and Al in modified Magnavaca's and Yoshida's solutions.
Total P and Fe are the total concentrations of P and chelated Fe provided in the control (−Al) nutrient solutions. Available P and Fe are the concentrations of soluble P and Fe measured in the Al-containing solutions using ICP-ES after centrifugation to pellet out precipitated P and Fe. %P and %Fe Decrease are the differences in soluble P and Fe concentrations between control and Al-treated nutrient solutions. Total Al is the concentration of Al added to the treatment solution (as AlCl3), and Soluble Al is the amount of soluble Al (not precipitated) in each nutrient solution as determined by centrifugation followed by ICP-ES analysis. %Soluble Al quantifies the percentage of the total Al added that is in a soluble state as determined by chemical analysis. The Al3+ activity values in the last column were predicted using the GEOCHEM-EZ speciation program based on chemical equilibrium constants for each nutrient solution.
| Solution | Total P | Available P | %P Decrease | Total Fe | Available Fe | %Fe Decrease | Total Al | Soluble Al | %Soluble Al | Al3+ Activity (Predicted with GEOCHEM-EZ) |
| μm | μm | % | μm | μm | % | μm | μm | % | μm | |
| Modified Magnavaca | 47.8 | 34.8 | 27.2 | 77 | 68.1 | 11.5 | 540 | 517 | 95.7 | 160 |
| Yoshida | 321 | 45.6 | 85.8 | 35.8 | 5.3 | 85.2 | 1,297 | 775 | 59.7 | 116 |
To address these problems, we developed an optimized nutrient solution, hereafter referred to as modified Magnavaca's solution, which minimizes the concentration of Al necessary to elicit significant levels of root growth inhibition in rice seedlings (Table II). We accomplished this by reducing the ionic strength of the nutrient solution and reducing the interactions between Al and other mineral ions. Geochem-EZ predictions and preliminary plant growth studies suggested that the optimal total concentration of Al required for screening of Al tolerance in rice would be 540 μm AlCl3 (approximately 60% less than Yoshida's solution), yielding an Al3+ activity of 160 μm in the modified Magnavaca's solution. Our solution also contained much lower levels of the ions that most strongly interact with Al3+ (SO42−H2PO4−) and used (2-hydroxyethyl)ethylenediaminetriacetic acid (HEDTA) as the Fe chelate, rather than citrate, which preferentially binds Al over Fe. ICP-ES analysis demonstrated that when the modified Magnavaca's +Al treatment solution was compared with the control (−Al) solution, only 4.3% of the total Al was precipitated (in contrast to 40% in Yoshida's solutions), available P was only reduced 27% (in contrast to 85.8% in Yoshida's solution), and available Fe was reduced by 11.5% (in contrast to 85.2% in Yoshida's solution; Table I).
Table II. Nutrient composition of the modified Magnavaca's solution optimized for rice Al tolerance screening.
Key differences between this solution and the standard rice Al tolerance screening Yoshida's solution include reduced P and sulfur concentrations and an Fe-HEDTA chelate, replacing the citrate Fe chelate used in Yoshida's solution.
| Compound | Concentration |
| KCl | 1 mm |
| NH4NO3 | 1.5 mm |
| CaCl | 1 mm |
| KH2PO4 | 45 μm |
| MgSO4 | 200 μm |
| Mg(NO3)2 | 500 μm |
| MgCl2 | 155 μm |
| MnCl24H2O | 11.8 μm |
| H3BO3 | 33 μm |
| ZnSO47H2O | 3.06 μm |
| CuSO45H2O | 0.8 μm |
| Na2MoO4H2O | 1.07 μm |
| Fe-HEDTA | 77 μm |
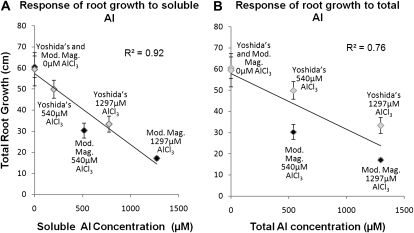
Plant growth experiments were conducted using seven diverse rice genotypes to investigate whether the two nutrient solutions had different effects on seedling root growth under control conditions (−Al) and whether there were differential responses to the total Al added to each solution. The average total root growth (TRG) of the seven genotypes after 3 d of growth in the two control solutions was virtually identical, 60.58 cm in modified Magnavaca solution and 59.47 cm in Yoshida's solution (Fig. 1A). However, when the same AlCl3 concentrations were used in the two treatment solutions, the average TRG of the seven genotypes was 40% to 50% less in the modified Magnavaca's solution than in the Yoshida's solution. At a total concentration of 540 μm AlCl3, TRG averaged 30.34 cm in the modified Magnavaca's solution (relative root growth [RRG] = 0.50) and 49.95 cm in the Yoshida's solution (RRG = 0.84). At the 1,297 μm AlCl3 concentration, root growth averaged 17.12 cm in modified Magnavaca's solution (RRG = 0.28) and 33.38 cm in the Yoshida's solution (RRG = 0.56; Fig. 1A). These results demonstrate that inhibition of TRG is not determined by the total amount of Al added to a solution but rather by the activity of available Al3+ in the solution. Figure 1 displays the correlation coefficients of TRG as a function of total Al (r2 = 0.40) added and available soluble Al (not precipitated; r2 = 0.86), demonstrating that available soluble Al is a much better predictor of root growth inhibition than total Al added.
Figure 1.
Mean root growth (±sd) of seven rice genotypes in Yoshida's (gray diamonds) and modified Magnavaca's (black diamonds) control and Al solutions. The Al concentrations represent previously reported concentrations for rice Al tolerance screening in Yoshida's (1,297 μm) and concentrations for modified Magnavaca's (540 μm) determined in this study. In control solutions (0 μm), root growth is identical. A, Total rootgrowth in response to the concentration of soluble Al in Yoshida's and modified Magnavaca nutrient solutions (r2 = 0.92). B, Total root growth in response to concentration of total Al in Yoshida's and modified Magnavaca's nutrient solutions (r2 = 0.76).
Al Tolerance Phenotyping Platform
RRG of the longest root is the most commonly used parameter for estimating Al tolerance in cereals. We compared estimates of Al tolerance based on RRG of the longest root and RRG of the total growth of the root system to determine whether the longest root measurement would serve as a useful proxy for estimating the inhibition of Al on total root growth of rice seedlings. Using 225 genetically diverse rice accessions (a subset of an association mapping panel; Zhao and McCouch, personal communication), the correlation coefficient for the relationship between RRG of the longest root and RRG of the total root system was r2 = 0.172 (Supplemental Fig. S1). Based on this analysis, it was determined that the RRG of the longest root was not a good proxy for RRG of the total root system because a genotype may appear tolerant based on longest root measurements when, in fact, total root growth is inhibited (Fig. 2). To obtain accurate estimations of total root growth, we used a custom root digital imaging system developed in our laboratories to quantify root length parameters for the thin, fibrous root systems of rice. The system was based on digital photography and semiautomatic measurement of individual primary, secondary, and tertiary roots using RootReader2D software (for details, see “Materials and Methods”). In this system, the length of the total root system can be reliably measured, and we are able to capture high-quality digital images of each root system.
Figure 2.
Example where growth of the longest root in an Al-grown (right) and a control-grown (left) rice seedling is similar but total root growth is significantly different. Images are of plants representative of the mean growth of the longest root in control (−Al) and treatment (+Al) solutions for genotype NSF4. The mean longest root growth was 1.8 ± 0.14 cm in control solution and 2.0 ± 0.18 cm in treatment solution. However, the mean total root growth was 50.29 ± 7.3 cm in control and 27.10 ± 2.47 cm in treatment. The mean longest root RRG was 1.11, although the mean total root RRG was only 0.54.
Comparison of Al Tolerance between Cereal Species
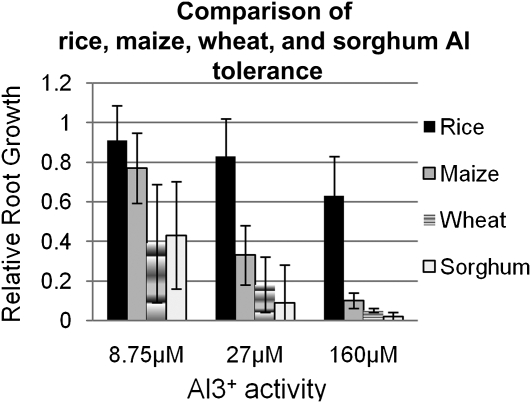
When Al tolerance was directly compared between diverse genotypes of maize, sorghum, wheat, and rice, at three Al3+ activity levels, 8.75, 27, and 160 μm, rice was consistently more tolerant than the other cereals, maize was intermediate, and sorghum and wheat were the most sensitive (Fig. 3). The genotypes used in this analysis were selected to represent the range of known Al tolerance within each species; that is, we selected varieties classified as Al sensitive, intermediate, and tolerant for each species to ensure adequate representation of variation within as well as between the species. At all Al3+ activities, the order of Al tolerance among the four cereal species was consistent: rice > maize > sorghum ≥ wheat.
Figure 3.
Average Al tolerance (RRG) of rice (n = 8), maize (n = 9), wheat (n = 8), and sorghum (n = 7) at three Al3+ activities ± sd.
To ensure that we were able to observe the full distribution of Al tolerance in each of the four cereal species, we evaluated RRG of the total root systems at three different concentrations of Al, based on previous studies. The use of suboptimal or superoptimal Al concentrations tends to mask the range of natural variation for Al tolerance that exists within each species (Supplemental Table S1). To avoid this, we used a free Al3+ activity of 8.75 μm that had been optimized for wheat (Sasaki et al., 2004), 27 μm as reported for sorghum and maize (Magalhaes et al., 2004), and, based on the results of this study, an Al3+ activity of 160 μm as optimal for rice.
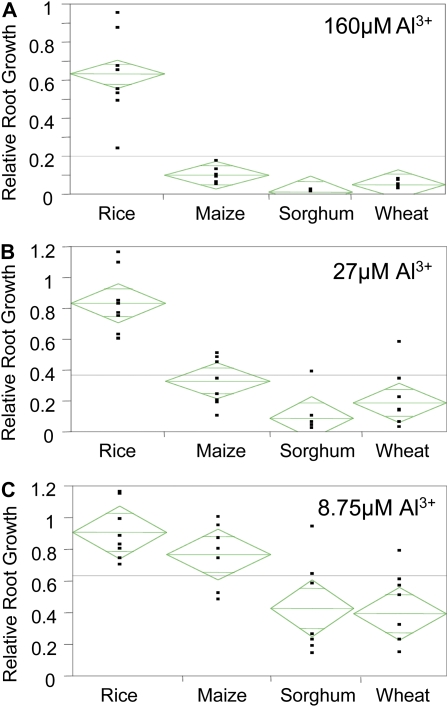
Figure 4 displays the mean RRG and sd observed among the accessions of wheat, sorghum, maize, and rice at each of the Al3+ activities employed. An Al3+ activity of 8.75 μm produced the highest sd and widest range of Al tolerance within wheat and sorghum. The eight wheat genotypes and seven sorghum genotypes screened at 8.75 μm Al3+ displayed similar means and ranges of variation. Maize and rice were both significantly more Al tolerant (P > 0.007) than wheat and sorghum at 8.75 μm Al3+. The RRG in rice and maize was not significantly different and was close to 1 (little or no inhibition of root growth; Fig. 4). Two rice genotypes exhibited increased root growth (RRG > 1) at 8.75 μm Al3+ compared with their root growth under control conditions. Although on average, rice and maize were both more Al tolerant than wheat and sorghum, there was overlap between the species, with the most tolerant genotypes of wheat and sorghum being more tolerant than the most sensitive genotypes of maize and rice.
Figure 4.
Phenotypic distribution of Al tolerance in rice, maize, sorghum, and wheat at three Al3+ activities: 160 μm (A), 27 μm (B), and 8.75 μm (C). [See online article for color version of this figure.]
At 27 μm Al3+ root growth was severely inhibited in wheat and sorghum and significantly reduced in maize, while minimal root growth inhibition was observed in rice. The most tolerant wheat variety was Atlas 66 (RRG = 0.58), which was the source of the Al tolerance allele in ALMT1 (Sasaki et al., 2004). In sorghum, root growth was inhibited by more than 90% in six of the varieties, while SC566 was considerably more tolerant than the other genotypes. SC566 is similar in Al tolerance to SC283, the donor of the tolerance allele for the sorghum Al tolerance gene, AltSB (Caniato et al., 2007; Magalhaes et al., 2007). The Al3+ activity of 27 μm produced a significant decrease in mean RRG in maize, but a clear distribution of Al tolerance was observed. At this activity, it is clear that rice is more tolerant than the other cereals screened, as the mean RRG for rice was 2.5 times greater than that of maize and four times that of wheat. The two most tolerant rice varieties at 27 μm Al3+, Cybonnet (RRG = 1.09) and Nipponbare (RRG = 1.16), demonstrated increased growth compared with the control. The least tolerant rice variety at 27 μm Al3+, China 1039 (RRG = 0.6), was more tolerant than nearly all the other genotypes of maize, sorghum, and wheat. Thus, at 27 μm Al3+, rice was significantly more tolerant than all the other species examined (P < 0.001).
The differences in tolerance between rice and the other species became even more apparent at 160 μm Al3+. Growth was essentially abolished in all sorghum and wheat genotypes screened and severely inhibited in all maize genotypes. Of the nine maize genotypes screened, root growth in all but two genotypes was inhibited over 90% (RRG < 0.1). The two most tolerant maize genotypes at 160 μm Al3+ were B57 (RRG = 0.13) and Cateto (RRG = 0.17). Cateto is a Brazilian variety bred for acid soils and is known to be one of the most Al-tolerant maize varieties (Piñeros et al., 2005). In 160 μm Al3+, the eight rice genotypes had a mean RRG of 0.63 ± 0.2 and a range of 0.25 to 0.95. At this Al3+ activity, the most susceptible rice variety, Kasalath (RRG = 0.25), shows significantly higher relative root growth than that of the most tolerant maize variety, Cateto (RRG = 0.17). These results clearly demonstrate that as a species, rice is significantly more Al tolerant than maize, sorghum, and wheat.
Investigation into the Role of Al Exclusion in Rice Al Tolerance
An Al tolerance diversity panel of 23 rice genotypes, representing the genetic and Al tolerance diversity of the Indica and Japonica varietal groups, was evaluated to determine if rice Al tolerance involves root apex Al exclusion, as it does in other cereals. Al tolerance was screened at 160 μm Al3+ and ranged from 0.15 to 0.97 RRG among all genotypes, with a mean value of 0.55 (sd = 0.21; Supplemental Table S2). The Japonica varietal group (n = 11) had a mean tolerance value of 0.69 (sd = 0.16) and RRG ranged from 0.34 to 0.97. The Indica varietal group (n = 12) was generally more susceptible than Japonica, with a mean tolerance value of 0.42 (sd = 0.18) and a range from 0.15 to 0.76.
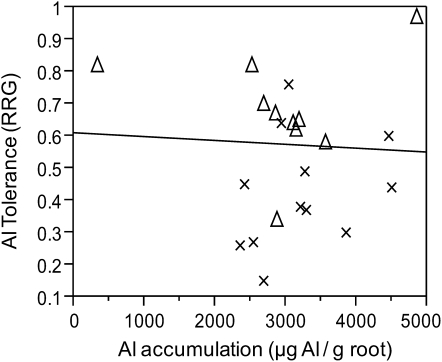
The mean root apex (1 cm) Al concentration among all rice genotypes was 3,027 μg Al g−1 (sd = 889) and ranged from 326 to 4,848. The mean root apex Al concentration was not significantly different between the Indica (3,217 μg Al g−1, sd = 724) and Japonica (2,875 μg Al g−1, sd = 1,065) varietal groups (Supplemental Table S2). The correlation of Al tolerance by Al exclusion across all rice genotypes demonstrated that there is no relation between Al exclusion and Al tolerance in rice (r2 = 0.002; Fig. 5). Similar results were obtained when each varietal group was analyzed separately: Indica (r2 = 0.01) and Japonica (r2 = 0.0).
Figure 5.
Correlation of root tip (1 cm) Al accumulation and Al tolerance across 23 genetically diverse rice genotypes. Genotypes were selected to represent the genetic and Al tolerance variation across the Indica (crosses) and Japonica (triangles) varietal groups. Note that there is no correlation between root tip Al accumulation and Al tolerance (r2 = 0.002).
Two maize controls were included in the above analysis for comparison between the two species, and each maize line was highly susceptible at 160 μm Al3+. The Al-tolerant maize line, Cateto, and the Al-sensitive maize line, B73, had RRG values of 0.17 and 0.13, respectively. These genotypes differed considerably with regard to Al exclusion: Cateto had a mean root apex Al concentration of 2,192 μg Al g−1 (se = 74.4) compared with 4,062 μg Al g−1 (se = 140) in B73. The mean root apex Al concentration of rice was greater than 900 μg Al g−1 higher than that of Cateto, which was much more susceptible at 160 μm Al3+ than any rice genotype. Three rice genotypes accumulated higher Al concentrations than the B73 genotype yet were between three and seven times more Al tolerant (Supplemental Table S2).
Investigation into the Role of Root Exudates in Rice Al Tolerance
Root exudation of citrate, malate, and phosphate, the three root excreted Al-binding ligands implicated in cereal Al tolerance, was quantified in 21 rice genotypes (10 Japonica and 11 Indica) evaluated under control (−Al) and treatment (+Al) conditions (Supplemental Table S2). Across all rice genotypes, the mean citrate exudation (in +Al) was 37.1 pmol plant−1 d−1 (sd = 39.8), the mean malate exudation was 47.4 pmol plant−1 d−1 (sd = 47.4), and the mean phosphate exudation was 102.4 pmol plant−1 d−1 (sd = 94.2). Regression analysis of root exudate by Al tolerance score (RRG) revealed no significant relationship between root exudation and Al tolerance for citrate, malate, or phosphate (Supplemental Fig. S2).
When the relationship between root exudates and Al tolerance was analyzed independently in each rice varietal group, it revealed that within the Indica group, there was a small correlation between Al tolerance and malate (r2 = 0.24) and phosphate exudation (r2 = 0.13; Fig. 6, A–C). In the more Al tolerant Japonica group, there was a negative correlation between Al tolerance and phosphate exudation (r2 = 0.18) and no correlation between citrate and malate exudation (Supplemental Fig. S2).
Figure 6.
Correlation of root exudates of citrate (A and D), malate (B and E), and phosphate (C and F) with Al tolerance (RRG) in the left column and root tip Al content in the right column for 11 genetically diverse Indica varieties. A significant negative correlation is observed between root citrate exudation and root tip Al content (D). However, there is no correlation between root citrate exudation and Al tolerance. There is a slight correlation between malate exudation and Al tolerance, although there is no relation between malate exudation and root tip Al exclusion (A). For the rest of the parameters, there is either no or very weak correlation.
In the tolerant maize variety Cateto, which has previously been reported to utilize an Al-activated citrate exudation Al tolerance mechanism (Piñeros et al., 2005), we observed Al-activated citrate exudation, and exudation rates were significantly higher in Cateto than in any rice variety. The citrate exudation rate of Cateto roots grown in treatment solution (+Al) was 288.3 pmol plant−1 d−1 (se = 66.1), compared with 76.4 pmol plant−1 d−1 (se = 8.8) when seedlings were grown under control (−Al) conditions. Under Al stress, the citrate exudation rate of Cateto was over six times that of any rice genotype; however, Cateto was more sensitive to Al than any rice variety (Supplemental Table S2).
Investigation into the Role of Root Exudates in Al Exclusion
When levels of organic acid exudation were compared with Al accumulation in root apices across all rice genotypes, we observed a slightly negative correlation between citrate exudation and root tip Al concentration (r2 = 0.06) and no relationship between malate or phosphate exudation and root tip Al accumulation (Supplemental Fig. S2).
When exudation levels were compared within each varietal group independently, there was a significantly negative correlation in the Indica group between citrate exudation (r2 = 0.47) and Al accumulation and a slightly negative correlation between malate (r2 = 0.07) and phosphate (r2 = 0.075) exudation and root tip Al accumulation (Fig. 6, D–F). Therefore, it appears that in the Indica varietal group, citrate exudation is associated with Al exclusion from the root apex, but this Al exclusion does not confer Al tolerance. In the Japonica varietal group, we observed no relationship between root exudation and Al accumulation, nor between either parameter and Al tolerance.
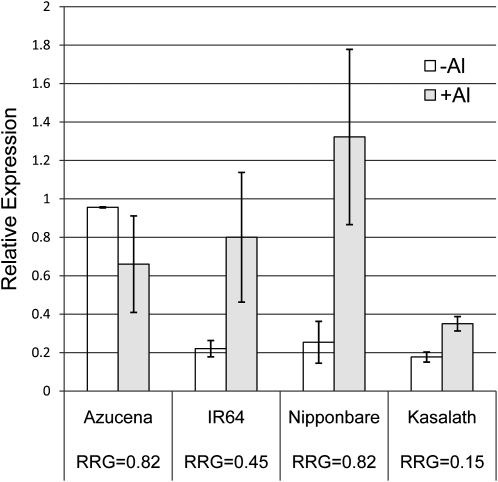
Gene Expression Analysis of the Rice MATE Homolog of SbMATE, the Sorghum Al Tolerance Gene
To investigate if the rice homolog (LOC_Os01g69010) of the sorghum MATE Al tolerance gene (SbMATE) is involved in rice Al tolerance, quantitative real-time (RT)-PCR was conducted in root tips (1 cm) of four diverse genotypes: Azucena (RRG = 0.82; tropical japonica), Nipponbare (RRG = 0.82; temperate japonica), IR64 (RRG = 0.45; indica), and Kasalath (RRG = 0.15; aus). Under control conditions, Azucena had significantly higher levels of root MATE expression than any of the other varieties (Fig. 7). Al treatment reduced MATE gene expression in Azucena, and no root citrate exudation was observed under either control or Al treatment, while Al treatment increased MATE gene expression and citrate exudation in Nipponbare, IR64, and Kasalath. Under Al stress, Nipponbare exhibited the highest MATE gene expression and Kasalath had the lowest. Nipponbare had significantly higher MATE gene expression than Azucena (P = 0.1), although both varieties were equally Al tolerant. Additionally, Azucena accumulated less Al (326 μg Al g−1) in the roots than any of the other varieties, nearly eight times less than that of Nipponbare (2,523 μg Al g−1). These findings provide strong evidence that expression of the rice MATE homolog is not associated with either Al exclusion or Al tolerance in any of the genotypes evaluated here.
Figure 7.
Relative gene expression determined using quantitative RT-PCR for the rice homolog of the sorghum Al tolerance gene, SbMATE, in roots of four rice genotypes that represent a wide range of Al tolerance. The Al tolerance (RRG) for each genotype is indicated below the name of each genotype.
DISCUSSION
Optimization of Nutrient Solution for Rice Al Tolerance Screening
One objective of this study was to address the problems encountered by the high concentrations of Al required for rice Al tolerance studies. Because Magnavaca's nutrient solution had been used successfully to screen for Al tolerance in sorghum and maize at relatively low levels of Al3+ (Magnavaca et al., 1987), we modified this solution so that it would be appropriate for screening rice at elevated Al concentrations while maintaining similar root growth as in Yoshida's solution under control (−Al) conditions.
Yoshida's rice solution is a complete and versatile hydroponic solution that was developed specifically for rice physiology experiments (Yoshida et al., 1976). It has been widely used to evaluate rice mineral nutrition, including toxicities to salt, Fe, and Al, as well as deficiencies of P (Nguyen et al., 2001; Lin et al., 2004; Shimizu et al., 2004; Dufey et al., 2009). The long history and functionality of the Yoshida's solution make it a natural first choice for Al tolerance screening in rice. However, because of the high ionic strength and high concentrations of mineral ions that complex Al in Yoshida's solution, there are very serious problems when it is used to evaluate Al tolerance in rice. These are exemplified by the clearly visible precipitate that forms in the Yoshida's +Al treatment solution. Some studies have avoided the problem of Al precipitation by screening seedlings in CaCl2 solution, which does not precipitate with Al and allows for reproducible Al3+ concentrations (Ma et al., 2002; Xue et al., 2006; Yamaji et al., 2009). However, a simple CaCl2 solution can only be used to screen very young seedlings, when the seed is still capable of providing all necessary mineral nutrients. The modified Magnavaca solution developed in this study can be used to screen Al tolerance in plants at all stages of development.
The precipitation issues confound the ability to quantify rice Al tolerance, as it is difficult to design a nutrient solution with reproducible levels of Al as well as the essential elements P, sulfur, and Fe, which can also impact root growth. Al3+ is highly reactive and readily precipitates with other essential elements; in the Yoshida's +Al solution, both P and Fe were reduced to such low levels that it was difficult to distinguish between root inhibition due to Al and that due to lack of P and Fe. P and Fe are typically present in nutrient solutions as PO4−Fe3+, and it has been well documented that different concentrations of P and/or Fe can lead to alterations in root growth and architecture (Lynch and Brown, 2001; Williamson et al. 2001; Lopez-Bucio et al., 2003; Ward et al., 2008). Furthermore, the use of citrate as the Fe-chelate in the Yoshida's solution is problematic, as citrate preferentially binds Al over Fe, leading to differences in Fe availability between the control and +Al Yoshida's solutions. In the modified Magnavaca's solution, soluble P concentrations were reduced 7-fold with respect to Yoshida's solution (from 322 to 45 μm), which is still well within the range of sufficient P concentrations for continuously flowing hydroponic solutions (Jones, 1997). Sulfate concentrations were reduced 16-fold (from 3.33 to 0.2 mm), the ionic strength was significantly reduced, and a Fe-HEDTA chelate was utilized to prevent Fe precipitation and citrate interaction with Al. ICP-ES analysis confirmed that the modified Magnavaca's solution has significantly reduced precipitation of P, Fe, and Al in the Al treatment solutions compared with the Yoshida's solution.
The differential root growth responses observed in +Al treatments between the two nutrient solutions were consistent with Geochem-EZ predictions. It is generally accepted that the primary rhizotoxic form of Al is Al3+; thus, when a large proportion of Al is precipitated in the Yoshida's solution, it becomes unavailable to affect root growth (Kochian et al., 2004a). The increased root growth inhibition in the modified Magnavaca's Al solution can be attributed to one or a combination of three factors: (1) less of the added Al is precipitated with sulfur and P compared with Yoshida's, leaving more Al in the active (rhizotoxic) form; (2) the citrate in Yoshida's solution added as an Fe chelate preferentially complexes with Al, whereas the modified Magnavaca's uses an HEDTA chelate, which chelates Fe preferentially over Al; and (3) the modified Magnavaca's solution has a lower overall ionic strength than the Yoshida's solution, which increases the activity coefficient (and hence the concentration of thermodynamically relevant ion in solution) of a trivalent ion. Also, as the nutrient solution ionic strength decreases, it prevents the roots from being protected from Al3+, as the Al ions have less competition for negatively charged sites within the root cell wall and root plasma membrane by decreasing the concentrations of other cations that can shield Al3+ from these negative sites.
Importance of Quantifying the Whole Root System in Al Tolerance Studies
Rice seedling root systems are fibrous and can have multiple primary, secondary, and tertiary roots within a few days after germination. There is also significant genetic variation in rice root architecture among varieties, ecotypes, and/or subpopulations. The phenotypic variation in root growth habit per se among varieties and ecotypes must be taken into consideration when determining Al tolerance. To date, published results on Al tolerance in maize, sorghum, and rice have all used the growth of the longest root(s) as the assay for Al tolerance (Wu et al., 2000; Nguyen et al., 2001, 2002, 2003; Piñeros and Kochian, 2001; Magalhaes et al., 2004; Xue et al., 2006, 2007). Although this approach has proven useful in assessing Al tolerance in other cereals, as demonstrated by the cloning of Al tolerance genes in wheat and sorghum, our results suggest that Al tolerance based on RRG of the longest root is not the best predictor of Al tolerance in rice.
Using a set of 225 diverse rice genotypes and the RootReader2D software, we determined that the correlation between the RRG of the longest root and the RRG of the total root system was weak (r2 = 0.17; Supplemental Fig. S1). Furthermore, in two quantitative trait locus (QTL) mapping studies where Al tolerance was evaluated based on both assays, we identified some of the same, but also some novel, major effect Al tolerance QTLs that were only detected by TRG-RRG (A. Famoso, K. Zhao, L. Kochian, and S. McCouch, personal communication). Our observations in this study are consistent with studies in maize, wheat, sorghum, soybean (Glycine max), sugarcane (Saccharum officinarum), and tobacco (Nicotiana tabacum), where all have reported severe inhibition of lateral roots in sensitive genotypes (Hetherington et al., 1988; Bushamuka and Zobel, 1998; Silva et al., 2001; Brichkova et al., 2007). We thus conclude that the RRG of the total root system is clearly a much better quantitative indicator of rice Al tolerance than RRG of the longest root, and our newly developed automated image capture and computational determination of growth of the total root system makes it feasible to use this parameter in large-scale genetic and physiological studies.
Comparison of Al Tolerance between Cereal Species
In this study, we demonstrated that young rice seedlings (3 d old) tolerate significantly higher concentrations of Al3+ than maize, sorghum, or wheat, consistent with the superior Al tolerance of rice observed in previous hydroponic Al3+ concentrations and field studies (Foy, 1988; Wu et al., 2000; Nguyen et al., 2001, 2002, 2003; Magalhaes et al., 2004; Sasaki et al., 2004). Yet, we know little about the genes and physiological mechanisms responsible for the high levels of Al tolerance in rice. Other cereals, such as rye (Secale cereale), have been reported to exhibit high levels of Al tolerance (Gallego and Benito, 1997). However, the Al concentrations in which rye has been screened are four times lower than those at which rice is screened (Gallego and Benito, 1997; Gallego et al., 1998; Li et al., 2000; Collins et al., 2008). This suggests that rice is a very useful model for characterizing the mechanisms conferring high levels of Al tolerance in cereals. Rice also has an abundance of genetic and genomic resources, including several sequenced genomes, high density of genotyping arrays, the availability of numerous immortal mapping populations, and extensive germplasm collections (www.gramene.org and www.irri.cgiar.org).
Despite the fact that sorghum has been previously demonstrated to exhibit higher Al tolerance than wheat (Sasaki et al., 2006; Caniato et al., 2007), in this study sorghum and wheat seedlings exhibited similar levels of Al tolerance after 3 d in Al solutions. A likely explanation for this discrepancy is the extended time in Al required to observe the Al tolerance response in sorghum (5–6 d; Magalhaes et al., 2007). Thus, the degree of Al tolerance observed for sorghum in our study is less than would be predicted if the plants were grown in Al solution for up to 6 d (Caniato et al., 2007; Magalhaes et al., 2007).
Rice Must Employ a Novel Al Tolerance Mechanism
Organic acid-mediated root tip Al exclusion has been reported in numerous plant species, explaining most of the phenotypic variation in wheat (Sasaki et al., 2004, 2006), sorghum (Magalhaes et al., 2007), and Arabidopsis (Hoekenga et al., 2003) and a portion of the variation in Al tolerance in maize (Piñeros et al., 2005). As a species, rice is two to five times more Al tolerant than wheat, sorghum, and maize, yet this study demonstrated that there is no significant correlation between Al exclusion from the root apex and root growth in Al. This indicates that the roots of tolerant rice varieties can continue to grow even with significant Al accumulation into the root tip. Thus, rice must employ unique mechanisms of Al tolerance not found in other cereal species.
Unlike Japonica, the more susceptible Indica varieties do exhibit a significant negative correlation between rates of citrate exudation and Al concentrations in the root tip (r2 = 0.47), although this response is not correlated with Al tolerance (RRG of the total root system). However, rates of root exudation of malate (r2 = 0.24) and phosphate (r2 = 0.13) showed a weak positive correlation with Al tolerance in Indica varieties, but not Al exclusion. These findings suggest that malate and/or phosphate exudation may function at least in part to chelate Al3+ within the apoplast of the root tip, rather than exclude Al3+ from entering the root tip. The primary function of root exudates in Al tolerance is believed to be the exclusion of Al from the root apex, but this alone is not responsible for the high levels of Al tolerance in rice. The clearest evidence for this comes from experiments where the wheat Al tolerance gene (ALMT1) was transformed into rice, resulting in Al-induced gene expression and enhanced malate exudation but no effect on Al tolerance (Sasaki et al., 2004). However, when the ALMT1 gene was transformed into barley (Hordeum vulgare), an Al-susceptible species, Al tolerance was increased by more than 100%. Multiple rice Al tolerance QTL studies have identified a region on chromosome 1 that is in close proximity to the rice MATE family member that is a homolog of the sorghum Al tolerance gene (SbMATE), leading to the hypothesis that this gene may be underlying these QTLs. SbMATE functions in sorghum Al tolerance as an Al-activated root citrate efflux transporter that excludes Al from the root tip, with differences in Al tolerance across sorghum genotypes directly related to gene expression (r2 = 0.98; Magalhaes et al., 2007).
Quantitative RT-PCR was conducted to determine if differences in rice MATE gene expression correlated with differences in rice Al tolerance in four genotypes with widely varying levels of Al tolerance. The highly susceptible Kasalath had the lowest MATE expression under Al stress, significantly less than Al-tolerant Nipponbare. However, Nipponbare and Azucena exhibit a similar level of Al tolerance, but MATE expression was significantly higher in Nipponbare under Al stress. Furthermore, Azucena accumulated less Al (326 μg Al g−1) than any other variety, nearly eight times less than that of Nipponbare (2,523 μg Al g−1). Based on the lack of correlation between rice MATE gene expression and Al exclusion, citrate exudation, and Al tolerance, we conclude that the rice homolog of the sorghum Al tolerance gene is not involved in mediating rice Al tolerance through Al-activated gene expression and root exclusion of Al.
In this study, one Al-tolerant maize line (Cateto) and one susceptible line (B73) were compared with rice in terms of Al accumulation, root exudation of organic acids and phosphate, and Al tolerance at 160 μm Al3+. Both maize genotypes were severely inhibited. However, our results were consistent with previously published results (Piñeros et al., 2005) reporting Al-activated citrate exudation and Al exclusion in the tolerant maize line and increased Al accumulation in the susceptible parent. At the high Al concentrations used in this study, RRG of Cateto was severely inhibited, showing levels of RRG similar to that of the susceptible maize line, B73, although Cateto accumulated less than half the Al of B73. This suggests that the level of Al accumulated by Cateto in 160 μm Al3+ was above the threshold at which root growth can occur in maize and that additional Al accumulation beyond this threshold does not further inhibit root growth. When previous studies of Al accumulation in wheat (Delhaize et al., 1993) and maize (Piñeros et al., 2005) are compared with results in this study, it appears that significant Al inhibition of root growth occurs at root tip Al concentrations around 1,000 μg Al g−1 root tip in wheat and maize. Delhaize et al. (1993) quantified Al inhibition of root growth over time in one Al-sensitive and one tolerant wheat variety, and a significant difference in Al tolerance was not observed until the susceptible variety accumulated over 1,000 μg Al g−1 in the root tip. Similarly, Piñeros et al. (2005) reported a nonlinear relationship between root tip Al accumulation and Al tolerance in six maize genotypes, two tolerant and four susceptible. The two tolerant genotypes, Cateto and Pioneer 3355, accumulated significantly different amounts of Al (495 and 900 μg Al g−1, respectively) and had Al tolerance values of 0.97 and 0.75 RRG, respectively. The four susceptible lines showed differences ranging from 1,250 and 2,225 μg Al g−1 in root tip Al accumulation, yet Al tolerance only ranged from 0.48 to 0.38. When comparing tolerant and susceptible maize lines, the tolerant line Pioneer 3355 that accumulated 900 μg Al g−1 was over 50% more Al tolerant than the susceptible line that accumulated 1,250 μg Al g−1. These results suggest that the relation between Al accumulation and Al tolerance in maize is not linear and, similar to wheat, a threshold is reached at around root tip Al concentrations of 1,000 μg Al g−1, where growth is significantly inhibited.
Based on these observations, it appears that rice, as a species, is capable of withstanding significantly higher Al concentrations both in the soil solution and the root tip than other cereals. All but one rice line was more tolerant than the most tolerant maize line (Cateto), yet all but one rice genotype accumulated more Al in the root apex. Some rice genotypes that accumulated 50% to 100% more Al in their root tips were two to five times more Al tolerant than other rice genotypes that accumulated less root tip Al (Supplemental Table S2). Based on the lack of correlation between rice Al exclusion and Al tolerance, and the relatively high levels of Al accumulation in rice compared with maize, we conclude that rice utilizes one or more novel Al tolerance mechanisms. At this time, we have little information regarding the nature of this new Al tolerance mechanism. Because the majority of the Al in the root tip resides in the apoplast (Kochian, 1995), it is logical to speculate that the root cell wall may play a role in the high level of Al tolerance observed in rice. Recent work from Jian Feng Ma's laboratory supports this speculation based on the map-based cloning of an Al-sensitive knockout mutant locus in rice (Huang et al., 2009). This resulted in the identification of two mutant genes, STAR1 and STAR2, which encode two interacting proteins that form an ATP-binding cassette transporter complex. Transport studies via the STAR1/STAR2 transporter complex in oocytes showed that the transporter mediates the efflux of UDP-Glc, presumably into the root apoplast, leading the authors to speculate that cell wall modification may play a role in rice Al tolerance. Furthermore, a study conducted by Yang et al. (2008) provided evidence that cell wall polysaccharides may be involved in rice Al tolerance.
It is known that the growing root tip is the site of Al toxicity (Ryan et al., 1993); however, the mechanism by which Al inhibits root growth in plants is still unclear. Based on observations that Al tolerance in wheat, sorghum, and maize is related to the plant's ability to exclude Al from the growing tip, but not from the mature root regions, researchers have inferred that Al in these species poisons proteins and/or structural components of the root tip that are critical to cell growth, elongation, and/or division. In rice, where there is no significant correlation between Al accumulation in the root tip and Al tolerance, it appears that the mechanism of toxicity must be categorically different than in other species. We hypothesize that at some point in evolution, the lineage leading to modern species of Oryza experienced a dramatic shift in its position within the landscape of plant response to Al, demonstrating greatly enhanced ability to grow under high concentrations of Al. If this hypothesis is true, identifying the genes/alleles underlying Al tolerance or susceptibility among rice varieties will provide limited insight into novel plant Al tolerance mechanisms. To fully understand the novelty of the mechanism(s) of Al toxicity and tolerance found in rice, it will be necessary to undertake very specific physiological, biochemical, and molecular experiments in a phylogenetic context. Thus, rice appears to hold the key to understanding how and when, over the course of evolution, a lineage of plants experienced a dramatic genetic change that led to enhanced levels of Al tolerance and will provide critical insights that are likely to help move this capability into other species that are critical to human survival.
MATERIALS AND METHODS
Plant Material
A set of seven to nine genotypes of rice (Oryza sativa), maize (Zea mays), wheat (Triticum aestivum), and sorghum (Sorghum bicolor) genotypes were used to compare Al tolerance between species. Rice seeds were obtained from S. McCouch and included the genotypes Azucena, BJ 1, China 1039, Cybonnet, IR64, Kasalath, Nipponbare, and Sabjaraj. Maize seeds were obtained from E. Buckler and included the genotypes B 164, B 57, Cateto, H 84, NC 264, NC 290A, NC 310, NC 328, and R 10. Wheat seeds were obtained from M. Sorrells and included the genotypes AC Reed, Atlas 66, Bob White, Caledonia, Cham 1, Opata, Roane, and Scout 66. Sorghum seeds were obtained from S. Kresovich and J. Magalhaes and included the genotypes BR007, BTX623, Cowley, IS3620C, SC452, SC566, and T309.
Plant Growth Conditions
Seeds were germinated in rolled germination paper at 26°C to 30°C for 3 to 5 d under dark conditions. Wheat and sorghum seeds were surface sterilized with 10% bleach and rice with 20% bleach for 15 to 20 min. Maize seeds were treated with a fungicide treatment of Captan400, Trilex, and Allegiance. Upon germination, seedlings were transferred to control (−Al) solutions for 24 h, then 20 uniform seedlings were photographed and root length was quantified using RootReader2D. Subsequently, 10 seedlings were transferred to fresh control solution and 10 seedlings to Al treatment solution. After 3 d in the respective treatments, roots were photographed and measured, mean root growth in control and +Al treatment was calculated for each genotype, and RRG was determined: RRG = treatment root growth/control root growth. Plants were grown in 9-L tubs with 48 plants per tub, and the plants were supported with eight foam strips (six plants per strip) with a slit cut into the foam to anchor the stem. Aeration was provided in all experiments, except for experiments comparing the nutrient solutions, in which only rice lines were compared. Plant growth chamber conditions for the maize diversity screen and species comparison experiments were 26°C (day)/23°C (night), while the rice diversity screening conditions were 30°C (day)/26°C (night). All experiments were conducted with 12-h days and a light intensity of 450 mmol photons m−2 s−1.
Nutrient Solutions
The control (−Al) Yoshida's nutrient solution was prepared as described previously (Yoshida et al., 1976), and the pH was adjusted to 4.0 with 1 n NaOH. The Yoshida treatment (+Al) solution was identical to the control but contained 35 μL L−1 (1,297 μm) AlCl3. The control (−Al) modified Magnavaca's nutrient solution was modified from Magnavaca et al. (1987). The treatment (+Al) modified Magnavaca's solution contained 540 μm AlCl3, added after pH adjustment to 7.8 with KOH to prevent Al precipitation, and the final pH was adjusted to 4.0 with 1 n HCl.
ICP-ES Analysis of Nutrient Solutions
ICP-ES elemental profiling was conducted on all elements, except nitrogen, in both nutrient solutions. To determine the available concentration of each element, 1 L of each nutrient solution per treatment was made and analyzed to confirm elemental composition. Four 50-mL samples of each nutrient solution per treatment were then collected and stored in the dark for 3 d under plant growth conditions to permit chemical equilibrium. Samples were then homogenized, and a 10-mL sample was collected for ICP analysis to calculate total elemental concentration after 3 d. The remaining 40 mL was centrifuged at 3,250g for 15 min, and 10 mL of supernatant was collected for ICP analysis to determine the amount of each element that was precipitated out of solution.
Chemical Speciation Analysis
Chemical speciation analysis was conducted according to Shaff et al. (2010) using the nutrient composition of Yoshida's nutrient solution (Yoshida et al., 1976) and the modified Magnavaca's solution presented here. The pH of all solutions was fixed at 4.0, and the predicted available activity of each element was determined through the primary distribution and case progress table output by the prediction of solid formation for each element.
Root Imaging and Measurements
A custom root-imaging system was used to accurately quantify total root length parameters from rice. For description of this system in detail, see www.plantmineralnutrition.net. The system utilizes digital photography and Java-based RootReader2D software (available at http://www.plantmineralnutrition.net/rootreader.htm). The imaging system consists of a Nikon D200 digital SLR camera with a 60-mm Macro lens, which was calibrated and aligned to a fixed focal plane scale of 120 pixels cm−1. Each plant was imaged with its root system spread out in a clear, solution-filled trough that was illuminated from below with a light box. Once the plants were photographed, the root images were converted from the RAW NEF file format to a 32-bit RGB TIFF file format using Nikon Capture NX software. The TIFF images were then converted from the RGB format to an eight-bit grayscale format using Adobe Photoshop. The grayscale images were then batch processed with RootReader2D software with a fixed threshold adjusted between 15 and 25 to maximize contrast and an error criterion of 6.0 pixels to optimize measurement accuracy. Total root system lengths were automatically measured, while individual roots were semiautomatically selected and measured with RootReader2D software.
Root Tip Al Content
Plants were grown as described above, and after 3 d of Al treatment, the first centimeter of the primary roots was collected (approximately three per rice plant) and bulked to 50 roots per replicate, with five replicates per genotype. Root tips were then dried in an oven at 60°C for 2 d. Dry weights were determined using a microgram balance (MT2; Mettler). Dry samples were digested with 100 μL of 50:50 ddH(NO3) and 70% perchloric acid, resuspended in 10.25 mL of 5% ddH(NO3), and analyzed using an inductively coupled argon plasma model 51000 emission spectrometer (Perkin-Elmer/Sciex).
Root Organic Acid Exudation
Seeds were germinated as described above, then 10 uniform seedlings were transferred to a plastic tube stopper with 2-mm holes drilled throughout the bottom and placed in 45 mL of 2.7 mm CaCl2 with and without 160 μm Al3+ treatment in a 50-mL Falcon tube (n = 3). After 48 h of growth, control plants were transferred to fresh CaCl2 solution and Al-treated plants were transferred to a CaCl2 solution containing 39 μm Al3+ for 24 h, and these solutions were processed and analyzed by capillary electrophoresis as described by Piñeros et al. (2005). The CaCl2 concentration was determined to best replicate the modified Magnavaca's ionic strength. It was necessary to grow the plants in CaCl2 and reduce the Al concentration in the sample collected to prevent noise and interference during capillary electrophoresis.
Quantitative RT-PCR
Plants were grown as described above, and after 3 d of Al or control treatment, the first centimeter of the primary roots was collected (approximately three per rice plant) and bulked to 50 roots per replicate, with two replicates per genotype, flash frozen in liquid nitrogen, and stored at −80C. RNA extraction, cDNA synthesis, and RT-PCR were conducted as described by Liu et al. (2009). Primer sequences for the MATE gene and the actin internal control were as follows: MATE, 5′-AGGAGATTCGTGCCGTCCGTGA-3’ and 5′-CGGCTTGACGCCCATGATGC-3’; Actin, 5′-ATCCTTGTATGCTAGCGGTCGA-3′ and 5′-ATCCAACCGGAGGATAGCATG-3′.
Statistical Analysis
ANOVA was conducted for comparison of Al tolerance between species, and correlation coefficients were determined using JMP version 7.0 (SAS Institute). Microsoft Excel 2007 was used to conduct regression analysis comparing longest root RRG and total root RRG. Mean Al tolerance of the accessions was used to estimate species Al tolerance.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Correlation of Al tolerance between longest root growth and total root growth.
Supplemental Figure S2. Correlation of root exudates and Al accumulation with Al tolerance.
Supplemental Table S1. Mean, sd, and range of Al tolerance for rice, maize, sorghum, and wheat.
Supplemental Table S2. Summary of Al tolerance, root tip Al accumulation, and root exudates in rice panel.
Supplementary Material
Acknowledgments
We thank the following undergraduate interns for their assistance in various aspects of phenotyping and/or greenhouse maintenance of plants: Crayton Montei, Laura Pursel, Vivian Li, Misty Carlisle, Sarah Villareal, Mandy Kain, and Melissa Major. We are grateful to Ed Buckler, Mark Sorrells, David Benscher, Steve Kresovich, and Jurandir Magalhaes for supplying maize, wheat, and sorghum seeds. We appreciate the proofreading and editing contribution of Elliot Heffner. We also thank Jennifer Thaler for use of her microbalance and Matthew Milner and Jiping Liu for assistance and advice on RT-PCR experiments.
References
- Brichkova GG, Shishlova AM, Maneshina TV, Kartel’ NA. (2007) The tolerance of tobacco genetically modified plants to aluminium. Tsitol Genet 41: 23–28 [PubMed] [Google Scholar]
- Bushamuka V, Zobel R. (1998) Tap, basal, and lateral root responses to a stratified acid, aluminum-toxic soil. Crop Sci 38: 416–421 [Google Scholar]
- Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, et al. (2007) Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet 3: 1745–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caniato FF, Guimaraes CT, Schaffert RE, Alves VM, Kochian LV, Borem A, Klein PE, Magalhaes JV. (2007) Genetic diversity for aluminum tolerance in sorghum. Theor Appl Genet 114: 863–876 [DOI] [PubMed] [Google Scholar]
- Champoux MC, Wang G, Sarkarung S, Mackill DJ, O'Toole JC, Huang N, McCouch S. (1995) Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular markers. Theor Appl Genet 90: 969–981 [DOI] [PubMed] [Google Scholar]
- Collins NC, Shirley NJ, Saeed M, Pallotta M, Gustafson JP. (2008) An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereal L.). Plant Physiol 179: 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dally AM, Second G. (1990) Chloroplast DNA diversity in wild and cultivated species of rice (genus Oryza, section Oryza): cladistic-mutation and genetic-distance analysis. Theor Appl Genet 80: 209–222 [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Larsen PB, Howell SH, Kochian LV. (1998) Aluminum resistance in the Arabidopsis mutant alr-104 is caused by an aluminum-induced increase in rhizosphere pH. Plant Physiol 117: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Craig S, Beaton CD, Bennet RJ, Jagadish VC, Randall PJ. (1993) Aluminum tolerance in wheat (Triticum aestivum L.) (I. Uptake and distribution of aluminum in root apices). Plant Physiol 103: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR. (1995) Aluminum toxicity and tolerance in plants. Plant Physiol 107: 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. (1999) Genomic control for association studies. Biometric 55: 997–1004 [DOI] [PubMed] [Google Scholar]
- Dufey I, Hakizimana P, Draye X, Lutts S, Bertin P. (2009) QTL mapping for biomass and physiological parameters linked to resistance mechanism to ferrous iron toxicity in rice. Euphytica 167: 143–160 [Google Scholar]
- Foy CD. (1988) Plant adaptation to acid, aluminum-toxic soils. Commun Soil Sci Plant Anal 19: 959–987 [Google Scholar]
- Gallego FJ, Benito C. (1997) Genetic control of aluminum tolerance in rye (Secale cereale L.). Theor Appl Genet 95: 393–399 [DOI] [PubMed] [Google Scholar]
- Gallego FJ, Calles B, Benito C. (1998) Molecular markers linked to the aluminum tolerance gene alt1 in rye (Secale cereale L.). Theor Appl Genet 97: 1104–1109 [Google Scholar]
- Garris AJ, McCouch SR, Kresovich S. (2003) Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington SJ, Asher CJ, Blamey FPC. (1988) Comparative tolerance of sugarcane, navybean, soybean and maize to aluminum toxicity. Aust J Agric Res 39: 171–176 [Google Scholar]
- Hoekenga OA, Vision TJ, Shaff JE, Monforte AJ, Lee GP, Howell SH, Kochian LV. (2003) Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta × Columbia) by quantitative trait locus mapping: a physiologically simple but genetically complex trait. Plant Physiol 132: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Mu J, Zhang H, Tao Y, Han B. (2006) Differentiation of a miniature inverted transposable element (MITE) system in Asian rice cultivars and its inference for a diphyletic origin of two sub-species of Asian cultivated rice. J Integr Plant Biol 48: 260–267 [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF. (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JB. (1997) Hydroponics: A Practical Guide for the Soilless Grower, Ed 2 CRC Press, Boca Raton, FL [Google Scholar]
- Kochian LV. (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46: 237–260 [Google Scholar]
- Kochian LV, Hoekenga OA, Pineros MA. (2004a) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Kochian LV, Pineros MA, Hoekenga OA. (2004b) The physiology, genetics and molecular biology of plant aluminum tolerance and toxicity. Lambers H, , Root Physiology: From Gene to Function. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 175–196 [Google Scholar]
- Li XF, Ma JF, Matsumoto H. (2000) Pattern of aluminum-induced secretion of organic acids differs between rye and wheat. Plant Physiol 123: 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley JM, Ludlow MM, McCouch S, O'Toole JC. (1996) Locating QTL for osmotic adjustment and dehydration tolerance in rice. J Exp Bot 47: 1427–1436 [Google Scholar]
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao D. (2004) QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 108: 143–160 [DOI] [PubMed] [Google Scholar]
- Liu J, Magalhaes JV, Shaff JE, Kochian LV. (2009) Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J 57: 389–399 [DOI] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA 103: 9578–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio J, Cruz-Ramirez A, Herrera-Estrella L. (2003) The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol 6: 280–287 [DOI] [PubMed] [Google Scholar]
- Lynch JP, Brown KM. (2001) Topsoil foraging: an architectural adaptation of plants to low phosphorus availability. Plant Soil 237: 225–237 [Google Scholar]
- Ma JF, Ryan PR, Delhaize E. (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6: 273–278 [DOI] [PubMed] [Google Scholar]
- Ma JF, Shen R, Zhao Z, Wissuwa M, Takeuchi Y, Ebitani T, Yano M. (2002) Response of rice to Al stress and identification of quantitative trait loci for Al tolerance. Plant Cell Physiol 43: 652–659 [DOI] [PubMed] [Google Scholar]
- Magalhaes JV, Garvin DF, Wang Y, Sorrells ME, Klein PE, Schaffert RE, Li L, Kochian LV. (2004) Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the Poaceae. Genetics 167: 1905–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JV, Liu J, Guimaraes CT, Lana UG, Alves VM, Wang YH, Schaffert RE, Hoekenga OA, Pineros MA, Shaff JE, et al. (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39: 1156–1161 [DOI] [PubMed] [Google Scholar]
- Magnavaca R, Gardner CO, Clark RB. (1987) Evaluation of inbred maize lines for aluminum tolerance in nutrient solution. Genetic Aspects of Plant Mineral Nutrition. Martinus Nijhoff, Dordrecht, The Netherlands, pp 255–265 [Google Scholar]
- Maron L, Pineros MA, Guimaraes CT, Magalhaes JV, Pleiman JK, Mao C, Shaff J, Belicuas SNJ, Kochian LV. (2010) Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J 61: 728–740 [DOI] [PubMed] [Google Scholar]
- Miyasaka SC, Buta JG, Howell RK. (1991) Mechanism of aluminum tolerance in snapbeans: root exudation of citric acid. Plant Physiol 96: 737–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BD, Brar DS, Bui BC, Nguyen TV, Pham LN, Nguyen HT. (2003) Identification and mapping of the QTL for aluminum tolerance introgressed from the new source, Oryza rufipogon Griff., into indica rice (Oryza sativa L.). Theor Appl Genet 106: 583–593 [DOI] [PubMed] [Google Scholar]
- Nguyen VT, Burow MD, Nguyen HT, Le BT, Le TD, Paterson AH. (2001) Molecular mapping of genes conferring aluminum tolerance in rice (Oryza sativa L.). Theor Appl Genet 102: 1002–1010 [Google Scholar]
- Nguyen VT, Nguyen BD, Sarkarung S, Martinez C, Paterson AH, Nguyen HT. (2002) Mapping of genes controlling aluminum tolerance in rice: comparison of different genetic backgrounds. Mol Genet Genomics 267: 772–780 [DOI] [PubMed] [Google Scholar]
- Parker DR, Kinraide TB, Zelazny LW. (1988) Aluminum speciation and phytotoxicity in dilute hydroxy-aluminum solutions. Soil Sci Soc Am J 52: 438–444 [Google Scholar]
- Pellet DM, Papernik LA, Kochian LV. (1996) Multiple aluminum-resistance mechanisms in wheat (roles of root apical phosphate and malate exudation). Plant Physiol 112: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros MA, Kochian LV. (2001) A patch-clamp study on the physiology of aluminum toxicity and aluminum tolerance in maize: identification and characterization of Al3+-induced anion channels. Plant Physiol 125: 292–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros MA, Shaff JE, Manslank HS, Alves VM, Kochian LV. (2005) Aluminum resistance in maize cannot be solely explained by root organic acid exudation: a comparative physiological study. Plant Physiol 137: 231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Donnelly P. (2001) Case-control for association in structured or admixed populations. Theor Popul Biol 60: 227–237 [DOI] [PubMed] [Google Scholar]
- Ryan P, Delhaize E, Jones D. (2001) Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 52: 527–560 [DOI] [PubMed] [Google Scholar]
- Ryan PR, DiTomaso JM, Kochian LV. (1993) Aluminum toxicity in roots: an investigation of spatial sensitivity and the role of the root cap. J Exp Bot 44: 437–446 [Google Scholar]
- Sasaki T, Ryan PR, Delhaize E, Hebb DM, Ogihara Y, Kawaura K, Noda K, Kojima T, Toyoda A, Matsumoto H, et al. (2006) Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol 47: 1343–1354 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Shaff JE, Schultz BA, Craft EJ, Clark RT, Kochian LV. (2010) GEOCHEM-EZ: a chemical speciation program with greater power and flexibility. Plant Soil 330: 207–214 [Google Scholar]
- Shimizu A, Yanagihara S, Kawasaki S, Ikehashi H. (2004) Phosphorus deficiency-induced root elongation and its QTL in rice (Oryza sativa L.). Theor Appl Genet 109: 1361–1368 [DOI] [PubMed] [Google Scholar]
- Silva IR, Smyth TJ, Raper CD, Carter TE, Rufty TW. (2001) Differential aluminum tolerance in soybean: an evaluation of the role of organic acids. Physiol Plant 112: 200–210 [DOI] [PubMed] [Google Scholar]
- von Uexkull HR, Mutert E. (1995) Global extent, development and economic impact of acid soils. Date RA, Grundon NJ, Raymet GE, Probert ME, , Plant-Soil Interactions at Low pH: Principles and Management. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 5–19 [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG. (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM. (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolt J. (1994) Soil Solution Chemistry: Applications to Environmental Science and Agriculture. John Wiley & Sons, New York [Google Scholar]
- Wu P, Liao CY, Hu B, Yi KK, Jin WZ, Ni J, He C. (2000) QTLs and epistatis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Theor Appl Genet 100: 1295–1303 [Google Scholar]
- Xu Y, This D, Pausch RC, Vonhof WM, Coburn JR, Comstock JP, McCouch SR. (2009) Leaf-level water use efficiency determined by carbon isotope discrimination in rice seedlings: genetic variation associated with population structure and QTL mapping. Theor Appl Genet 18: 1065–1081 [DOI] [PubMed] [Google Scholar]
- Xue Y, Jiang L, Su N, Wang JK, Deng P, Ma JF, Zhai HQ, Wan JM. (2007) The genetic basic and fine-mapping of a stable quantitative-trait loci for aluminium tolerance in rice. Planta 227: 255–262 [DOI] [PubMed] [Google Scholar]
- Xue Y, Wan J, Jiang L, Wang C, Liu L, Zhang YM, Zhai H. (2006) Identification of quantitative trait loci associated with aluminum tolerance in rice. Euphytica 150: 37–45 [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Li YY, Zhang YJ, Zhang SS, Wu YR, Wu P, Zheng SJ. (2008) Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol 146: 602–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JA, Gomez KA. (1976) Laboratory Manual for Plant Physiological Studies of Rice, Ed 3 International Rice Research Institute, Manila, Philippines [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zhang WH, Ryan PR, Tyerman SD. (2001) Malate-permeable channels and cation channels activated by aluminum in the apical cells of wheat roots. Plant Physiol 125: 1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, et al. (2007) An Arabidopsis example of association mapping in structured samples. PLoS Genet 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.