Abstract
Symbiotic associations between plants and fungi are a dominant feature of many terrestrial ecosystems, yet relatively little is known about the signaling, and associated transcriptome profiles, that define the symbiotic metabolic state. Using the Epichloë festucae-perennial ryegrass (Lolium perenne) association as a model symbiotic experimental system, we show an essential role for the fungal stress-activated mitogen-activated protein kinase (sakA) in the establishment and maintenance of this mutualistic interaction. Deletion of sakA switches the fungal interaction with the host from mutualistic to pathogenic. Infected plants exhibit loss of apical dominance, premature senescence, and dramatic changes in development, including the formation of bulb-like structures at the base of tillers that lack anthocyanin pigmentation. A comparison of the transcriptome of wild-type and sakA associations using high-throughput mRNA sequencing reveals dramatic changes in fungal gene expression consistent with the transition from restricted to proliferative growth, including a down-regulation of several clusters of secondary metabolite genes and up-regulation of a large set of genes that encode hydrolytic enzymes and transporters. Analysis of the plant transcriptome reveals up-regulation of host genes involved in pathogen defense and transposon activation as well as dramatic changes in anthocyanin and hormone biosynthetic/responsive gene expression. These results highlight the fine balance between mutualism and antagonism in a plant-fungal interaction and the power of deep mRNA sequencing to identify candidate sets of genes underlying the symbiosis.
Most, if not all, plants in natural ecosystems form symbiotic associations with fungi that can range in space and time across a continuum from mutualism to antagonism (Clay and Schardl, 2002; Parniske, 2004). While much is known about the signals and mechanisms that lead to pathogenic interactions between fungi and plants, much less is known about how fungi maintain mutualistic interactions and the molecular mechanisms that control symbiont growth within the host (Kogel et al., 2006). Symbiotic associations between endophytes of Epichloë/Neotyphodium species (epichloë endophytes; family Clavicipitaceae) and cool season grasses (family Poaceae, subfamily Pooideae) provide an ideal experimental system to investigate plant-fungus symbiotic interactions. In these associations (symbiota), the fungus occupies a specialized niche, the aerial grass tissues, in which it has access to nutrients in the apoplastic space and a means of dissemination by transmission through the host seeds (Schardl et al., 2004). In return, the fungus promotes host growth and persistence through improved nutrient acquisition and enhanced protection from a range of biotic and abiotic stresses, including drought, disease, and animal herbivory. Protection from herbivory has been shown to be associated with fungal synthesis of various biologically active metabolites, including peramine, lolines, indole-diterpenes, and ergot alkaloids (Schardl, 2001).
In epichloë endophyte-grass associations, hyphae display a highly restricted growth pattern, systemically colonizing all aerial host tissues but remaining within host intercellular spaces, aligned parallel to the leaf axis and seldom branching (Christensen et al., 2002, 2008). Hyphal growth is closely coordinated with host growth, such that hyphae only grow during periods of host leaf growth but remain metabolically active throughout the life of the host (Tan et al., 1997; Christensen et al., 2002, 2008). This highly regulated control of endophyte growth within the grass is indicative of signaling between host and symbiont. Recent work using the Epichloë festucae-perennial ryegrass (Lolium perenne) association as a model experimental system has highlighted how finely balanced the interaction is between mutualism and antagonism. Disruption of components of the reactive oxygen species (ROS)-generating NADPH oxidase (Nox) complex, including NoxA, NoxR, and RacA, led to a breakdown of the mutualistic interaction, with endophyte mutants displaying unrestricted growth in planta and inducing host stunting and premature senescence (Takemoto et al., 2006; Tanaka et al., 2006, 2008). However, little is known about the signaling pathways required to maintain a mutualistic symbiotic interaction.
One of the most important signal transduction pathways for responding to environmental cues is the stress-activated mitogen-activated protein (MAP) kinase pathway. This pathway has been shown to be important for the virulence of many phytopathogenic fungi. For example, Cryphonectria parasitica MAP kinase 1 (cpmk1) is required for optimal virulence on chestnut (Castanea sativa; Park et al., 2004), and Cochliobolus heterostrophus high osmolarity glycerol (hog1) is required for full virulence on maize (Zea mays; Igbaria et al., 2008). Similarly, Botrytis cinerea stress-activated kinase (bcsak1) mutants display greatly reduced pathogenicity and are unable to penetrate unwounded bean (Phaseolus vulgaris) leaves (Segmüller et al., 2007). Previously, we isolated a sakA mutant of E. festucae and showed that it was essential for growth under conditions of temperature and osmotic stress in culture (Eaton et al., 2008).
The objective of this study was to determine the symbiotic phenotype of the E. festucae sakA mutant and to analyze the transcriptome of wild-type and mutant symbiota using deep mRNA sequencing to identify candidate sets of genes associated with the different symbiotic states.
RESULTS
Deletion of E. festucae sakA Disrupts the Mutualistic Association with Perennial Ryegrass
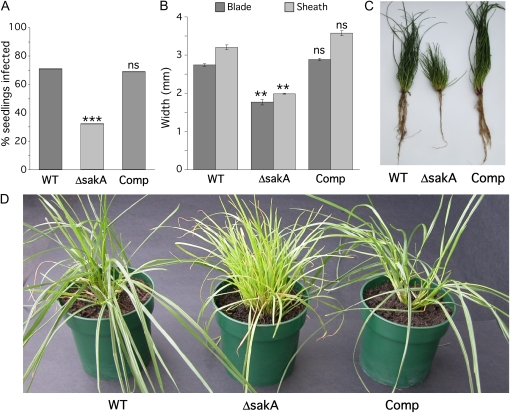
The E. festucae sakA gene, encoding a stress-activated MAP kinase, has previously been shown to be essential for growth under stress conditions in culture (Eaton et al., 2008). To investigate whether this gene is also important in the symbiotic interaction between E. festucae and perennial ryegrass, endophyte-free perennial ryegrass seedlings were inoculated with the wild type and the ΔsakA mutant. Examination of seedlings 6 weeks after inoculation revealed that this mutant displays significantly reduced ability to establish systemic colonization of perennial ryegrass (Fig. 1A). Those plants in which ΔsakA colonization was established displayed a stunted phenotype characterized by the production of a large number of very thin tillers (Fig. 1, B–D). These plants had poorly developed root systems with few lateral roots (Fig. 1C) and prematurely senesced 2 to 3 months after inoculation. Plants infected with a ΔsakA/sakA complemented strain displayed a wild-type interaction phenotype (Fig. 1D; Eaton et al., 2008).
Figure 1.
Symbiotic phenotype of the E. festucae ΔsakA mutant. A, Infection rate analysis for perennial ryegrass inoculated with E. festucae wild-type (WT), ΔsakA mutant, and complemented (Comp) strains. Bars represent average infection rates after multiple trials (n = 265, 282, and 200 for wild-type, ΔsakA, and Comp strains, respectively). Statistical significance in comparison with the wild-type strain was determined using the χ2 test (ns, not significant; ** 0.01 ≥ P > 0.001; *** P < 0.001). B, Average tiller widths of plants infected with wild-type, ΔsakA, and Comp strains. Measurements were taken just above the ligule on the blade (dark shading) and just below the ligule on the sheath (light shading). Error bars are se for two independent experiments (n = 50 for all strains). Reductions in blade and sheath widths were determined using a one-tailed Student's t test. C and D, Tiller and root phenotypes of perennial ryegrass infected with wild-type, ΔsakA, and Comp strains. These photographs were taken 12 weeks after inoculation. Yellowing of ΔsakA mutant-infected tillers indicates the onset of senescence.
Infection with the ΔsakA Mutant Alters Host Plant Development
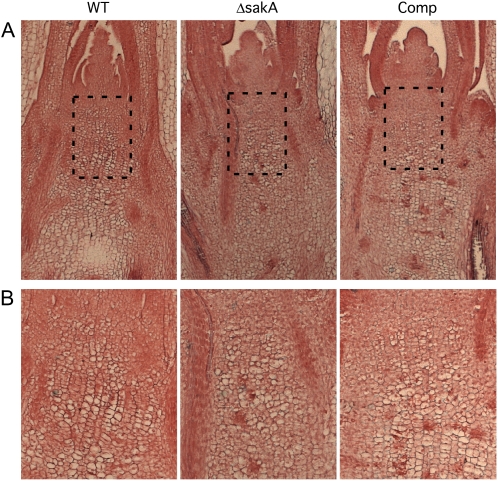
Examination of the base of tillers infected with the ΔsakA mutant revealed dramatic changes in the development of the host plant, with the formation of bulb-like swellings (Fig. 2). Light microscopic examination of longitudinal sections through the swollen region in ΔsakA-infected plants revealed that the host cells, just below the host shoot apical meristem, were much less organized and no longer formed the linear cell files found in wild-type associations (Fig. 3). These cells were irregular in size and shape, preventing tight packing into cell files, likely explaining the observed swollen tissue phenotype.
Figure 2.
Phenotype of perennial ryegrass tillers infected with the E. festucae ΔsakA mutant. Photographs were taken of the base of perennial ryegrass plants infected with wild-type E. festucae (WT), ΔsakA mutant, and complemented (Comp) strains 10 weeks after inoculation. The boxed region of the ΔsakA-infected plant was removed and is magnified below. Bulges can be seen at the bases of ΔsakA-infected tillers, and there is an almost complete loss of anthocyanin pigmentation.
Figure 3.
Cellular organization of perennial ryegrass shoot apical meristems infected with the ΔsakA mutant. A, Light micrographs of Alcian blue/Safranin O-stained longitudinal sections through the meristem region of tillers infected with E. festucae wild-type (WT), ΔsakA mutant, and complemented (Comp) strains. Cells below the shoot apical meristem in ΔsakA mutant associations do not form linear cell files and appear disorganized and less uniform in size and shape compared with cells from the same region in wild-type associations. B, Enlargement of boxed regions in the micrographs in A.
Plants infected with the ΔsakA mutant displayed almost complete loss of the photoprotective anthocyanins (Steyn et al., 2002), which normally accumulate at the base of perennial ryegrass tillers, suggesting a change in the biosynthesis or breakdown of these pigments (Fig. 2).
Infection with the ΔsakA mutant also induced changes in host vasculature development (Supplemental Fig. S1). The frequency of cross-vein branching between parallel vascular bundles in the ΔsakA mutant associations was significantly greater than in wild-type associations.
The ΔsakA Mutant Elicits a Host Defense Response
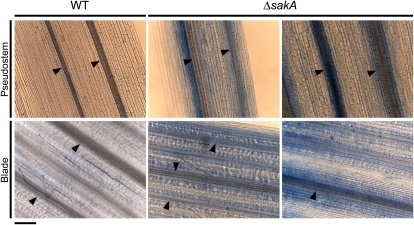
Given that ΔsakA mutant-infected plants displayed premature senescence, we investigated whether the ΔsakA mutant induced any host defense response. Incubation of blade and pseudostem tissue with lactophenol trypan blue (Koch and Slusarenko, 1990) revealed diffuse staining throughout the ΔsakA mutant-infected tissue, particularly close to host vascular tissue, but very little staining of the wild-type-infected tissue (Fig. 4). This diffuse pattern of staining is indicative of a systemic defense response to the ΔsakA mutant, a result consistent with the long period of time (2–3 months) over which the ΔsakA mutant-infected plants senesce compared with wild-type-infected plants.
Figure 4.
Host defense response to the E. festucae ΔsakA mutant. Light micrographs show lactophenol trypan blue-stained sections from pseudostem and blade tissues infected with wild-type E. festucae (WT) and the ΔsakA mutant. Plants infected with the ΔsakA mutant show diffuse staining throughout the tissue, particularly surrounding host vascular bundles. Arrowheads indicate the positions of host vascular bundles. Bar = 100 μm.
The ΔsakA Mutant Displays Unrestricted Growth in Planta
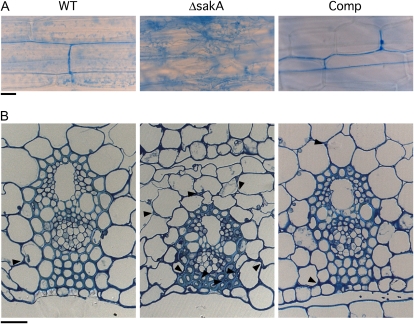
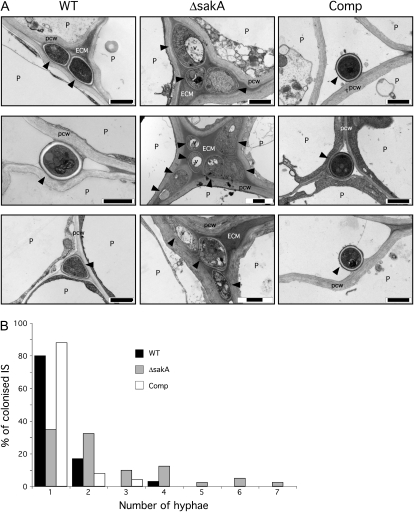
Examination of the growth of the ΔsakA mutant in planta, using aniline blue to stain the fungal cells, revealed dramatic changes in host colonization compared with that seen in wild-type associations (Fig. 5A). ΔsakA mutant hyphae were rarely aligned parallel to the leaf axis and appeared hyperbranched, with an apparent increase in fungal biomass when compared with wild-type associations, in which hyphae grow parallel to the leaf axis and seldom branch. ΔsakA mutant hyphae also colonized the host vascular bundles, a feature rarely seen in wild-type associations (Fig. 5B). Examination of hyphal morphology by transmission electron microscopy revealed marked differences between the wild-type and ΔsakA mutant strains (Fig. 6). Multiple ΔsakA hyphae were often found within host intercellular spaces, in contrast to the generally single hyphae seen in wild-type associations, and these appeared larger and more irregular in size and shape than wild-type hyphae. The ΔsakA mutant hyphae were also highly vacuolated, a feature seldom seen in wild-type associations. No differences in vacuolation were observed between the wild-type and ΔsakA mutant strains grown in culture (Supplemental Fig. S2), suggesting that the change seen in planta is induced by the physiological conditions within the host. The extracellular matrix surrounding the ΔsakA mutant hyphae also appeared electron dense compared with the relatively clear space surrounding wild-type hyphae (Fig. 6).
Figure 5.
In planta phenotype of the E. festucae ΔsakA mutant. A, Light micrographs of aniline blue-stained hyphae growing in the leaf sheaths of 8-week-old perennial ryegrass plants infected with E. festucae wild-type (WT), ΔsakA mutant, and complemented (Comp) strains. Bar = 30 μm. B, Light micrographs of toluidine blue-stained transverse sections showing vascular bundles in the pseudostem of wild-type-infected, ΔsakA-infected, and Comp-infected perennial ryegrass. ΔsakA mutant hyphae can be seen within the vascular bundle complex. Arrowheads indicate the positions of fungal hyphae. Bar = 20 μm.
Figure 6.
E. festucae ΔsakA hyphal morphology and density in planta. A, Transmission electron micrographs of cross sections showing E. festucae hyphae growing in the intercellular spaces of host pseudostem tissue. Arrowheads indicate fungal hyphae. ECM, Extracellular matrix; P, plant cell; pcw, plant cell wall; v, vacuole. Bars = 1 μm. B, Graph showing the number of hyphae in intercellular spaces (IS) colonized by the wild-type (WT), ΔsakA, and complemented (Comp) strains. n = 30, 40, and 26 for wild-type, ΔsakA, and Comp strains, respectively.
The ΔsakA Mutant Appears Unable to Penetrate the Host Cuticle
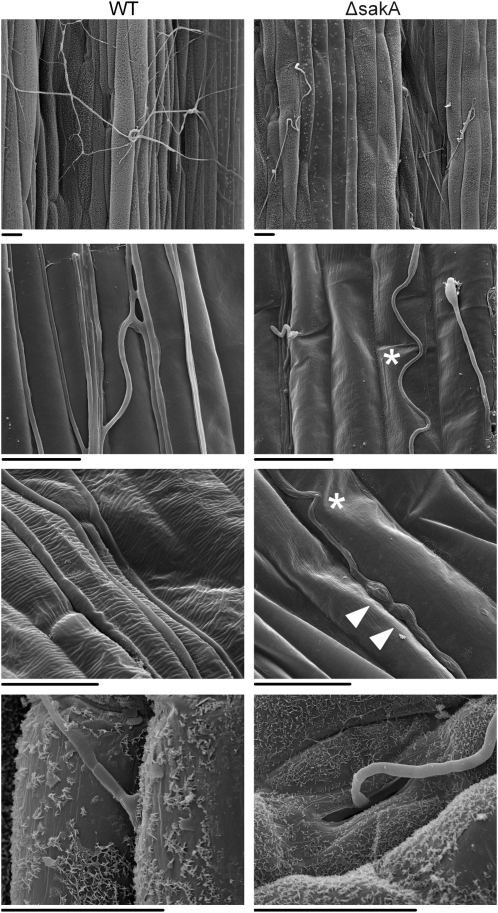
E. festucae hyphae are frequently observed growing on the surface of blade and pseudostem tissue of infected plants as epiphyllous hyphae. Examination of blade tissue from infected plants grown under axenic growth conditions by scanning electron microscopy revealed that few ΔsakA epiphyllous hyphae were seen compared with the wild-type strain (Fig. 7). Those ΔsakA epiphyllous hyphae that were seen frequently contained swollen compartments and often appeared convoluted compared with the relatively straight hyphae of the wild-type strain. Examination of the sites from which the epiphyllous hyphae exited the host tissues revealed that in contrast to wild-type hyphae, which appeared to emerge through the host cuticle, ΔsakA mutant hyphae were only observed to exit through host stomatal pores, suggesting that they are unable to penetrate the host cuticle (Fig. 7).
Figure 7.
Epiphyllous growth of the E. festucae ΔsakA mutant. Scanning electron micrographs show epiphyllous hyphae of wild-type E. festucae (WT) and the ΔsakA mutant growing on the surface of infected perennial ryegrass blade tissue. Photographs were taken 9 weeks after inoculation. Arrowheads indicate swollen compartments. Asterisks indicate convoluted hyphae. Bars = 20 μm.
Production of ROS Is Increased in ΔsakA Mutant Associations
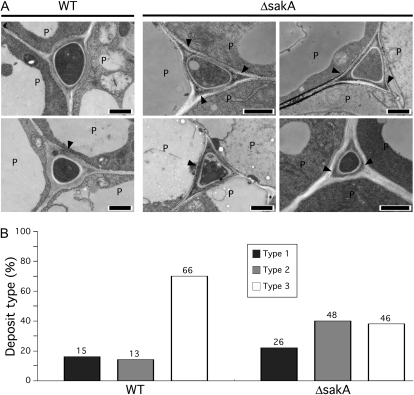
The ΔsakA mutant has previously been shown to produce elevated levels of the ROS hydrogen peroxide in culture (Eaton et al., 2008), so ROS levels were also examined in planta. Using transmission electron microscopy to detect deposits of cerium perhydroxides, which are formed as a result of cerium chloride reacting with hydrogen peroxide, electron-dense precipitates were detected in the fungal extracellular matrix and along the host cell walls surrounding both wild-type and ΔsakA hyphae in meristematic tissue (Fig. 8). Just single hyphae of the ΔsakA mutant were observed in this tissue, compared with multiple hyphae in the pseudostem (Fig. 6), presumably because of the compact nature of the meristematic tissue. The number of infected intercellular spaces containing precipitate on the fungal extracellular matrix or host cell wall was greater in ΔsakA mutant associations than in wild-type associations (Fig. 8), suggesting that more hydrogen peroxide is present around the ΔsakA mutant than in wild-type hyphae.
Figure 8.
ROS production in perennial ryegrass infected with the E. festucae ΔsakA mutant. A, Transmission electron micrographs showing hydrogen peroxide localization in meristematic tissue infected with the wild type (WT) or the ΔsakA mutant. Cerium chloride-reactive deposits indicative of hydrogen peroxide production are indicated by arrowheads. P, Plant cell. Bars = 1 μm. B, Distribution of cerium chloride-reactive deposits in meristematic tissue infected with the wild type or the ΔsakA mutant. Type 1, Deposits in the fungal extracellular matrix; type 2, deposits in the fungal extracellular matrix and host cell wall; type 3, no deposits detected. The number of intercellular spaces of each type is given above each column.
Molecular Dissection of the ΔsakA-Plant Interaction Phenotype by High-Throughput mRNA Sequencing
To dissect the molecular basis for the dramatic differences in plant interaction phenotypes between the wild-type and ΔsakA mutant strains, the transcriptomes of these two plant-fungus associations were analyzed by high-throughput mRNA sequencing (Kim et al., 2007). Sample identification was confirmed by mapping reads to the hygromycin phosphotransferase (hph) reporter gene present in the mutant sample: no hits were identified with the wild-type sample, while 191 hits were found for the ΔsakA mutant (χ2 = 187, degrees of freedom = 1, P < 0.001). Differences in gene expression were determined using a map-and-count strategy. We found a 3.6-fold increase in read counts for the ΔsakA relative to the wild-type sample, reflecting the increased fungal biomass observed in this association. To confirm the accuracy of this method, differential gene expression tests were carried out between the two wild-type lanes and between the two ΔsakA lanes. No differentially expressed genes were identified in either of these sets of technical replicates. Conversely, the fungal and plant genes that were differentially expressed between the wild-type and ΔsakA lanes are described in more detail below (Supplemental Tables S1 and S2).
Differentially Expressed Fungal Genes
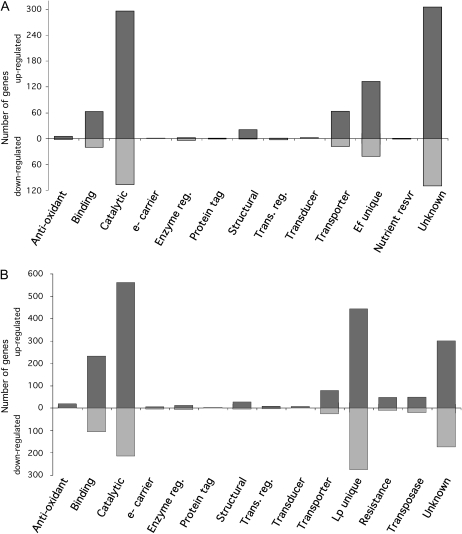
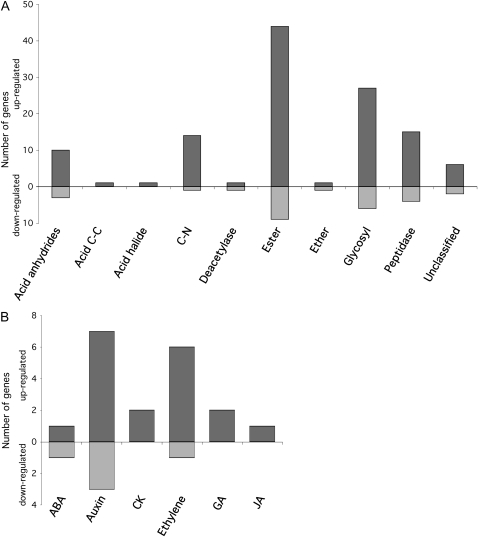
Approximately 11% of genes in the E. festucae sequence set were differentially expressed between the two samples, with 894 genes up-regulated and 308 genes down-regulated in the ΔsakA sample relative to the wild-type sample (Fig. 9A). These genes were organized into molecular function Gene Ontology (GO) categories based on their primary function (Supplemental Figs. S3A, S4A, and S5A; Supplemental Table S1). A number of genes for degradative enzymes, including peptidases, glycosyl hydrolases, and ester hydrolases, and several transporter proteins were up-regulated in the ΔsakA sample, consistent with a transition of the fungal endophyte from restricted to proliferative growth (Fig. 10A). The up-regulation of translation-associated genes, including those for ribosomal proteins and translation initiation factors, as well as cell wall and extracellular matrix protein-encoding genes, is consistent with this growth transition. The dramatic change in expression of a number of transcription factor genes provides some explanation for the large number of differentially expressed genes. Interestingly, 14.5% of the differentially expressed fungal genes had no significant BLASTX hits to the GenBank database and were classified as E. festucae unique genes. This is particularly interesting given that some of these genes show large (greater than 50-fold) differences in expression (Supplemental Table S1), suggesting that they are important for regulating the symbiotic relationship.
Figure 9.
Differentially expressed fungal and plant genes organized by molecular function GO categories. Organization of all E. festucae (A) and perennial ryegrass (B) genes differentially expressed between the wild-type-infected and ΔsakA mutant-infected samples is by molecular function GO categories. Bars show the number of genes within each category that are up- or down-regulated in the ΔsakA mutant-infected sample relative to the wild-type-infected sample. Categories are as follows: antioxidant activity (GO:0016209), binding (GO:0005488), catalytic activity (GO:0003824), electron carrier activity (e- carrier; GO:0009055), E. festucae unique genes (Ef unique), enzyme regulator activity (Enzyme reg.; GO:0030234), perennial ryegrass unique genes (Lp unique), nutrient reservoir activity (GO:0045735), protein tag (GO:0031386), disease resistance genes (Resistance; non-GO category), structural molecule activity (GO:0005198), transcription regulator activity (Trans. reg.; GO:0030528), molecular transducer activity (Transducer; GO:0060089), transporter activity (GO:0005215), transposase activity (GO:0004803), and molecular function unknown (Unknown; non-GO category).
Figure 10.
Differentially expressed fungal hydrolases and plant hormone-associated genes. A, Organization of E. festucae hydrolases differentially expressed between the wild-type-infected and ΔsakA mutant-infected samples by GO category. Categories are as follows: hydrolases acting on acid anhydrides (GO:0016817), acid carbon-carbon bonds (Acid C-C; GO:0016822), acid halide bonds (GO:0016824), carbon-nitrogen (but not peptide) bonds (C-N; GO:0016810), ester bonds (GO:0016788), ether bonds (GO:0016801), glycosyl bonds (GO:0016798); peptidase activity (GO:0008233), deacetylase activity (GO:0019213), and unclassified hydrolase activity. B, Number of perennial ryegrass hormone-associated genes differentially expressed between the wild-type-infected and ΔsakA mutant-infected samples. ABA, Abscisic acid; CK, cytokinin; GA, gibberellic acid; JA, jasmonic acid.
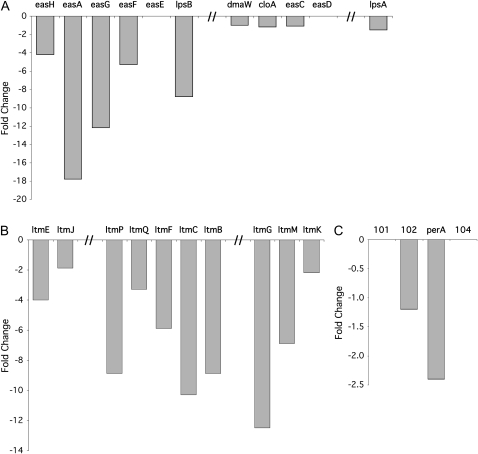
Some of the most striking changes in expression were found for secondary metabolism genes, where nearly all genes required for ergot alkaloid biosynthesis (Fig. 11A), all of the genes required for indole-diterpene biosynthesis (Fig. 11B), and the perA gene, which is necessary for the production of the anti-insect feeding deterrent peramine (Fig. 11C), were down-regulated in the ΔsakA sample. Interestingly, a gene adjacent to perA, encoding a major facilitator superfamily transporter, was also found to be down-regulated. In addition to the genes involved in these well-characterized secondary metabolism clusters, a further 19 cytochrome P450 monooxygenases, four nonribosomal peptide synthetases, and three polyketide synthases, key secondary metabolism genes, also showed altered expression between the two samples.
Figure 11.
Changes in expression of E. festucae secondary metabolism genes. A, Fold change in expression of the ergot alkaloid biosynthetic genes in the ΔsakA mutant-infected sample relative to the wild-type strain-infected sample. Genes for ergot alkaloid synthesis (eas) are spread across three clusters as indicated. B, Fold change in lolitrem (ltm) gene expression in the ΔsakA mutant-infected sample relative to the wild-type strain-infected sample. Genes for lolitrem biosynthesis are spread across three linked clusters interspersed with transposon relic DNA (Young et al., 2006). C, Fold change in expression of perA and EF102 in the ΔsakA mutant-infected sample relative to the wild-type strain-infected sample. perA and EF102, a putative major facilitator superfamily transporter, are proposed to form a two-gene cluster, with EF102 predicted to be involved in the transport of peramine.
To identify potentially novel gene clusters, sets of coregulated genes were categorized. Cluster sizes with three or more up-regulated genes, or two or more down-regulated genes, are outliers under the simulated size distribution (95% confidence interval). The empirical data set contains 67 clusters in this size range, which is statistically unlikely when compared with simulated data sets (P < 0.001). Cluster sizes with five or more up-regulated genes, or four or more down-regulated genes, are found in fewer than one in 20 simulated data sets (P = 0.05). We identified six such clusters in the empirical data set, of which three contained genes showing higher expression and three contained genes showing lower expression in the mutant. One of these down-regulated clusters corresponds to the indole-diterpene gene cluster (Fig. 11B). The additional two down-regulated putative clusters corresponded to a putative nitrogen metabolism cluster and a cluster containing the novel endophyte gene Nc25 (Johnson et al., 2003; Supplemental Fig. S6). Nc25 is a novel endophyte gene that is highly expressed in planta and thought to regulate the production of an uncharacterized oligopeptide (Johnson et al., 2003, 2007). It is the most abundant transcript in the wild-type sample, representing 3.35% of all fungal reads, but it drops to 0.06% of the fungal reads in the ΔsakA sample, where the most abundant transcript encodes a chitinase (2.3% of all fungal reads; Li et al., 2004). The three up-regulated gene clusters are predicted to have roles in sugar metabolism, cell wall integrity, and hydrolytic processes (Supplemental Fig. S7).
Differentially Expressed Plant Genes
Given the lack of a reference perennial ryegrass genome sequence, it is difficult to determine the proportion of perennial ryegrass genes that show differential expression between the wild-type and ΔsakA samples. However, approximately 17% of genes within our de novo assembled EST library displayed differential expression, and if we assume that the perennial ryegrass genome size is around 25,000 genes, as in Brachypodium distachyon (International Brachypodium Initiative, 2010), then this EST library contains approximately 60% of the total predicted perennial ryegrass gene set. The differentially expressed plant genes were organized into molecular function GO categories based on their primary function (Fig. 9B; Supplemental S3B, S4B, S5B, and S8; Supplemental Table S2). These differentially expressed genes include classes indicative of a stressed physiological state, including transposases (Grandbastien, 1998), and a variety of disease resistance genes. In addition, genes encoding a number of components of the RNA-induced silencing complex, including DICER and numerous ARGONAUTE proteins, showed altered expression. Many genes for kinases and hydrolytic enzymes were up-regulated in the mutant, suggestive of changes to global phosphorylation and protein degradation (Supplemental Fig. S8). Differential expression of genes associated with DNA and histone modification, including DNA methyltransferases, histone acetyltransferases, histone deacetylases, and histone demethylases, suggests that there may be widespread changes to chromatin structure in the mutant. A large number of genes (n = 720) displaying no BLASTX hits to the GenBank database were found to be differentially expressed between the two samples. These are referred to as perennial ryegrass unique, but due to a lack of availability of annotated grass genome data, we cannot determine whether they are also found in other grasses. Interestingly, genes for antioxidant enzymes were dramatically up-regulated in the ΔsakA sample compared with the wild-type sample, with 27 genes, including those for peroxidases, catalases, glutathione S-transferases, thioredoxins, and glutaredoxins, displaying elevated expression levels (Fig. 9B). This is likely in response to the elevated ROS levels seen in these mutant associations (Fig. 8). Genes for protein degradation, including ubiquitin, multiple ubiquitin ligases, and numerous F-box proteins, were also differentially expressed (Ho et al., 2006). Many of the changes in expression seen for the ΔsakA sample are consistent with the dramatically altered development displayed by ΔsakA mutant-infected plants. For example, genes for two novel cell wall-loosening expansin proteins were up-regulated in the mutant association, suggesting that these proteins may play a key role in the organization of host cells below the shoot meristem (Fig. 3). There were also considerable changes in the expression of plant hormone-associated genes (Fig. 10B). The most obvious of these was the up-regulation of genes for ethylene biosynthesis or response to ethylene, a potent inducer of plant senescence (Grbić and Bleecker, 1995). A number of genes involved in auxin signaling also showed differential expression, including the auxin influx carrier AUX1, whose expression was up-regulated in the ΔsakA sample. This suggests dramatic interference with host auxin signaling by the ΔsakA mutant. There were also changes in expression of a few abscisic acid-, cytokinin-, and jasmonic acid-associated genes, implying that the ΔsakA mutant induces changes to all major host hormone signaling pathways.
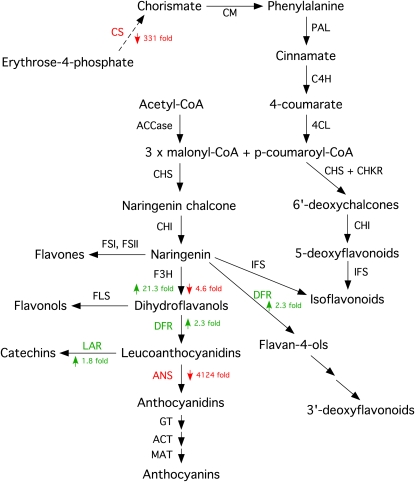
A dramatic reduction in expression of anthocyanidin synthase (ANS; 4,124-fold) provides a good explanation for the loss of anthocyanin pigmentation displayed by ΔsakA-infected plants (Fig. 12). ANS catalyzes the second to last step in the anthocyanin biosynthetic pathway, converting colorless leucoanthocyanidins into colored anthocyanidins (Turnbull et al., 2000). There was also down-regulation of chorismate synthase, which catalyzes the last seven steps in the shikimate pathway to generate chorismate (Kitzing et al., 2004). These results suggest that the loss of anthocyanin pigmentation in ΔsakA mutant-infected plants may be a consequence of both reduced flux through the anthocyanin biosynthetic pathway due to reduced Phe availability and dramatically reduced conversion of leucoanthocyanidins into anthocyanidins by ANS.
Figure 12.
Differential expression of perennial ryegrass anthocyanin biosynthetic genes. The scheme shows changes in the expression of anthocyanin biosynthetic genes between wild-type-infected and ΔsakA mutant-infected samples. Genes up-regulated in the ΔsakA mutant-infected sample, and their fold changes in expression, are shown in green. Genes down-regulated in the ΔsakA mutant-infected sample, and their fold changes in expression, are shown in red. CS, Chorismate synthase; CM, chorismate mutase; PAL, Phe ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:CoA ligase; CHS, chalcone synthase; CHKR, chalcone ketide reductase; CHI, chalcone isomerase; IFS, isoflavone synthase; DFR, dihydroflavonol 4-reductase; ACCase, acetyl-CoA carboxylase; FSI, flavone synthase I; FSII, flavone synthase II; F3H, flavanone 3β-hydroxylase; FLS, flavonol synthase; LAR, leucoanthocyanidin reductase; GT, glucosyltransferase; ACT, anthocyanin acyltransferase; MAT, malonyltransferase. Adapted from Springob et al. (2003).
DISCUSSION
Interactions between plants and fungi span a broad continuum from mutualistic to pathogenic associations. Establishment and maintenance of these associations involves a complex interplay of plant and fungal genes that act to either maintain mutualism or enable/prevent pathogenicity. We show here that deletion of the E. festucae stress-activated MAP kinase (sakA) gene triggers a transition from a highly restricted, mutualistic association to a proliferative, pathogenic-like association with perennial ryegrass. Plants infected with the ΔsakA mutant not only display stunted growth and premature senescence but also show previously unseen, dramatic changes in development. Using a high-throughput mRNA sequencing approach to compare the transcriptomes of the wild-type and perturbed ΔsakA mutant associations, we have been able to identify genes putatively involved in controlling the symbiosis.
The reduced ability of the ΔsakA mutant to successfully colonize its host is consistent with reduced virulence displayed by stress-activated MAP kinase mutants of the phytopathogenic fungi B. cinerea and C. heterostrophus (Segmüller et al., 2007; Igbaria et al., 2008). This reduction in the ability of the ΔsakA mutant to colonize perennial ryegrass may be due to the osmotic environment within host tissues, as this mutant has previously been shown to be osmosensitive (Eaton et al., 2008). Interestingly, no other E. festucae symbiotic mutants characterized to date, including ΔnoxA, ΔnoxR, and ΔracA (Takemoto et al., 2006; Tanaka et al., 2006, 2008), displayed a reduction in colonization ability.
Infection of perennial ryegrass with the ΔsakA mutant led to a loss of apical dominance, with increased tillering and stunting of host growth, similar to the host interaction phenotypes displayed by the E. festucae nox mutants (Takemoto et al., 2006; Tanaka et al., 2006, 2008). These plants also displayed poorly developed root systems with few lateral roots. This loss of apical dominance and reduced lateral root growth would be consistent with a reduction in auxin signaling (Casimiro et al., 2001; McSteen, 2009). However, while transcriptome data do show changes in the expression of auxin-associated genes, it appears that the stunted phenotype is not simply due to down-regulation of auxin signaling. Many auxin response genes are up-regulated in the mutant association, suggesting that other factors also play a role in generating the stunted host phenotype. In contrast, increased expression of the auxin influx transporter AUX1 provides an explanation for the altered vasculature development displayed by ΔsakA mutant-infected plants. These plants show increased formation of cross-veins, which are thought to provide alternative paths for solute and water transport in cases of leaf damage, linking parallel vascular bundles in blade and sheath tissue (Blackman, 1971). Increase of these cross-veins is thought to be triggered by auxin transport (Sakaguchi and Fukuda, 2008).
Hyphal growth of the ΔsakA mutant appeared unrestricted, with hyphae hyperbranched and seldom exhibiting the parallel alignment to the leaf axis that typifies wild-type E. festucae. Multiple, irregularly sized and shaped hyphae were often found colonizing single host intercellular spaces. In addition, hyphae were found within host vascular bundles, a feature seldom seen in wild-type associations (Christensen et al., 2002). These hyphae are likely acting as nutrient sinks, removing nutrients directly from the host phloem. The up-regulation of transporters for uptake of host-derived nutrients and degradative enzymes to break them down is consistent with the switch from restricted to proliferative growth. The finding of more than 3.6 times as many fungal transcriptome reads in the ΔsakA mutant-infected sample (relative to the total number of reads) is consistent with the dramatic increase in fungal biomass in planta displayed by this mutant. This is undoubtedly aided by the increased expression of genes involved in the translational machinery, as evidenced from the transcriptome data.
The switch from highly restricted, mutualistic growth to proliferative growth appears to have also greatly affected fungal secondary metabolism, with dramatic down-regulation of genes required for the production of indole-diterpenes, ergot alkaloids, and peramine. This down-regulation of secondary metabolism genes is likely to be a direct consequence of the energy requirements for proliferative growth.
In contrast to the proliferative growth displayed by the ΔsakA mutant within host tissues, epiphyllous growth of this mutant was drastically reduced. While this reduction in epiphyllous growth was unexpected, it is not without explanation. The apparent inability of the ΔsakA mutant to penetrate the host cuticle is likely due to an inability to establish sufficient turgor pressure, or alternatively to produce degradative enzymes, for penetration (Wang et al., 2005; Noguchi et al., 2007). This would limit epiphyllous growth to those few hyphae that encounter and grow out of host stomatal openings, analogous to how some phytopathogenic fungi enter host tissues (Willmer and Fricker, 1996).
The up-regulation of expression of a large number of disease resistance genes (n = 47), including those for the classic pathogenicity response proteins (PR1, PR4b, PR5, and PR10), and numerous nucleotide-binding site Leu-rich repeat-type resistance proteins in the ΔsakA mutant-infected sample is consistent with the host defense-like phenotypes observed, including premature senescence, diffuse staining with lactophenol trypan blue, and the accumulation of electron-dense extracellular matrix around ΔsakA mutant hyphae in planta (Koga et al., 1993; Bolwell et al., 2001; Lam et al., 2001). The highly vacuolated nature of the ΔsakA mutant hyphae in planta may also be the result of a plant defense response, or it may simply be due to the osmotic environment within the host tissues (Dixon et al., 1999). The observed increased expression of genes for cell wall proteins and the translational machinery is consistent with the responses of pathogenic fungi to host defense processes (Bhabhra and Askew, 2005). Ethylene is a likely mediator of the host defense response, given its known role in senescence (Grbić and Bleecker, 1995) and the observation here that levels of ethylene biosynthetic genes and ethylene response genes were found to be elevated in the ΔsakA mutant-infected plant. Interestingly, a large number of genes for plant transposase proteins were up-regulated in the ΔsakA mutant-infected plant. Previous work has shown a link between transposon activation and a stressed physiological state (Wessler, 1996; Grandbastien, 1998).
The ΔsakA mutant-infected plants also displayed dramatic changes in development, with the formation of bulb-like structures at the base of the tillers, loss of anthocyanin pigmentation, and changes to vasculature development. The up-regulation of two genes for novel cell wall-loosening expansins may explain why the cells located in the true stem are disordered in mutant compared with wild-type associations. The loss of anthocyanin pigmentation in ΔsakA mutant-infected plants is likely due to decreased expression of genes encoding key enzymes in the shikimate (chorismate synthase) and anthocyanin (ANS) biosynthetic pathways. The down-regulation (331-fold) of chorismate synthase has the potential to reduce levels of chorismate and in turn Phe to feed into the anthocyanin biosynthetic (phenylpropanoid) pathway. Similarly, the down-regulation of ANS (4,124-fold) is likely to be responsible for the dramatically reduced levels of the anthocyanidins and the potential diversion of their proximate precursors, the leucoanthocyanidins, into the catechin/tannin pathway. This is of particular importance given that tannins are known to precipitate proteins, act as metal ion chelators, and have antioxidant properties (Hagerman et al., 1998; Karamac, 2009).
This study demonstrates the ability of high-throughput mRNA sequencing to identify putative gene clusters based on similar changes in gene expression, providing an alternative to the use of microarrays to identify gene clusters by comparing wild-type with mutant expression levels, especially because microarrays cannot be designed for nonsequenced organisms (Bok et al., 2006). We identified five putative novel gene clusters in E. festucae that are predicted to be involved in various cellular processes ranging from sugar metabolism to maintenance of cell integrity. Of particular interest was a putative nitrogen metabolism gene cluster (Hagerman et al., 1998; Karamac, 2009) whose genes are down-regulated in the ΔsakA mutant association, as nitrogen metabolism is known to play an important role in fungal pathogenicity (Pellier et al., 2003; Divon et al., 2005; Krappmann and Braus, 2005). The putative cluster containing the novel endophyte gene Nc25 was also interesting, as it revealed coregulation of Nc25 with a kexin protein and two other proteins of unknown function, suggesting that they may all be involved in the synthesis and processing of the predicted novel oligopeptide (Johnson et al., 2003, 2007).
This study highlights the power of combining genetics with high-throughput transcriptome analysis to analyze the effect of a single gene deletion on a plant-fungus symbiotic interaction to identify the candidate sets of genes associated with mutualistic and disrupted symbiotic states. While many of the effects observed may well be due to a breakdown in the symbiosis rather than a direct effect of the sakA deletion itself, use of such mutants provides a powerful approach to gain insight into the symbiosis. We anticipate that this approach will be widely used to analyze many other symbiotic plant-fungus interactions.
MATERIALS AND METHODS
Study Organisms
Epichloë festucae strain Fl1 was used for all experimental analyses described in this study. The reference genome for E. festucae strain E2368 is available at http://www.endophyte.uky.edu. All plant analysis was performed using perennial ryegrass (Lolium perenne ‘Samson’).
Biological Material and Growth Conditions
E. festucae cultures were grown on 2.4% potato dextrose agar or in 2.4% potato dextrose broth at 22°C. Endophyte-free perennial ryegrass seedlings were surface sterilized and then germinated on 3% water agar plates. Seedlings were inoculated using the method of Latch and Christensen (1985). Plants were grown and screened for infection as described previously (Tanaka et al., 2005). For examination of epiphyllous growth, seedlings were grown on Murashige and Skoog-phytoagar (0.75% [w/v] phytoagar, 0.43% [w/v] Murashige and Skoog medium) under sterile conditions. Fungal strains used in this study are listed in Supplemental Table S3.
Tissue Fixation, Wax Embedding, Sectioning, and Staining
Plant tissue was fixed in fresh 3.7% (v/v) formaldehyde, 5% (v/v) glacial acetic acid, and 50% ethanol for 4 h and then dehydrated through a graded ethanol series. Ethanol was then replaced with Histoclear (National Diagnostics) by passage through a graded Histoclear:ethanol series. The Histoclear was then gradually replaced with Paraplast X-tra (McCormick Scientific) and embedded in 100% Paraplast X-tra. Ten- to 12-μm sections were cut from the Paraplast X-tra-embedded samples using a Leica RM 2145 Rotary Microtome and mounted onto polylysine-coated slides. Sections were stained with Alcian blue/Safranin O, incubated in Histoclear to remove the Paraplast X-tra, and then dehydrated by incubation in isopropanol. Slides were air dried and then incubated in Alcian blue/Safranin O for 15 min, air dried again, and coverslips were mounted in Entellan (Merck).
Light and Fluorescence Microscopy
Vacuoles were visualized in culture by staining hyphae with FM4-64 (16.4 mm in MilliQ water; Invitrogen) at 22°C for 28 to 30 h. Fluorescence was then observed using an Olympus IX71 microscope with a ×63 oil-immersion objective and a numerical aperture of 1.4. Photographs were taken with a Hamamatsu ORCA-ER C4742-80 digital CCD camera and analyzed using Metamorph software (Molecular Devices).
For examination of fungal growth in planta, longitudinal epidermal peels from leaf sheath tissue or transverse sections from pseudostem tissue were stained with aniline blue or toluidine blue, respectively, and viewed using a Zeiss Axiophot light microscope, and images were recorded using a Leica DCF320 digital camera. For examination of host cell death, blade and pseudostem tissues were stained with lactophenol trypan blue as described previously (Koch and Slusarenko, 1990) and viewed as described above.
Electron Microscopy
For examination of hyphal structure in planta, 0.5- to 1-mm-thick sections of pseudostem tissue were fixed in 3% glutaraldehyde and 2% formaldehyde in 0.1 m phosphate buffer, pH 7.2, and then transverse sections were prepared for transmission electron microscopy as described by Spiers and Hopcroft (1993). Samples were examined using a Philips CM10 transmission electron microscope, and images were recorded using a SIS Morada digital camera. For detection of hydrogen peroxide in planta, a modified cytochemical method was used (Briggs et al., 1975; Bestwick et al., 1997). Samples were incubated in CeCl3, fixed, and prepared as described previously (Tanaka et al., 2006). Leaf samples examined by scanning electron microscopy were incubated in fixative (3% glutaraldehyde and 2% formaldehyde in 0.1 m phosphate buffer, pH 7.2) for a minimum of 24 h at room temperature. Samples were washed three times with 0.1 m phosphate buffer (pH 7.2) and dehydrated by passage through a graded ethanol series. Samples were then critical point dried using liquid CO2, mounted onto aluminum specimen support stubs, sputter-coated with gold, and observed using an FEI Quanta 200 scanning electron microscope.
Statistical Analysis
The statistical significance of differences in tiller widths and vasculature branching between strains was analyzed using Student's t test. The statistical significance of differences between infection rates of the different strains was analyzed using the χ2 test. P values were calculated using R (version 2.9.2; http://www.r-project.org/).
cDNA Sequencing, Assembly, and Analysis
mRNA libraries were generated, and a 60-nucleotide paired-end run was performed according to Illumina guidelines (Supplemental Protocol S1). Libraries were sequenced in four flow cell lanes of an Illumina Genome Analyser IIx (two for the wild-type sample and two for the ΔsakA mutant sample), and the raw results were processed using version 1.4 of the Illumina analysis pipeline. Using custom in-house software, reads were dynamically trimmed 5′ of the point where their quality scores exceeded a defined error threshold (i.e. P = 0.05, or Illumina quality score Q ≈ 13, or a one in 20 chance that the associated base call is incorrect). Trimmed reads smaller than 25 nucleotides in length were ignored in all downstream analyses. We obtained 42,150,292 and 42,919,413 60-nucleotide read pairs for the wild-type and ΔsakA samples, respectively. Following quality trimming, forward and reverse reads had a mean length of 47.7 and 44.7 nucleotides for the wild type and 49.0 and 46.0 nucleotides for the ΔsakA mutant, respectively.
De Novo Transcript Assembly
To identify novel transcripts for perennial ryegrass (a nonmodel organism with no reference genome), a de novo transcript assembly was generated with ABySS (version 1.1.0) for trimmed reads pooled from all four lanes using an algorithm based on de Bruijn graph theory. Only paired-end data for which both reads had lengths of 25 nucleotides or greater were used. ABySS was tuned using a grid of combinations from the following parameter set: k = 25 to 59, c = 2 to 12, e = 2 to 5, n = 2 to 10. The assembly that maximized N50 (i.e. a measure approximating the median contig size) was chosen for downstream analysis, and only contigs of greater than 500 bp were analyzed further. The run maximizing N50 (k = 33, c = 10, e = 3, n = 5) produced 132,295 contigs with N50 of 733 bp, mean length of 269 bp, and a maximum length of 11,242 bp. Contigs were compared individually with the E. festucae genome reference using a local installation of BLASTN, and nonmapping contigs were sorted into an EST library putatively representing perennial ryegrass transcripts. Of the 21,077 contigs greater than 500 bp in size, 5,075 mapped to the E. festucae genome, while 16,002 were putatively specific to perennial ryegrass. Only the latter contigs were analyzed further. At this stage, the library is also expected to contain sequences from bacterial and non-E. festucae fungal contaminants. The EST library is available from the authors on request.
Read Mapping, Counting, and Statistics
Trimmed reads were mapped individually for the wild-type and ΔsakA mutant lanes to poly(A) stripped reference sequences: the hph reporter gene, the set of annotated gene models for E. festucae (EfM2; n = 12,199), gene/EST sequences for perennial ryegrass (as available on GenBank in January 2010; n = 2,193), and the de novo-assembled EST library for perennial ryegrass described above (n = 16,002). Reads were mapped with the gap-aware mapping software, bwa (version 0.5.5), using the Burrows-Wheeler alignment algorithm. Reads mapping to each sequence were counted using custom in-house software (code available on request). Paired-end matches were counted as only a single hit. Statistical analysis of differential gene expression was performed using the R package, DEGseq (version 1.0.4). We applied Fisher's exact test with a significance cutoff of q = 0.05, which yields an experiment-wide false discovery rate of 5% (i.e. the equivalent of a single statistical test with P = 0.05; Storey and Tibshirani, 2003). Fold difference was calculated using methods described in Supplemental Protocol S2. Only genes where expression between the wild-type and mutant sample was statistically significantly different and either halved or doubled were considered further (i.e. analysis was focused on statistically and biologically meaningful changes in gene expression).
Functional Mapping by Homology
To identify possible gene functions and to confirm plant/fungal provenance, the set of annotated gene models for E. festucae, the GenBank sequences for perennial ryegrass, and the perennial ryegrass EST library were compared with public databases. Each sequence was mapped to a local installation of the National Center for Biotechnology Information nonredundant protein database (nr; January 3, 2010) using BLASTX. BLAST results were parsed manually. The unique identifier, species name, and E value of the top BLASTX hit were recorded. Putative function was predicted based on consensus from multiple BLASTX hits.
Gene Cluster Analysis
Statistically significant runs of up- or down-regulated genes were determined as described in Supplemental Protocol S2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Vasculature of plants infected with the E. festucae ΔsakA mutant.
Supplemental Figure S2. Fluorescence micrographs of FM4-64-stained E. festucae vacuoles.
Supplemental Figure S3. Differentially expressed fungal and plant genes organized by catalytic activity ontology.
Supplemental Figure S4. Differential expression of fungal and plant oxidoreductase genes.
Supplemental Figure S5. Differential expression of fungal and plant transferase genes.
Supplemental Figure S6. Putative gene clusters down-regulated in ΔsakA mutant association.
Supplemental Figure S7. Putative gene clusters up-regulated in ΔsakA mutant association.
Supplemental Figure S8. Differential expression of perennial ryegrass hydrolase genes.
Supplemental Table S1. Differentially expressed fungal genes organized by GO categories.
Supplemental Table S2. Differentially expressed plant genes organized by GO categories.
Supplemental Table S3. Biological materials.
Supplemental Protocol S1. Transcriptome generation.
Supplemental Protocol S2. Transcriptome analysis.
Supplementary Material
Acknowledgments
We acknowledge the support of the following individuals for genome sequencing and assembly as well as gene modeling: Jerzy W. Jaromczyk, Elissaveta G. Arnaoudova, Venu-Gopal Puram, Wayne E. Beach, Jennifer S. Webb, and Jennifer L. Wiseman (University of Kentucky); Bruce A. Roe, Simone L. Macmil, Steven Kenton, James White, Hongshing Lai, Chunmei Qu, Yanbo Xing, Ping Wang, Mounir Elharam, and Jennifer Lewis (University of Oklahoma); and Anar K. Khan (AgResearch). We also thank Isabelle Jourdain and Yvonne Rolke (IMBS, Massey University), Mike Christensen, Wayne Simpson, and Anouck de Bonth (AgResearch), Doug Hopcroft and Michael Loh (Manawatu Microscopy and Imaging Centre, Massey University), and Daniel Peterson and Colleen Eaton (IMBS, Massey University) for technical assistance. We also thank Yvonne Rolke for comments on the manuscript.
References
- Bestwick CS, Brown IR, Bennett MH, Mansfield JW. (1997) Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseolicola. Plant Cell 9: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabhra R, Askew DS. (2005) Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med Mycol (Suppl 1) 43: S87–S93 [DOI] [PubMed] [Google Scholar]
- Blackman E. (1971) Morphology and development of cross veins in leaves of bread wheat (Triticum aestivum L.). Ann Bot (Lond) 35: 653–665 [Google Scholar]
- Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. (2006) Genomic mining for Aspergillus natural products. Chem Biol 13: 31–37 [DOI] [PubMed] [Google Scholar]
- Bolwell PP, Page A, Pislewska M, Wojtaszek P. (2001) Pathogenic infection and the oxidative defences in plant apoplast. Protoplasma 217: 20–32 [DOI] [PubMed] [Google Scholar]
- Briggs RT, Drath DB, Karnovsky ML, Karnovsky MJ. (1975) Localization of NADH oxidase on the surface of human polymorphonuclear leukocytes by a new cytochemical method. J Cell Biol 67: 566–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen MJ, Bennett RJ, Ansari HA, Koga H, Johnson RD, Bryan GT, Simpson WR, Koolaard JP, Nickless EM, Voisey CR. (2008) Epichloë endophytes grow by intercalary hyphal extension in elongating grass leaves. Fungal Genet Biol 45: 84–93 [DOI] [PubMed] [Google Scholar]
- Christensen MJ, Bennett RJ, Schmid J. (2002) Growth of Epichloë/Neotyphodium and p-endophytes in leaves of Lolium and Festuca grasses. Mycol Res 106: 93–106 [Google Scholar]
- Clay K, Schardl C. (2002) Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat 160: S99–S127 [DOI] [PubMed] [Google Scholar]
- Divon HH, Rothan-Denoyes B, Davydov O, Di Pietro A, Fluhr R. (2005) Nitrogen-responsive genes are differentially regulated in planta during Fusarium oxysporum f. sp. lycopersici infection. Mol Plant Pathol 6: 459–470 [DOI] [PubMed] [Google Scholar]
- Dixon KP, Xu JR, Smirnoff N, Talbot NJ. (1999) Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11: 2045–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton CJ, Jourdain I, Foster SJ, Hyams JS, Scott B. (2008) Functional analysis of a fungal endophyte stress-activated MAP kinase. Curr Genet 53: 163–174 [DOI] [PubMed] [Google Scholar]
- Grandbastien MA. (1998) Activation of plant retrotransposons under stress conditions. Trends Plant Sci 3: 181–187 [Google Scholar]
- Grbić V, Bleecker AB. (1995) Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J 8: 595–602 [Google Scholar]
- Hagerman AE, Riedl KM, Jones A, Sovik KN, Ritchard NT, Hartzfeld PW, Riechel TL. (1998) High molecular weight plant polyphenolics (tannins) as biological antioxidants. J Agric Food Chem 46: 1887–1892 [DOI] [PubMed] [Google Scholar]
- Ho MS, Tsai PI, Chien CT. (2006) F-box proteins: the key to protein degradation. J Biomed Sci 13: 181–191 [DOI] [PubMed] [Google Scholar]
- Igbaria A, Lev S, Rose MS, Lee BN, Hadar R, Degani O, Horwitz BA. (2008) Distinct and combined roles of the MAP kinases of Cochliobolus heterostrophus in virulence and stress responses. Mol Plant Microbe Interact 21: 769–780 [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Johnson LJ, Johnson RD, Schardl CL, Panaccione DG. (2003) Identification of differentially expressed genes in the mutualistic association of tall fescue grass with Neotyphodium coenophialum. Physiol Mol Plant Pathol 63: 305–317 [Google Scholar]
- Johnson RD, Borchert S, Christensen MJ, Johnson LJ, Koulman A, van Gils MJ, Bryan GT. (2007) A gene identified from Neotyphodium lolii is expressed only in planta and regulates the biosynthesis of a putative oligopeptide secondary metabolite. Popay A, Thom ER, , Proceedings of the 6th International Symposium on Fungal Endophytes of Grasses. Grasslands Research and Practice Series No. 13. New Zealand Grassland Association, Christchurch, New Zealand, pp 485–489 [Google Scholar]
- Karamac M. (2009) Chelation of Cu(II), Zn(II), and Fe(II) by tannin constituents of selected edible nuts. Int J Mol Sci 10: 5485–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. (2007) Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science 316: 1481–1484 [DOI] [PubMed] [Google Scholar]
- Kitzing K, Auweter S, Amrhein N, Macheroux P. (2004) Mechanism of chorismate synthase: role of the two invariant histidine residues in the active site. J Biol Chem 279: 9451–9461 [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Christensen MJ, Bennett RJ. (1993) Incompatibility of some grass/Acremonium endophyte associations. Mycol Res 97: 1237–1244 [Google Scholar]
- Kogel KH, Franken P, Hückelhoven R. (2006) Endophyte or parasite: what decides? Curr Opin Plant Biol 9: 358–363 [DOI] [PubMed] [Google Scholar]
- Krappmann S, Braus GH. (2005) Nitrogen metabolism of Aspergillus and its role in pathogenicity. Med Mycol (Suppl 1) 43: S31–S40 [DOI] [PubMed] [Google Scholar]
- Lam E, Kato N, Lawton M. (2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411: 848–853 [DOI] [PubMed] [Google Scholar]
- Latch GCM, Christensen MJ. (1985) Artificial infection of grasses with endophytes. Ann Appl Biol 107: 17–24 [Google Scholar]
- Li HM, Sullivan R, Moy M, Kobayashi DY, Belanger FC. (2004) Expression of a novel chitinase by the fungal endophyte in Poa ampla. Mycologia 96: 526–536 [PubMed] [Google Scholar]
- McSteen P. (2009) Hormonal regulation of branching in grasses. Plant Physiol 149: 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi R, Banno S, Ichikawa R, Fukumori F, Ichiishi A, Kimura M, Yamaguchi I, Fujimura M. (2007) Identification of OS-2 MAP kinase-dependent genes induced in response to osmotic stress, antifungal agent fludioxonil, and heat shock in Neurospora crassa. Fungal Genet Biol 44: 208–218 [DOI] [PubMed] [Google Scholar]
- Park SM, Choi ES, Kim MJ, Cha BJ, Yang MS, Kim DH. (2004) Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol Microbiol 51: 1267–1277 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2004) Molecular genetics of the arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 7: 414–421 [DOI] [PubMed] [Google Scholar]
- Pellier AL, Laugé R, Veneault-Fourrey C, Langin T. (2003) CLNR1, the AREA/NIT2-like global nitrogen regulator of the plant fungal pathogen Colletotrichum lindemuthianum is required for the infection cycle. Mol Microbiol 48: 639–655 [DOI] [PubMed] [Google Scholar]
- Sakaguchi J, Fukuda H. (2008) Cell differentiation in the longitudinal veins and formation of commissural veins in rice (Oryza sativa) and maize (Zea mays). J Plant Res 121: 593–602 [DOI] [PubMed] [Google Scholar]
- Schardl CL. (2001) Epichloë festucae and related mutualistic symbionts of grasses. Fungal Genet Biol 33: 69–82 [DOI] [PubMed] [Google Scholar]
- Schardl CL, Leuchtmann A, Spiering MJ. (2004) Symbioses of grasses with seedborne fungal endophytes. Annu Rev Plant Biol 55: 315–340 [DOI] [PubMed] [Google Scholar]
- Segmüller N, Ellendorf U, Tudzynski B, Tudzynski P. (2007) BcSAK1, a stress-activated mitogen-activated protein kinase, is involved in vegetative differentiation and pathogenicity in Botrytis cinerea. Eukaryot Cell 6: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers AG, Hopcroft DH. (1993) Black canker and leaf spot of Salix in New Zealand caused by Glomerella miyabeana (Colletotrichum gloeosporioides). Eur J Forest Pathol 23: 92–102 [Google Scholar]
- Springob K, Nakajima J, Yamazaki M, Saito K. (2003) Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 20: 288–303 [DOI] [PubMed] [Google Scholar]
- Steyn WJ, Wand SJE, Holcroft DM, Jacobs G. (2002) Anthocyanins in vegetative tissues: a proposed function in photoprotection. New Phytol 155: 349–361 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Tanaka A, Scott B. (2006) A p67(Phox)-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell 18: 2807–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan YY, Spiering M, Christensen MJ, Saunders K, Schmid J. (1997) In planta metabolic state of Neotyphodium endophyte mycelium assessed through use of the GUS reporter gene in combination with hyphal enumeration. Bacon CW, Hill NS, , Neotyphodium/Grass Interactions; Plenum Press, New York, pp 85–87 [Google Scholar]
- Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. (2006) Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic association. Plant Cell 18: 1052–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Takemoto D, Hyon GS, Park P, Scott B. (2008) NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol Microbiol 68: 1165–1178 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Tapper BA, Popay A, Parker EJ, Scott B. (2005) A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Mol Microbiol 57: 1036–1050 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Sobey WJ, Aplin RT, Hassan A, Firmin JL, Schofield CJ, Prescott AG. (2000) Are anthocyanidins the immediate products of anthocyanidin synthase? Chem Commun (Camb) 2473–2474 [Google Scholar]
- Wang ZY, Jenkinson JM, Holcombe LJ, Soanes DM, Veneault-Fourrey C, Bhambra GK, Talbot NJ. (2005) The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe grisea. Biochem Soc Trans 33: 384–388 [DOI] [PubMed] [Google Scholar]
- Wessler SR. (1996) Turned on by stress: plant retrotransposons. Curr Biol 6: 959–961 [DOI] [PubMed] [Google Scholar]
- Willmer CM, Fricker M. (1996) Stomata, Ed 2. Chapman & Hall, London [Google Scholar]
- Young CA, Felitti S, Shields K, Spangenberg G, Johnson RD, Bryan GT, Saikia S, Scott B. (2006) A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet Biol 43: 679–693 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.