Abstract
Plants grown under a canopy recognize changes in light quality and modify their growth patterns; this modification is known as shade avoidance syndrome. In leaves, leaf blade expansion is suppressed, whereas petiole elongation is promoted under the shade. However, the mechanisms that control these responses are largely unclear. Here, we demonstrate that both auxin and brassinosteroid (BR) are required for the normal leaf responses to shade in Arabidopsis (Arabidopsis thaliana). The microarray analysis of leaf blades and petioles treated with end-of-day far-red light (EODFR) revealed that almost half of the genes induced by the treatment in both parts were previously identified as auxin-responsive genes. Likewise, BR-responsive genes were overrepresented in the EODFR-induced genes. Hence, the auxin and BR responses were elevated by EODFR treatment in both leaf blades and petioles, although opposing growth responses were observed in these two parts. The analysis of the auxin-deficient doc1/big mutant and the BR-deficient rot3/cyp90c1 mutant further indicates that auxin and BR were equally required for the normal petiole elongation response to the shade stimulus. In addition, the spotlight irradiation experiment revealed that phytochrome in leaf blades but not that in petioles regulated petiole elongation, which was probably mediated through regulation of the auxin/BR responses in petioles. On the basis of these findings, we conclude that auxin and BR cooperatively promote petiole elongation in response to the shade stimulus under the control of phytochrome in the leaf blade.
Light provides important information for plants to control organ growth. When plants are under a canopy of shade cast by neighboring plants, various growth and developmental responses, such as promotion of stem elongation, are elicited. This phenomenon is known as the shade avoidance syndrome (SAS). SAS directs the reallocation of energy resources from the storage organs to the stalk organs in order to escape from the surrounding competitors at the expense of growth of the storage organs (McLaren and Smith, 1978).
Under shade conditions, the ratio of red light (R) to far-red light (FR) is substantially decreased (Kasperbauer, 1971). This change in the light quality is recognized by phytochrome photoreceptors (Whitelam and Smith, 1991; Devlin et al., 1999; Franklin et al., 2003). Phytochrome exists in two photointerconvertible forms: the FR-absorbing Pfr form and the R-absorbing Pr form. The dynamic equilibrium between Pfr and Pr depends on the ratio of R to FR (R:FR; Neff et al., 2000). The low R:FR under the shade shifts the equilibrium of phytochrome toward Pr and triggers SAS. Treatment with a pulse of FR at the end of the light period (EODFR) also induces SAS, because EODFR eliminates Pfr during the subsequent dark period (Smith, 1982).
Arabidopsis (Arabidopsis thaliana) contains five phytochrome-encoding genes, PHYA to PHYE (Sharrock and Quail, 1989; Clack et al., 1994). The SAS response is primarily mediated by the phytochrome B holoprotein (phyB) encoded by PHYB (Robson et al., 1993). Transcriptomic analyses revealed that the expression of many genes is altered in response to the low R:FR treatment (Devlin et al., 2003). Furthermore, a number of reports have described the involvement of various phytohormones, such as auxin, brassinosteroid (BR), ethylene, and GA, in SAS (Kim et al., 1998; Neff et al., 1999; Kanyuka et al., 2003; Djakovic-Petrovic et al., 2007; Tao et al., 2008; Pierik et al., 2009; Sorin et al., 2009).
Among the various phytohormones, the role of auxin in SAS has been explored intensively. For example, the hypocotyl response to low R:FR and the constitutive SAS phenotype in the phyB mutant are suppressed in auxin-resistant mutants, such as axr1-12, shy2-1D, and axr2-1 (Kim et al., 1998; Steindler et al., 1999; Pierik et al., 2009). Furthermore, the endogenous auxin level is increased in response to low R:FR through de novo synthesis (Tao et al., 2008). Several studies have reported that the auxin transport inhibitor N-1-naphthylphthalamic acid (NPA) reduces the hypocotyl SAS response (Steindler et al., 1999; Pierik et al., 2009).
In contrast to the hypocotyl, less is known about the SAS signaling pathway in the leaf. The Arabidopsis leaf consists of a leaf blade and a petiole. The petiole physically supports the leaf blade in the position that is best suited for light reception. Hence, the regulation of petiole elongation should be important to maximize photosynthesis. Accordingly, petiole elongation is promoted by shade stimuli, such as low R:FR and EODFR (Vince-Prue et al., 1976; Nagatani et al., 1991). On the other hand, the growth of leaf blades is suppressed under low R:FR conditions (McLaren and Smith, 1978). Hence, phyB regulates the growth in the leaf blade and the petiole in opposite manners (Kozuka et al., 2005). Although ethylene, auxin, and GA have been shown to be involved in the shade-promoted petiole elongation (Hisamatsu et al., 2005; Djakovic-Petrovic et al., 2007; Pierik et al., 2009), little progress has been made in determining the mechanisms of the petiole response to shade.
In this study, we examined the roles of auxin and BR in the leaf SAS. Microarray analyses revealed that many auxin- and BR-responsive genes showed altered expression in response to EODFR in both the leaf blade and the petiole. The involvement of auxin and BR was further confirmed using loss-of-function mutants of DOC1/BIG, which encodes the calossin-like protein involved in auxin transport (Gil et al., 2001; Yamaguchi et al., 2007), and ROT3/CYP90C1, which encodes a BR biosynthesis enzyme (Kim et al., 2005; Ohnishi et al., 2006). A spotlight irradiation experiment further demonstrated that the shade stimulus was perceived by phytochrome in the leaf blade to control petiole elongation. An inhibitor analysis suggested that auxin transport was involved in this blade-petiole communication. Collectively, this work provides a mechanistic basis for the processing of light stimuli and subsequent regulation of leaf responses under the shade.
RESULTS
Light Regulation of Leaf Growth
To investigate the effects of light on leaf growth, we compared the sizes of the leaf blade and petiole under different light conditions (Fig. 1). Arabidopsis seedlings grown for 7 d under continuous white light (70 μmol m−2 s−1) were further grown under the same light conditions or were placed under a 10-h-light (120 μmol m−2 s−1)/14-h-dark cycle for 20 d with or without the EODFR (50 μmol m−2 s−1 for 5 min) treatment. Consequently, the sizes of the leaf blades and petioles under the light/dark cycles were indistinguishable from those under continuous white light (Fig. 1). In contrast, the EODFR treatment substantially promoted petiole elongation, whereas it suppressed leaf blade expansion, as expected from previous reports (Vince-Prue et al., 1976; McLaren and Smith, 1978; Nagatani, et al., 1991; Kozuka et al., 2005).
Figure 1.
Effects of the light treatments on the growth of leaf blades and petioles. Leaf blade area (left) and petiole length (right) were determined in the wild-type seedlings grown under continuous white light (cWL; 70 μmol m−2 s−1) or short days without (−) or with (+) EODFR (50 μmol m−2 s−1 for 5 min). The short days consisted of 10-h-white-light (100 μmol m−2 s−1)/14-h-dark cycles. Fourth leaves were used for the size measurement (n ≥ 15, mean ± sd). Asterisks indicate significant differences from the white light controls (* P < 0.05, Student's t test).
Transcriptomic Analysis of EODFR Responses in Leaf Blades and Petioles
To gain insight into the mechanisms of the growth responses of the leaf to the shade, we examined the gene expression profiles under different light conditions using the Affymetrix ATH1 GeneChips. Because the leaf blade and the petiole responded to EODFR in opposite manners (Fig. 1), the leaf blade and petiole were analyzed separately. The Arabidopsis young plants were grown under continuous white light (70 μmol m−2 s−1) for 19 d and then subjected to the light treatments shown in Figure 2A. Namely, the plants were further grown under white light (70 μmol m−2 s−1) for 2 h, placed in the dark for 2 h, or incubated in the dark for 2 h after a pulse of FR (5 min at 50 μmol m−2 s−1). Total RNAs prepared from either leaf blades or petioles were hybridized to the gene chips to determine the gene expression profiles.
Figure 2.
Microarray analysis of differential gene expression across three light conditions. A, The light regimes. An FR pulse (50 μmol m−2 s−1 for 5 min) was given at the end of the day. cWL, Continuous white light. B, Flow chart showing the number of genes that were statistically and robustly induced or repressed in response to the light treatments.
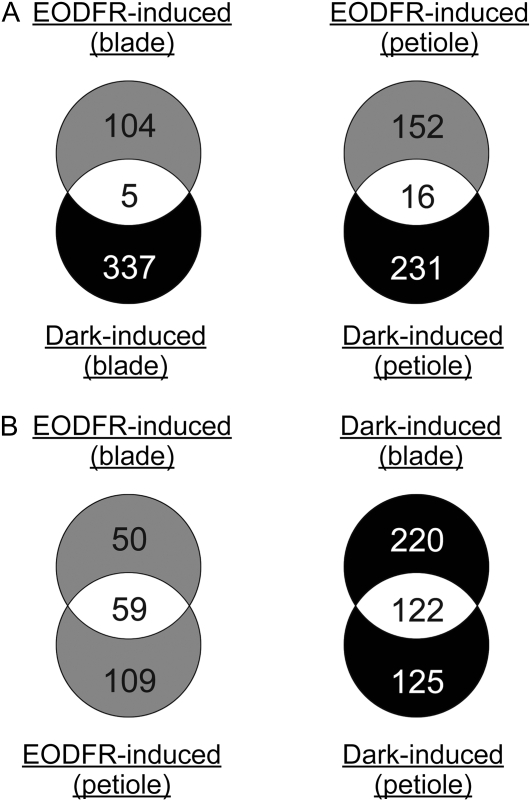
The transcriptomic data were processed as shown (Fig. 2B). The genes that were expressed differentially across the three light conditions were determined using the one-way standard ANOVA model (P < 0.05). Among them, the EODFR-responsive genes were defined as genes that showed at least a 2-fold difference between the dark and EODFR conditions. The dark-responsive genes, which exhibited differential expression between the continuous white light and dark conditions, were defined in the same way. The numbers of genes found in the leaf blade and petiole are shown (Fig. 2B). Although both induced and repressed genes were found in each category, there were less of the latter than the former (Fig. 2B). Hence, we focused on the induced genes, which are referred to as the EODFR- and dark-induced genes.
We found a small number of overlapping genes between the EODFR- and dark-induced genes, indicating that the gene expression profiles were altered quite differently by the inactivation of phytochrome (EODFR) and that of the other photoperception systems (dark). In contrast, many overlapping genes were found between the leaf blade and petiole (Fig. 3B). Nevertheless, 50 and 109 EODFR-induced genes were specific to the leaf blade and the petiole, respectively. The genes are listed in Supplemental Data Sets S1 to S4.
Figure 3.
Comparative analysis of EODFR- and dark-induced genes in leaf blades and petioles. A, Venn diagrams illustrating the different responses to the EODFR and dark treatments. B, Venn diagrams illustrating the different responses in leaf blades and petioles. The numbers of genes belonging to each category are shown.
Correlation between the EODFR-Induced Genes and Phytohormone-Regulated Genes
In the whole seedling, many phytohormone-responsive genes, especially auxin-responsive genes, are induced in response to low R:FR (Devlin et al., 2003; Carabelli et al., 2007). Hence, we asked whether the phytohormone-responsive genes were differentially expressed in response to EODFR in the leaf. For this purpose, the sets of genes responsive to abscisic acid, ethylene precursor (1-aminocyclopropane-1-carboxylic acid), BR, cytokinin, GA, auxin, and jasmonate were cross-referenced with the EODFR-induced genes (Nemhauser et al., 2006).
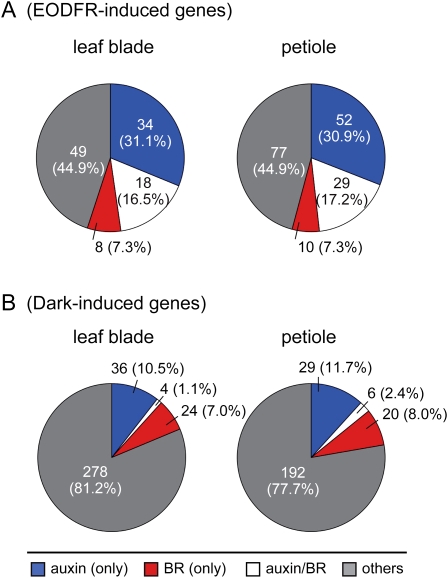
The auxin-responsive genes were overrepresented in the EODFR-induced genes both in the leaf blade (Fisher's exact test, P < 2.2 × 10−16; Supplemental Table S1) and in the petiole (P < 2.2 × 10−16; Supplemental Table S2). Indeed, 52 and 81 auxin-responsive genes, including the IAA, GH3, and SAUR families, were up-regulated by EODFR in the leaf blade and petiole, respectively (Fig. 4A). The lists of those genes are shown in Supplemental Data Set S5.
Figure 4.
The percentages of auxin- and BR-responsive genes found in the EODFR-induced (A) and dark-induced (B) genes. Pie charts show the numbers and percentages (in parentheses) of genes that are induced only by auxin (blue), only by BR (red), and by both auxin and BR (white). Auxin- and BR-responsive genes were defined according to Nemhauser et al. (2006).
We also found that the BR-responsive genes were significantly overrepresented in the EODFR-induced genes in both the leaf blade and petiole (Fig. 4A). In particular, genes that are dually controlled by auxin and BR were overrepresented in the EODFR-induced genes (Fig. 4A). Notably, this tendency was not observed for the dark-induced genes, although similar numbers of BR-responsive genes were found in this category (Fig. 4B; Supplemental Data Set S6).
The Leaf Response to the Shade Stimulus in Auxin- and BR-Related Mutants
To investigate the physiological relevance of auxin and BR in the leaf SAS, we employed a genetic approach using doc1 and rot3 mutants. We reasoned that severe auxin-deficient mutants, such as axr1 and axr2 (Lincoln et al., 1990), are unsuitable for this purpose. Hence, the less severe doc1 mutant was chosen (Gil et al., 2001; Yamaguchi et al., 2007). Likewise, the rot3 mutant, which exhibits a milder dwarf phenotype (Tsuge et al., 1996; Kim et al., 2005) than other mutants such as cpd and bri1 (Szekeres et al., 1996; Li and Chory, 1997), was used.
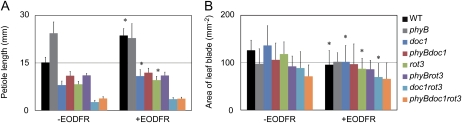
Arabidopsis seedlings grown for 7 d under continuous white light were further grown under a 10-h-light/14-h-dark cycle for 20 d with or without the EODFR treatment as described in Figure 1. As expected from the above reports, the doc1 and rot3 single mutants had shorter petioles than the wild type under both light conditions (Fig. 5A). Importantly, the petiole length was increased by 56% of the EODFR control in the wild type (Fig. 5A), whereas the same treatment was less effective in the doc1 (35%) and rot3 (23%) mutants. Hence, the responsiveness of the petiole to EODFR was decreased by the doc1 and rot3 mutations. Furthermore, an additive responsiveness phenotype was not observed in the doc1rot3 double mutant (34%), indicating that DOC1/BIG and ROT3/CYP90C1 are involved in the same pathway for the regulation of petiole elongation.
Figure 5.
The growth response to EODFR in the phyB, doc1, and rot3 single and multiple mutants. The petiole lengths (A) and areas of the leaf blade (B) of the matured fourth rosette leaves were determined in the wild type (WT), phyB, doc1, phyBdoc1, rot3, phyBrot3, doc1rot3, and phyBdoc1rot3. These seedlings were grown under short days without (−) or with (+) EODFR as described in Figure 1. Fourth leaves were used for the size measurement (n ≥ 15, mean ± sd). Asterisks indicate significant differences from the respective EODFR controls (* P < 0.05, Student's t test).
We also measured the area of the leaf blade in the mutants. However, the doc1 mutation never affected the leaf blade area regardless of the light conditions (Fig. 5B). The rot3 mutant had a slightly decreased leaf blade area in both light conditions. Although the leaf size was further reduced in the doc1rot3 mutant, this mutant retained the normal responsiveness to EODFR. Hence, neither DOC1/BIG nor ROT3/CYP90C1 was required for the leaf blade response to the shade stimulus.
The phyB mutant exhibits constitutive SAS due to permanent loss of phyB Pfr (Nagatani et al., 1991; Whitelam and Smith, 1991; Devlin et al., 1999; Franklin et al., 2003). We confirmed that the introduction of the phyB mutation into the wild-type background increased the petiole length by 60% of the wild-type length without EODFR (Fig. 5A). In the same way, the phyB mutation increased the petiole length by 36%, 34%, and 37% in the doc1, rot3, and doc1rot3 mutant backgrounds, respectively (Fig. 5A). These numbers were consistent with the effects of EODFR observed in the respective genotypes. Hence, the doc1 and rot3 mutations significantly suppressed the petiole elongation induced by the phyB mutation.
Similar to the petiole response, the constitutive leaf blade SAS was observed in the phyB mutant (Fig. 5B). Hence, the phyB Pfr suppressed SAS not only in the petiole but also in the leaf blade. Furthermore, similar effects were observed in all of the genetic backgrounds with the phyB mutation. These results genetically confirmed that the leaf blade SAS was independent of the functions of DOC1/BIG and ROT3/CYP90C1.
Gene Expression Response to the Shade Stimulus in Auxin- and BR-Related Mutants
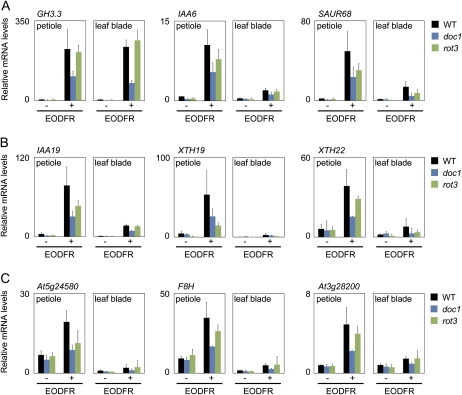
To elucidate the molecular basis of the physiological phenotypes of the doc1 and rot3 mutants, we examined the expression levels of the auxin- and BR-responsive genes under different light conditions. The 19-d-old wild-type, doc1, and rot3 plants were subjected to the same light treatments used for Figure 2A and analyzed by quantitative reverse transcription (RT)-PCR analysis. The genes that are induced only by auxin (GH3.3, IAA6, and SAUR68; Fig. 6A), by both auxin and BR (IAA19, XTH19, and XTH22; Fig. 6B), and only by BR (At5g24580, F8H, and At3g28200; Fig. 6C) were selected for this analysis. According to the microarray analysis, their responses to EODFR were much stronger in the petiole than in the leaf blade (Supplemental Data Set S5).
Figure 6.
Up-regulation of the auxin- and BR-responsive genes in response to EODFR. The mRNA levels were determined in the leaf blade and petiole of the wild-type (WT), doc1, and rot3 plants treated with (+) or without (−) EODFR as described in Figure 2A. A, Auxin-responsive genes: GH3.3, IAA6, and SAUR68. B, Auxin- and BR-responsive genes: IAA19, XTH19, and XTH22. C, BR-responsive genes: At5g24580, F8H, and At3g28200. The transcription levels were quantified by real-time RT-PCR and normalized to UBQ10 (see “Materials and Methods”). Data are expressed in relative units and represented as means ± sd (n = 3).
In wild-type plants, all of the genes except At3g28200 exhibited clear responses to EODFR in both the leaf blade and petiole. The discrepancy between the RT-PCR and the microarray results might be due to the higher sensitivity of the real-time RT-PCR technique. It was noteworthy that the expression levels relative to the internal control (for details, see “Materials and Methods”) were generally higher in the petiole than in the leaf blade in both light conditions (Fig. 6A). Hence, auxin and BR might be maintained at higher levels in the petiole than in the leaf blade independent of the light conditions.
We then examined the gene expression responses in the doc1 mutants. In this mutant, the expression of the auxin-responsive (Fig. 6A) and auxin/BR-responsive (Fig. 6B) genes was reduced in the leaf blade and petiole regardless of the light conditions. Interestingly, the expression of BR-responsive genes was also reduced in doc1, although doc1 has never been regarded as a BR-deficient mutant (Fig. 6C). Furthermore, this phenotype became more pronounced when the plants were treated with EODFR.
The rot3 mutant had similar changes in gene expression to the doc1 mutant. Namely, the expression of all genes, including auxin-responsive (Fig. 6A), auxin/BR-responsive (Fig. 6B), and BR-responsive (Fig. 6A) genes, was more or less reduced in both the leaf blade and the petiole regardless of the light conditions. Interestingly, the effects of rot3 on some genes, such as At5g24580, were more pronounced when the plants were treated with EODFR. Taken together, the DOC1/BIG and ROT3/CYP90C1 functions were equally required for the normal response of the auxin- and BR-responsive genes to the shade.
Photoperceptive Sites for the Petiole Response to EODFR
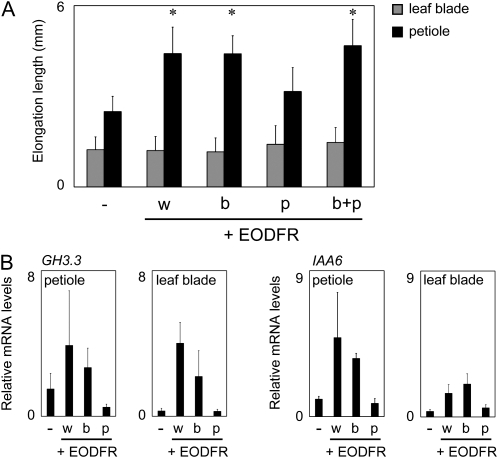
The light signals perceived by phyB in a certain part of the leaf could be delivered to the other parts by way of phytohormones, such as auxin. Hence, we compared the effects of spotlight FR irradiation applied separately to the leaf blade and petiole. Fourteen-day-old young plants grown on agar plates under continuous white light (50 μmol m−2 s−1) were placed under a 10-h-light (50 μmol m−2 s−1)/14-h-dark cycle for 2 d. A spotlight FR pulse (2,000 μmol m−2) was then given on the fifth rosette leaves at the end of each light period. The lengths of the leaf blades and petioles were measured before and after the 48-h treatment, and the increments were determined.
The whole plant irradiation with FR effectively promoted petiole elongation (Fig. 7A). However, the effect of FR on the leaf blade expansion, which was clearly observed after the 20-d treatment (Fig. 1), was not observed in this short-term experiment (Fig. 7A). Interestingly, the spotlight given on the leaf blade was as effective for petiole elongation promotion as the whole plant irradiation (Fig. 7A). In striking contrast, irradiation of the petiole was much less effective. Hence, the photoperceptive site for the regulation of petiole elongation was the leaf blade but not the petiole itself.
Figure 7.
Effect of FR spotlight irradiation on the leaf blades and petioles. Fifth rosette leaves of 14-d-old plants grown under continuous white light were treated without (−) or with a FR pulse on the whole plants (w), leaf blade (b), petiole (p), or leaf blade plus petiole (b+p). A, Increases in the petiole (black bars) and blade (gray bars) lengths. The increments were determined after 2-h rounds of the EODFR treatment. Data are represented as means ± sd (n ≥ 15). Asterisks indicate significant differences from the respective EODFR controls (* P < 0.05, Student's t test). B, GH3.3 and IAA6 mRNA levels in the leaf blade and petiole in response to a FR pulse given as described in A. The mRNA levels were quantified by real-time RT-PCR and normalized to UBQ10 (see “Materials and Methods”). Data are represented as means ± sd (n =3).
Because auxin is basipetally transported in maize (Zea mays) coleoptiles (Iino and Carr, 1982), we speculated that auxin might mediate the signal from the leaf blade to the petiole. Hence, we examined whether expression of auxin-responsive genes in the petiole was under the control of phytochrome in the leaf blade. For this purpose, the fifth rosette leaves of 14-d-old young plants grown under continuous white light (50 μmol m−2 s−1) were treated with or without spotlight FR irradiation and further incubated in the dark for 2 h before RNA extraction. The expression of auxin-responsive genes, such as GH3.3 and IAA6, was then examined by quantitative real-time RT-PCR (Fig. 7B). Consequently, the spotlight given on the leaf blade effectively induced gene expression not only in the leaf blade but also in the petiole. In contrast, petiole irradiation was not effective at all.
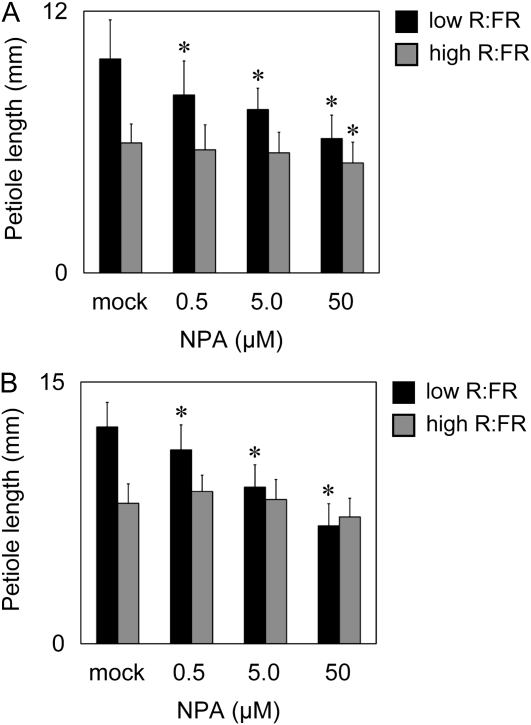
Effect of NPA on the Petiole Elongation Response
To investigate the involvement of auxin transport in the petiole response, we examined the effects of the auxin transport inhibitor NPA. The fifth rosette leaves were excised from 14-d-old plants and placed on an agar plate under R light (10 μmol m−2 s−1) supplemented with (low R:FR) or without (high R:FR) FR light (90 μmol m−2 s−1) for 48 h. We found that the promoting effect of low R:FR on petiole elongation disappeared at NPA concentrations above 50 μm (Fig. 8A). In contrast, almost no inhibitory effect of NPA was observed under the high R:FR condition. Similar results were obtained with intact plants sprayed with an NPA solution (Fig. 8B). Taken together, normal auxin transport within the leaf was important for the promotion of petiole elongation in the shade.
Figure 8.
Effect of NPA on petiole elongation under different light conditions. A, The leaves were excised and grown on agar plates containing NPA at various concentrations for 48 h under high (R [10 μmol m−2 s−1]) or low R:FR (R [10 μmol m−2 s−1], FR [90 μmol m−2 s−1]) conditions. B, The wild-type seedlings were sprayed with the NPA solution and then grown for 48 h as in A. The fifth rosette leaves were used (n ≥ 15, mean ± sd). Asterisks indicate significant differences from the mock controls (* P < 0.05, Student's t test).
DISCUSSION
Global Transcriptional Analysis during Leaf SAS
The present microarray analysis provides, to our knowledge, the first comprehensive genome-wide evidence for the responses of the leaf blade and petiole to shade. Although similar efforts have been made at the whole seedling level (Devlin et al., 2003; Carabelli et al., 2007), SAS responses in different parts within a certain organ had never been compared. Consequently, we identified 50 and 109 genes that were specifically induced in the leaf blade and petiole by EODFR, respectively (Fig. 3). At the same time, we identified 59 genes that were up-regulated both in the leaf blade and petiole. These numbers are comparable to the numbers of genes that responded to the shade stimulus in the whole seedling (Carabelli et al., 2007).
Interestingly, many transcription factor genes were induced by EODFR. Namely, 25 and 35 transcription factors were induced in the leaf blade and petiole, respectively (Supplemental Data Set S7). The SAS response has been proposed to be under the control of a few key transcriptional factors, which activate various target genes and other transcription factors (Carabelli et al., 1996). Indeed, the ATHB2 gene, which encodes a homeodomain ZIP transcription factor, and the HFR1 gene, which encodes a basic helix-loop-helix transcription factor, were up-regulated by the shade stimulus to promote and suppress hypocotyl elongation, respectively (Steindler et al., 1999; Sessa et al., 2005). We inspected the list of the EODFR-induced genes for transcription factors that may be involved in the leaf SAS (Supplemental Data Set S7). Consequently, we found that ATHB2 and HFR1 exhibited the most dramatic responses to EODFR compared with other genes (Supplemental Data Set S7). Although we analyzed other transcription factors, none of them showed a strong response. We confirmed those results by real-time RT-PCR (Supplemental Fig. S1). Hence, ATHB2 and HFR1 may act as key regulators of SAS not only in the seedling but also in the leaf.
According to our microarray data, certain EODFR-induced genes, such as IAA6, SAUR68, XTH19, XTH22, F8H, and At3g28200, were responsive to the light treatment only in the petiole (Supplemental Data Set S7). Nevertheless, the real-time RT-PCR analysis showed that all of them were responsive in both the leaf blade and petiole (Fig. 6). This is probably because the expression levels were too low in the leaf blade to be properly analyzed by the microarray analysis. Indeed, the expression levels in the real-time RT-PCR analysis were three or more times lower in the leaf blade than in the petiole for these genes. Likewise, YUC2, YUC8, and YUC9 genes, which responded to EODFR only in the leaf blade in the microarray analysis (Supplemental Data Set S7), exhibited the response at lower expression levels in the petiole (Supplemental Fig. S1).
Up-Regulation of Auxin Signals in the Leaf SAS
Many auxin-responsive genes were included in the EODFR-induced genes in both the leaf blade and petiole (Fig. 4). Hence, the auxin response was broadly elevated under shade conditions in the leaf. The physiological relevance of this phenomenon was confirmed by the observation that the petiole response to EODFR was reduced in the doc1 mutant (Fig. 5). Furthermore, the promoted petiole cell elongation observed in the phyB mutant was suppressed by the doc1 mutation (Supplemental Fig. S2). Hence, auxin was implicated in the regulation of petiole cell elongation by the shade stimuli. It should be noted that the petiole elongation response to EODFR was not completely abolished in the doc1 mutant. This is probably because the doc1 mutant exhibits a milder phenotype than the severe auxin response mutants such as axr1 (Gil et al., 2001).
The above conclusion should not be surprising, because a tight relationship between phytochrome and auxin has been reported previously. For example, the dominant gain-of-function mutation of SHY2/IAA3, which is defective in auxin-induced growth and gene expression (Tian et al., 2002), suppresses the hypocotyl elongation phenotype of the hy2 phytochrome-deficient mutant (Kim et al., 1996). Our group and others (Pierik et al., 2009) confirmed that the promotion of petiole elongation by EODFR is impaired in the auxin-insensitive mutants, such as axr1 and axr2 (data not shown).
In contrast to the petiole, the up-regulation of auxin-responsive genes was accompanied by the suppression of leaf blade expansion in response to EODFR (Fig. 4A). Hence, the auxin signals apparently suppressed the expansion of the leaf blade. It is noteworthy that the XTH genes, XTH19 and XTH22, were strongly up-regulated in the petiole (Fig. 6B) but not in the leaf blade (Supplemental Data Set S7), although these genes are typical auxin-responsive genes (Nemhauser et al., 2006). Because the XTH family is involved in cell wall loosening (Rose et al., 2002), this expression pattern could explain how the up-regulation of auxin signals promotes the growth only in the petiole. We speculate that an unidentified tissue-specific factor plays an important role in this type of growth regulation.
The above results gave rise to the possibility that the level of auxin in the leaf blade and petiole would be increased under the shade conditions. Previously, it has been reported that the shade stimulus increases free auxin levels in seedlings (Tao et al., 2008). Our microarray data and RT-PCR analysis indicate that the expression of YUC2, YUC8, and YUC9, which are involved in auxin biosynthesis (Zhao et al., 2001; Sugawara et al., 2009), was increased in response to the EODFR treatment (Supplemental Fig. S1). However, no significant change was observed in the auxin levels in both the leaf blade and petiole under different light conditions (Supplemental Fig. S3). Hence, the auxin levels might be regulated differentially between seedlings and leaves in response to the shade stimulus. A more detailed analysis is necessary to characterize the spatial distribution of the auxin levels.
If the auxin levels did not change in response to the shade stimulus, how is the large number of auxin-responsive genes up-regulated? One possible explanation is that local changes in auxin levels, which would not be detected by the conventional auxin measurement method, triggered the change in gene expression and cell elongation response. Alternatively, sensitivity of the tissue to auxin rather than the auxin level itself might be increased in response to the shade stimulus. Indeed, such a phenomenon has been reported repeatedly in various plant tissues (Palme et al., 1991). Furthermore, a member of the TIR1/AFB auxin-binding proteins, AFB1, which is involved in the transcriptional response to auxin (Dharmasiri et al., 2005), was up-regulated in response to EODFR (Supplemental Data Set S7). It remains unclear whether this is the main reason why the expression of auxin-responsive genes was induced.
Involvement of BR in the Leaf SAS
In addition to auxin, BR-responsive genes were significantly overrepresented in the EODFR-induced genes in both the leaf blade and petiole (Fig. 4). Accordingly, the petiole elongation response to EODFR was reduced in the rot3 mutant, suggesting that a normal BR level mediated by ROT3/CYP90C1 is required for this response. These observations are consistent with the previous reports that BR-deficient mutants and seedlings treated with a BR biosynthesis inhibitor show a light-grown phenotype in darkness (Szekeres et al., 1996; Asami et al., 2001). Because a homolog of ROT3/CYP90C1, CYP90D1, redundantly catalyzes the same reaction in the BR biosynthetic pathway (Kim et al., 2005; Ohnishi et al., 2006), it is not surprising that a residual response to EODFR was observed in the rot3 mutant.
Like the doc1 mutant, the rot3 mutation suppressed the cell elongation phenotype in the phyB mutant (Supplemental Fig. S2). Hence, normal BR levels are required for the response of petiole cell elongation to the shade stimulus. However, it remains unclear how the BR responses are related to the leaf blade expansion. As is the case with auxin, the BR-responsive genes were up-regulated in both the leaf blade and petiole in response to EODFR (Fig. 4), whereas the leaf blade expansion was suppressed in the same conditions (Fig. 1). It is noted here that the expression of the XTH19 and XTH22 genes, which are not only auxin-responsive but also BR-responsive genes (Nemhauser et al., 2006), was strongly up-regulated in the petiole (Fig. 6B). Hence, the tissue-specific nature of the BR-responsive genes is probably responsible for mechanisms by which the BR signals differentially regulate the growth of the leaf blade and the petiole.
It is possible that the BR levels are increased under the shade condition. However, this might not be true. The expression level of ROT3/CYP90C1, which is one of the key BR biosynthesis enzymes (Kim et al., 2005; Ohnishi et al., 2006), was altered neither in the leaf blade nor in the petiole in response to EODFR (Supplemental Fig. S2). These observations are consistent with a report that the active BR levels were not changed during the seedling deetiolation process in pea (Pisum sativum; Symons and Reid, 2003). In addition, the BRI1 gene encoding a BR receptor protein (Li and Chory, 1997) was found in the EODFR-induced genes (Supplemental Data Set S7), indicating that sensitivity of BR might be increased by EODFR.
Coordinated Action of Auxin and BR Responses in the Petiole
The doc1 and rot3 mutations affected petiole elongation in similar manners (Fig. 5). Furthermore, the petiole phenotype of these mutants was more pronounced when the plants were treated with EODFR. Interestingly, an additive effect was not observed when these two mutations were combined with respect to responsiveness to EODFR (Fig. 5). Hence, auxin and BR appeared to act cooperatively to promote the petiole elongation under the shade.
The similarity of the two mutants was further observed at the level of cell elongation. Namely, the cell elongation observed in the phyB mutant was partially suppressed by the doc1, rot3, and doc1rot3 mutants to very similar extents (Supplemental Fig. S2). In addition, the doc1 and rot3 mutations affected the expression of both auxin- and BR-responsive genes (Fig. 6). Hence, many auxin- and/or BR-responsive genes might be controlled by both auxin and BR in the leaf (see below).
The above idea is not surprising, considering the previously reported tight coordination of the functions of auxin and BR. For example, auxin plays crucial roles in certain BR-induced growth responses (Mandava, 1988). Auxin and BR share many target genes, such as IAA, GH3, and SAUR (Goda et al., 2004; Nemhauser et al., 2004). Hence, the petiole might be one of the sites where the coordinated action of these two hormones plays a key physiological role. However, it is not likely that phytochrome acts in the petiole to trigger those responses. As discussed below, this response was under the control of phytochrome in the leaf blade but not in the petiole.
Blade-Petiole Communication in the Leaf SAS
Our spotlight irradiation experiment clearly demonstrated that phytochrome in the leaf blade but not that in the petiole controlled the gene expression and elongation responses in the petiole (Fig. 8). Because the Arabidopsis petiole has a stem-like structure, the relationship between the leaf blade and the petiole might be analogous to the relationship between the cotyledon and the hypocotyl. Indeed, leaf phytochrome is known to regulate elongation of the stem (Black and Shuttleworth, 1974; Casal and Smith, 1988a, 1988b).
It is intriguing that auxin transport might play a role in blade-petiole communication. Indeed, the auxin transport inhibitor NPA specifically suppressed the petiole elongation response to EODFR (Fig. 8). In addition, auxin biosynthetic genes were induced in the leaf blade more intensely than in the petiole (Supplemental Fig. S1). Taken together, we speculate that newly synthesized auxin in the leaf blade accumulates in the petiole to induce the auxin/BR cooperative responses. The details of the signaling network that governs plant morphogenesis and the responses to the various light stimuli will be ultimately revealed through functional genomic analyses at the organ and tissue levels in combination with bioinformatics.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) wild type, the PHYB-deficient mutant phyB-9 (Chory et al., 1989), the DOC1/BIG-deficient mutant doc1-1 (Gil et al., 2001), and the ROT3-deficient mutant rot3-1 (Tsuge et al., 1996) were in the Columbia background. The phyBrot3 double mutant was established by crossing phyB-9 with rot3-1 (Tsukaya et al., 2002). The phyBdoc1 double mutant was established by crossing phyB-9 with doc1-1. The rot3doc1 double mutant was established by crossing rot3-1 with doc1-1. The phyBrot3doc1 triple mutant was established by crossing phyBrot3 with phyBdoc1.
PCR-Based Genotyping
The phyB-9 and doc1-1 genomic sequences in the candidate individuals were examined by the derived cleaved amplified polymorphic sequence method (Neff et al., 1998). The phyB-9 mutation was identified using the primers 5′-CATGGGATCTATTGCGTCTTTAGCAATGGC-3′ and 5′-AGAAGAAGTGTGATGGCAAACAACCAGAGC-3′. The PCR product was resolved on a 3.0% agarose gel after digestion with the restriction endonuclease SacI. The doc1-1 mutation was identified using the primers 5′-CTTTTACGTCCAGTGGCAGTAATTTCATGG-3′ and 5′-ACACGGTGACCCCGGTGGCAAACTTTAGCG-3′, in combination with digestion with HhaI. The rot3-1 mutation was identified using the primers 5′-TGGCTCTGGTTACGTCTACG-3′ and 5′-GGAAAGCTGAAGTCTATGGA-3′.
Growth Conditions
Plants were sown on rockwool and grown at 22°C under white fluorescent light as described previously (Tsukaya et al., 1991). For cultivation of plants on plates, seeds were sterilized in a solution of NaClO and incubated for 3 d in sterile distilled water at 4°C in darkness. The seeds were then sown on MS0 (Tsukaya et al., 1991) plates and grown under continuous white fluorescent light (50 μmol m−2 s−1) at 22°C.
Light Treatment
White light was provided from cool-white fluorescent tubes (FL20SSW/18; Toshiba). R and FR light were provided from light-emitting diodes with maximum wavelengths at 660 nm (IS-Series; CCS Inc.) and 735 nm (IS-Series; CCS Inc.), respectively. For the spotlight FR irradiation experiments, plants were grown on agar plates for 14 d under continuous white light. FR light provided by the light-emitting diodes was guided through an acrylic fiber as described previously (Tanaka et al., 2002).
Leaf Size Measurement
The leaf size was determined in the fourth rosette leaves treated with or without EODFR. The leaves were numbered from the first rosette leaves that emerged after the cotyledons to true leaves. For the size measurement, leaves were excised at the basal part of the petiole and flattened by lightly pressing on a sheet of paper. The leaves were then scanned to measure the petiole length and leaf blade area using the ImageJ software (http://rsb.info.nih.gov/ij/). For the spotlight irradiation experiments, the petiole and blade lengths were determined both before and after the light treatment in the fifth rosette leaves of 14-d-old plants. In this case, the lengths were measured with a caliper square. Because the leaf blade tapers into the petiole in the Arabidopsis rosette leaf, the middle point of the transition zone was defined as the petiole-blade boundary.
Microarray Experiment
Biological triplicates for each light condition were subjected to the microarray analysis. Plants were sown on rockwool and grown at 22°C under continuous white fluorescent light. Total RNA was separately prepared from leaf blades and petioles using the Sepazol RNA I Super Kit (Nacalai Tesque) following the manufacturer's instructions. Third and fourth leaves were collected from at least 20 plants. The samples were further purified using the RNeasy Mini Kit (Qiagen). ATH1 GeneChips were used for the expression analysis using 10 μg each of total RNA sample following the manufacturer's instructions (Affymetrix).
Microarray Data Analysis
Array data were processed and analyzed with the Microarray Suites 5.0 software (Affymetrix) and GeneSpring 7.3 (Agilent). Data values were normalized per chip to the 50th percentile and per gene to the median of the measurement in white light. To verify the reliability of the data, genes classified as “present” or “marginal” in at least three of nine samples (triplicate samples in the three light conditions) were selected and used for further analysis. Subsequently, after the removal of the data from synthetic control probes, subsets of the triplicate samples with statistically different or similar expression levels among the three light treatments were compared. The one-way standard ANOVA model with the false discovery rate (P < 0.05; Benjamini and Hochberg, 1995) method was performed. Among them, any genes that did not display at least a 2-fold change across the three light conditions were discarded. Lists of the remaining genes in the leaf blades and petioles were considered as robust sets of differentially expressed genes in response to EODFR and dark (Fig. 2A).
RT-PCR Analyses
Total RNA was prepared from leaf blades and petioles separately using the Sepazol RNA I Super Kit (Nacalai Tesque) for real-time RT-PCR analysis. Third and fourth leaves were collected from at least 20 plants. cDNA was synthesized with the oligo(dT) primer using the SuperScript First-Strand Synthesis System (Invitrogen) and Deoxyribonuclease I, Amplification Grade (Invitrogen), following the manufacturer's instructions. Real-time PCR was carried out as described previously (Mochizuki et al., 2008). The primers used for PCR are listed in Supplemental Table S3.
NPA Treatment
Seedlings were grown under continuous white light conditions for 14 d. The aquatic solution of NPA with 0.01% (v/v) Triton X-100 was sprayed onto intact seedlings. The basal part of the petioles of the excised fifth rosette leaves was vertically inserted into the plates supplemented with the NPA solution in dimethyl sulfoxide (DMSO). Stock solutions of NPA at concentrations of 0.5, 5.0, and 50 mm were prepared in DMSO. Each stock solution was diluted 1,000-fold with distilled water and used for the feeding experiments. As a control, plants were also treated with 0.1% (v/v) DMSO and 0.01% (v/v) Triton X-100.
Sequence data from this study can be found in the GenBank data libraries under the following accession numbers: At4g16780 (ATHB2), At5g47370 (HFR1), At1g52830 (IAA6), At3g15540 (IAA19), At2g23170 (GH3.3), At1g29490 (SAUR68), At4g13260 (YUC2), At4g28720 (YUC8), At1g04180 (YUC9), At4g30290 (XTH19), At5g57560 (XTH22), At4g36380 (ROT3/CYP90C1), At3g02260 (DOC1/BIG), At4g05320 (UBQ10), At1g70560 (TAA1), At5g24580, At5g22940 (F8H), and At3g28200. The microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE17845.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantitative real-time RT-PCR analysis.
Supplemental Figure S2. Length and number of petiole cells.
Supplemental Figure S3. Free IAA levels in leaf blades and petioles.
Supplemental Table S1. Overlap between the hormone-responsive and EODFR-induced genes in leaf blades.
Supplemental Table S2. Overlap between the hormone-responsive and EODFR-induced genes in petioles.
Supplemental Table S3. Primer sequences used for the real-time RT-PCR analysis.
Supplemental Data Set S1. Lists of the EODFR-induced genes in leaf blades and petioles.
Supplemental Data Set S2. Lists of the EODFR-repressed genes in leaf blades and petioles.
Supplemental Data Set S3. Lists of the dark-induced genes in leaf blades and petioles.
Supplemental Data Set S4. Lists of the dark-repressed genes in leaf blades and petioles.
Supplemental Data Set S5. Lists of the auxin- and BR-responsive genes in the EODFR-induced genes.
Supplemental Data Set S6. Lists of the auxin- and BR-responsive genes in the dark-induced genes.
Supplemental Data Set S7. Lists of the EODFR-induced genes in leaf blades and petioles.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We thank Dr. G.T. Kim (Dong-A University, Korea) for helpful discussions and N. Mochizuki (Kyoto University, Japan) for technical advice. We also thank S. Oyama (RIKEN, Japan) for assistance with the GeneChip experiment and M. Kojima (RIKEN, Japan) for support with the IAA measurement.
References
- Asami T, Mizutani M, Fujioka S, Goda H, Min YK, Shimada Y, Nakano T, Takatsuto S, Matsuyama T, Nagata N, et al. (2001) Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J Biol Chem 276: 25687–25691 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57: 289–300 [Google Scholar]
- Black M, Shuttleworth JE. (1974) Role of cotyledons in photocontrol of hypocotyl extension in Cucumis sativus L. Planta 117: 57–66 [DOI] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. (1996) Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA 93: 3530–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, Ruberti I. (2007) Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 21: 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Smith H. (1988a) Persistent effects of changes in phytochrome status on internode growth in light-grown mustard: occurrence, kinetics and locus of perception. Planta 175: 214–220 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Smith H. (1988b) The loci of perception for phytochrome control of internode growth in light-grown mustard: promotion by low phytochrome photoequilibria in the internode is enhanced by blue-light perceived by the leaves. Planta 176: 277–282 [DOI] [PubMed] [Google Scholar]
- Chory J, Peto CA, Ashbaugh M, Saganich R, Pratt L, Ausubel F. (1989) Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1: 867–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC. (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol 119: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LA, Pierik R. (2007) DELLA protein function in growth responses to canopy signals. Plant J 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC. (2003) Phytochromes B, D, and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131: 1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, Palme K, Estelle M, Chory J. (2001) BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev 15: 1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu T, King RW, Helliwell CA, Koshioka M. (2005) The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol 138: 1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino M, Carr DJ. (1982) Sources of free IAA in the mesocotyl of etiolated maize seedlings. Plant Physiol 69: 1109–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanyuka K, Praekelt U, Franklin KA, Billingham OE, Hooley R, Whitelam GC, Halliday KJ. (2003) Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J 35: 57–70 [DOI] [PubMed] [Google Scholar]
- Kasperbauer MJ. (1971) Spectral distribution of light in a tobacco canopy and effects of end-of-day light quality on growth and development. Plant Physiol 47: 775–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Soh MC, Kang BJ, Furuya M, Nam HG. (1996) Two dominant photomorphogenic mutations of Arabidopsis thaliana identified as suppressor mutations of hy2. Plant J 9: 441–456 [DOI] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG. (1998) Photomorphogenic development of the Arabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J 15: 61–68 [DOI] [PubMed] [Google Scholar]
- Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H. (2005) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 41: 710–721 [DOI] [PubMed] [Google Scholar]
- Kozuka T, Horiguchi G, Kim GT, Ohgishi M, Sakai T, Tsukaya H. (2005) The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant Cell Physiol 46: 213–223 [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. (1990) Growth and development of the axr1 mutant of Arabidopsis. Plant Physiol 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava NB. (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39: 23–52 [Google Scholar]
- McLaren JS, Smith H. (1978) Phytochrome control of the growth and development of Rumex obtusifolious under simulated canopy light environments. Plant Cell Environ 1: 61–67 [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc Natl Acad Sci USA 105: 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A, Chory J, Furuya M. (1991) Phytochrome B is not detectable in the hy3 mutant of Arabidopsis, which is deficient in responding to end-of-day far-red light treatments. Plant Cell Physiol 32: 1119–1122 [Google Scholar]
- Neff MM, Fankhauser C, Chory J. (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. (1998) dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al. (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: 1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Szatmari AM, Watanabe B, Fujita S, Bancos S, Koncz C, Lafos M, Shibata K, Yokota T, Sakata K, et al. (2006) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18: 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palme K, Hesse T, Moore I, Campos N, Feldwisch J, Garbers C, Hesse F, Schell J. (1991) Hormonal modulation of plant growth: the role of auxin perception. Mech Dev 33: 97–106 [DOI] [PubMed] [Google Scholar]
- Pierik R, Djakovic-Petrovic T, Keuskamp DH, de Wit M, Voesenek LA. (2009) Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and DELLA proteins in Arabidopsis. Plant Physiol 149: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PH, Whitelam GC, Smith H. (1993) Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in phytochrome B. Plant Physiol 102: 1179–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Braam J, Fry SC, Nishitani K. (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757 [DOI] [PubMed] [Google Scholar]
- Smith H. (1982) Light quality, photoperception, and plant strategy. Annu Rev Plant Physiol Plant Mol Biol 33: 481–518 [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martinez-García JF. (2009) ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 59: 266–277 [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H. (2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 106: 5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Reid JB. (2003) Hormone levels and response during de-etiolation in pea. Planta 216: 422–431 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kaushmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Nakamura S, Mochizuki N, Nagatani A. (2002) Phytochrome in cotyledons regulates the expression of genes in the hypocotyl through auxin-dependent and -independent pathways. Plant Cell Physiol 43: 1171–1181 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Kozuka T, Kim GT. (2002) Genetic control of petiole length in Arabidopsis thaliana. Plant Cell Physiol 43: 1221–1228 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Ohshima T, Naito S, Chino M, Komeda Y. (1991) Sugar-dependent expression of the CHS-A gene for chalcone synthase from petunia in transgenic Arabidopsis. Plant Physiol 97: 1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince-Prue D, Guttridge CG, Buck MW. (1976) Photocontrol of petiole elongation in light-grown strawberry plants. Planta 131: 109–114 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Smith H. (1991) Retention of phytochrome-mediated shade avoidance responses in phytochrome-deficient mutants of Arabidopsis, cucumber and tomato. EMBO J 139: 119–125 [Google Scholar]
- Yamaguchi N, Suzuki M, Fukaki H, Morita-Terao M, Tasaka M, Komeda Y. (2007) CRM1/BIG-mediated auxin action regulates Arabidopsis inflorescence development. Plant Cell Physiol 48: 1275–1290 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.