Abstract
ATP-binding cassette (ABC) transporters represent a large family in plants, but the functions of most of these transporters are unknown. Here we report a gene, AtSTAR1, only encoding an ATP-binding domain of a bacterial-type ABC transporter in Arabidopsis (Arabidopsis thaliana). AtSTAR1 is an ortholog of rice (Oryza sativa) OsSTAR1, which has been implicated in aluminum (Al) tolerance. Knockout of AtSTAR1 resulted in increased sensitivity to Al and earlier flowering. Unlike OsSTAR1, AtSTAR1 was expressed in both the roots and shoots and its expression was not induced by Al or other stresses. Investigation of tissue-specific localization of AtSTAR1 through β-glucuronidase fusion revealed that AtSTAR1 was predominantly expressed at outer cell layers of root tips and developing leaves, whose localization is also different from those of OsSTAR1. However, introduction of OsSTAR1 into atstar1 mutant rescued the sensitivity of atstar1 to Al, indicating that AtSTAR1 has a similar function as OsSTAR1. Furthermore, we found that AtSTAR1 may interact with ALS3, a transmembrane-binding domain in Arabidopsis to form a complex because introduction of OsSTAR1, a functional substitute of AtSTAR1, into als3 mutant resulted in the loss of OsSTAR1 protein. All these findings indicate that AtSTAR1 is involved in the basic detoxification of Al in Arabidopsis.
ATP-binding cassette (ABC) proteins constitute a large, diverse, and ubiquitous superfamily. In the genomes of rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana), there are 121 and 120 members estimated, respectively (Garcia et al., 2004). In a model legume, Lotus japonicus, there are at least 91 ABC proteins (Sugiyama et al., 2007). Although the function of most these proteins are unknown, recent studies have shown that plant ABC proteins not only are involved in the transport of hormones, lipids, metals, secondary metabolites, and xenobiotics, but also contribute to plant-pathogen interactions and the modulation of ion channels (Rea, 2007; Verrier et al., 2008).
Most plant ABC proteins are characterized by having both a nucleotide-binding domain (NBD) and a transmembrane domain (TMD), forming full-size (two NBDs and two TMDs) or half-size (one NBD and one TMD) proteins. However, a few ABC proteins only contain NBD or TMD. Recently, some of these types of proteins have been reported to be implicated in the tolerance to aluminum (Al) toxicity, which is a major limiting factor for plant growth and production on acid soils (von Uexkull and Mutert, 1995). Larsen et al. (2005) reported that an Arabidopsis gene, ALS3, which encodes only TMD, is involved in Al tolerance. Knockout of ALS3 resulted in increased sensitivity to Al (Larsen et al., 2005). ALS3 is primarily expressed in leaf hydathodes and the phloem throughout the plants, along with root cortex following Al treatment. However, ALS3 has not been functionally characterized. Also it is unknown whether ALS3 functions alone or requires NBDs to form a complex.
More recently, a rice gene, STAR1 (for sensitive to Al rhizotoxicity), which encodes only NBD of an ABC transporter, was reported to be involved in Al tolerance (Huang et al., 2009). Knockout of this gene resulted in hypersensitivity to Al (Ma et al., 2005; Huang et al., 2009). STAR1 interacts with STAR2, a TMD protein, to form a complex, which functions as a bacterial-type ABC transporter. The complex of STAR1/STAR2 is localized at the membrane of vesicles in the root cells and transports UDP-Glc (Huang et al., 2009). UDP-Glc may be used for the modification of the cell wall, thereby detoxifying Al, although the exact mechanism remains to be elucidated (Huang et al., 2009). The expression of both STAR1 and STAR2 is up-regulated by Al and controlled by ART1, a C2H2 zinc-finger transcription factor (Yamaji et al., 2009).
STAR1 is single-copy gene in the rice genome and its putative orthologous genes were found in maize (Zea mays), Arabidopsis, grape (Vitis vinifera), and moss (Physcomitrella patens; Huang et al., 2009). However, it is unclear whether STAR1 represents a universal Al tolerance gene in plant species or is only required for Al tolerance in rice. In this study, we characterized AtSTAR1, an ortholog of rice STAR1, in a dicot species Arabidopsis. We found that AtSTAR1 is also required for Al tolerance in Arabidopsis, but the expression patterns and localization are different from those of OsSTAR1. Furthermore, we found the flowering time in a T-DNA insertion mutant of AtSTAR1 is altered.
RESULTS
Structure of AtSTAR1 Gene
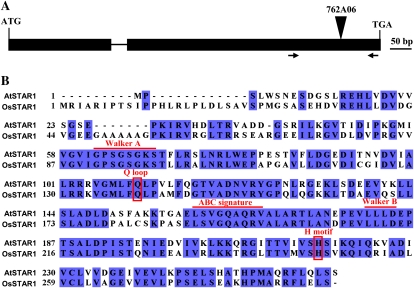
A BLAST search using OsSTAR1 protein sequence resulted in identification of a unique homolog, AtSTAR1 (At1g67940), in the Arabidopsis genome. AtSTAR1 has two exons and one intron, encoding a 263-amino acid protein (Fig. 1A). AtSTAR1 shows 62% identity and 77% similarity with OsSTAR1 (Fig. 1B). Similar to OsSTAR1, AtSTAR1 is characterized by conserved domains of a NBD of an ABC transporter, such as Walker A, Q loop, ABC signature, Walker B, and H motif (Fig. 1B).
Figure 1.
Gene structure of AtSTAR1 in Arabidopsis (A) and sequence similarity with rice OsSTAR1 (B). The T-DNA insertion position of AtSTAR1 in a line (762A06) was shown on the top (A). Arrows show a primer pair for detection of AtSTAR1 expression by RT-PCR. B, Alignment of AtSTAR1 and OsSTAR1. Lines above the sequence indicate the conserved domains, and boxes show the conserved amino acids of two motifs. [See online article for color version of this figure.]
Phenotype of AtSTAR1 Knockout Line
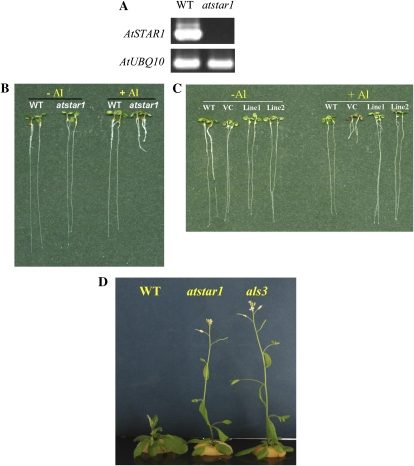
To examine the biological function of AtSTAR1 in Arabidopsis, we obtained a knockout line (762A06, atstar1) that has a T-DNA insertion at the coding sequence of AtSTAR1 (724 bp from ATG; Fig. 1A). Reverse transcription (RT)-PCR analysis indicated that the full-length transcript of AtSTAR1 was absent in atstar1 (Fig. 2A). When the wild-type Arabidopsis and atstar1 knockout line were grown in the absence of Al, their root growth was similar (Fig. 2B). However, in the presence of 2 μm Al, the roots of the knockout line were severely inhibited, whereas those of the wild type were hardly affected (Fig. 2B).
Figure 2.
Phenotype of knockout line atstar1. A, mRNA expression of AtSTAR1 in the wild type (WT) and atstar1 knockout line. RT-PCR analysis was performed to detect the mRNA expression of AtSTAR1 (35 cycles) and the internal control AtUBQ10 (25 cycles). B, Al sensitivity in the wild type and the knockout line. Seedlings were grown in a nutrient solution containing 0 or 2 μm Al at pH 5.0 for 7 d. C, Complementation test of the atstar1 knockout line. The wild type and atstar1 transformed with empty vector (VC) and AtSTAR1 (two lines), respectively, were grown in a nutrient solution containing 0 or 2 μm Al at pH 5.0 for 7 d. D, Flowering time of atstar1, als3, and the wild type. Plants were grown in a nutrient solution for 35 d.
To confirm that disruption of AtSTAR1 was responsible for the Al sensitivity in the knockout line, a complementation test was performed. A 3.7-kb DNA fragment harboring 1.7-kb AtSTAR1 promoter and the candidate gene was introduced into the knockout line by Agrobacterium-mediated transformation method. Examination of two independent T2 transgenic lines carrying AtSTAR1 showed that their tolerance to Al toxicity was recovered to a similar level to that of the wild type, whereas the tolerance of the vector control plants to Al was not (Fig. 2C). The growth of all lines was similar in the absence of Al (Fig. 2C). These results indicate that Al-sensitive phenotype in the knockout line was caused by loss of function of AtSTAR1 gene.
When the wild type and the knockout line were cultivated to the mature stage, we further found that the flowering time of the knockout line was almost 4 d earlier than that of the wild type (28.6 ± 1.1 d for knockout line versus 32.3 ± 1.2 d for the wild type; Fig. 2D). In addition, we found that a knockout line of ALS3 also showed early flowering like atstar1 (Fig. 2D).
Physiological Characterization of atstar1 Knockout Line
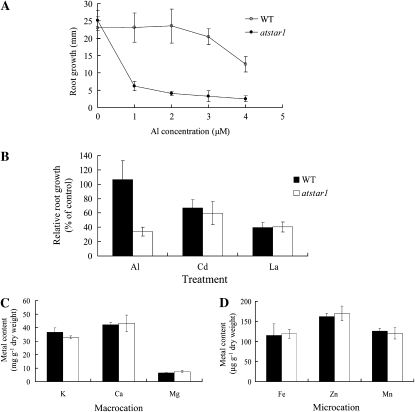
We further physiologically characterized atstar1 knockout line in terms of dose response, metal specificity, and uptake of other cations. A dose-response experiment showed that the root growth of atstar1 was inhibited by as high as 75% even at 1 μm Al, at which concentration the roots of the wild type were not inhibited (Fig. 3A). With increasing Al concentrations in the external solution, the Al-induced root inhibition was also observed in the wild type, but the inhibition was much lower at either Al concentration (Fig. 3A).
Figure 3.
Physiological characterization of the knockout line atstar1. A, Dose response to Al in the wild type (WT) and atstar1. Seedlings were grown in a nutrient solution containing 0, 1, 2, 3, or 4 μm Al at pH 5.0 for 7 d. Root growth after the treatment was measured with a ruler. Data are means ± sd (n = 8–11). B, Specificity of atstar1 to Al. Seedlings of the wild type and atstar1 were grown in a nutrient solution containing 0, 2 μm Al, 2 μm Cd, or 0.1 μm La at pH 5.0 for 7 d. Relative root growth expressed as (root length with metal treatment/root length without metal) × 100 is shown. Data are means ± sd (n = 21–30). C and D, Shoot concentration of macrocations (C) and microcations (D). Both the wild type and atstar1 were grown in one-thirtieth-strength Hoagland nutrient solution for 38 d. Data are means ± sd (n = 3).
To investigate whether atstar1 is specifically sensitive to Al stress or not, the sensitivity to other toxic metals, including cadmium (Cd) and lanthanum (La), was compared between the wild type and the knockout line. In the presence of 2 μm Cd or 0.1 μm La, the root growth inhibition caused by these metals was similar between the wild type and atstar1 (Fig. 3B), although the difference in Al tolerance was significantly evident. These results indicate that the sensitivity of atstar1 to Al was highly specific.
We also compared the uptake of some essential cations between the wild type and atstar1 grown hydroponically in the absence of Al. Among cations tested, including potassium, calcium, magnesium, iron, zinc, and manganese, there was no difference in the concentration of these cations in the shoots between the wild type and atstar1 (Fig. 3, C and D).
Expression Pattern of AtSTAR1
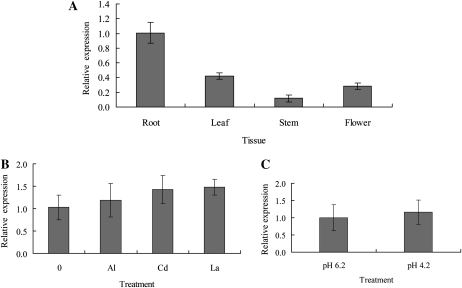
The tissue-specific expression pattern of AtSTAR1 mRNA was examined with quantitative real-time PCR. AtSTAR1 was expressed in the roots as well as in the leaves, stems, and flowers of Arabidopsis (Fig. 4A), but the expression level in the roots was relatively higher (Fig. 4A).
Figure 4.
mRNA expression pattern of AtSTAR1 in Arabidopsis. A, Quantitative real-time RT-PCR analysis of AtSTAR1 expression in various tissues. AtUBQ10 was used as an internal control. The data were normalized to AtSTAR1 expression in roots. Data are means ± sd (n = 3). B, Quantitative real-time RT-PCR analysis of AtSTAR1 expression in response to different metals. Seedlings were grown on a nutrient solution consisting of one-thirtieth-strength Hoagland nutrient solution (NH4H2PO4 omitted) plus 1 mm CaCl2 at pH 5.0 for 7 d and then exposed to the same nutrient solution containing 0, 3 μm Al, 3 μm Cd, or 3 μm La at pH 5.0 for 6 h and then the roots were sampled for RNA isolation. AtUBQ1 was used as an internal control. The data were normalized to AtSTAR1 expression under no metal treatment. Data are means ± sd (n = 3). C, Quantitative real-time RT-PCR analysis of AtSTAR1 expression in response to low pH. Forty-day-old plants were exposed to one-tenth-strength Hoagland nutrient solution at pH 6.2 or pH 4.2 for 8 h and then the roots were sampled for RNA isolation. AtUBQ1 was used as an internal control. The data were normalized to AtSTAR1 expression at pH 6.2. Data are means ± sd (n = 3).
The expression of AtSTAR1 mRNA in response to Al or other stresses was also examined. Exposure to Al, Cd, or La did not affect the expression of AtSTAR1 in the roots (Fig. 4B). Low pH treatment also did not change the expression (Fig. 4C).
Localization of AtSTAR1
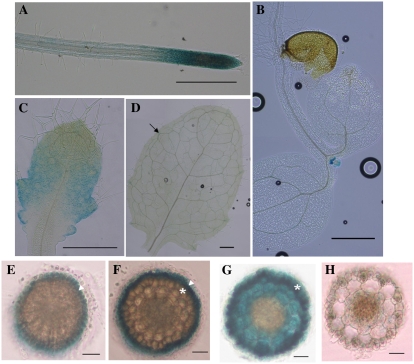
To investigate the tissue-specific localization of AtSTAR1 in Arabidopsis, a DNA fragment consisting of 2.3-kb promoter and the full genomic sequence of AtSTAR1 was fused in frame to the GUS reporter gene and the construct containing the fused gene was transformed into Arabidopsis wild-type plants. GUS analysis in the transgenic plants revealed that AtSTAR1 was expressed in both the roots and shoots (Fig. 5). This result is consistent with the expression pattern of AtSTAR1 (Fig. 4A). In the roots, AtSTAR1 was predominantly expressed in the root tip region, including root caps, meristem, and elongation zone (Fig. 5A). In the shoots, GUS activity was strongly detected in newly generated leaves but not observed at cotyledon leaves (Fig. 5B). High GUS activity was also observed at developing young leaves, whereas in older leaves GUS expression was weak and became confined to epithem tissue of hydathodes (Fig. 5, C and D).
Figure 5.
GUS analysis of AtSTAR1 expression. T2 transgenic plants expressing pAtSTAR1:AtSTAR1-GUS were used for AtSTAR1 expression analysis. A to D, GUS activity in the roots (A), cotyledon and newly generated leaves (B), developing leaves (C), and old leaves (D). Arrow indicates GUS expression in hydathode of an elder leaf. Scale bars for A to D: 500 μm. E to H, Cellular localization of GUS expression in roots. E, Root cap. Triangle indicates the outer cell layers. F, Meristem region. Triangle indicates the lateral root caps and asterisk indicates the epidermal cells. G, Elongation zone. Asterisk indicates the epidermal cells. H, Mature zone. Scale bars for E to H: 30 μm.
To further examine the cellular localization of AtSTAR1 in the root tip region, we made cross sections through the GUS-stained roots. Results showed the cell specificity of localization of AtSTAR1 differed with root regions. In the root cap and meristem region, AtSTAR1 was mainly expressed at the outer cell layers; the outer cell layers of root caps (Fig. 5E), lateral root caps of root meristem (Fig. 5F). However, at the elongation zone, AtSTAR1 was expressed in the epidermal cells, cortex, and endodermis, although the expression was relatively higher in the epidermal cells (Fig. 5G). The expression of AtSTAR1 in the mature root region was much weaker compared with that of root tips (Fig. 5H).
Rescue of the Al Sensitivity in atstar1 by OsSTAR1
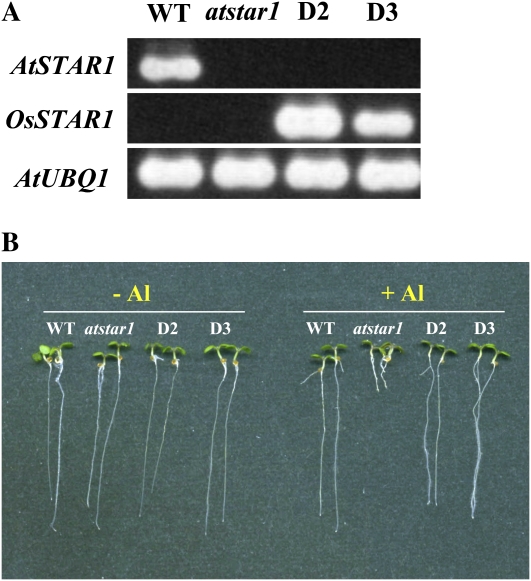
To investigate whether rice OsSTAR1 has similar function to AtSTAR1 in Arabidopsis, we performed a complementation test by introducing OsSTAR1 into atstar1 mutant under the control of cauliflower mosaic virus 35S promoter. RT-PCR analysis showed that OsSTAR1 was expressed, whereas AtSTAR1 expression was absent in the two independent OsSTAR1;atstar1 transgenic lines (Fig. 6A). Al tolerance test showed that introduction of OsSTAR1 into atstar1 rescued the Al sensitivity of the knockout line, being a similar tolerance level to the wild type (Fig. 6B). These results indicate that OsSTAR1 and AtSTAR1 have similar function in conferring Al tolerance.
Figure 6.
Complementation of atstar1 by OsSTAR1. A, RT-PCR analysis of AtSTAR1 and OsSTAR1 expression in the wild type (WT), atstar1 knockout line, and OsSTAR1;atstar1 transgenic lines (D2 and D3). B, Al sensitivity in OsSTAR1;atstar1 lines. Seedlings of the wild type, atstar1, and OsSTAR1;atstar1 lines were grown in a nutrient solution containing 0 or 2 μm Al at pH 5.0 for 7 d. [See online article for color version of this figure.]
Possible Interaction between AtSTAR1 and ALS3
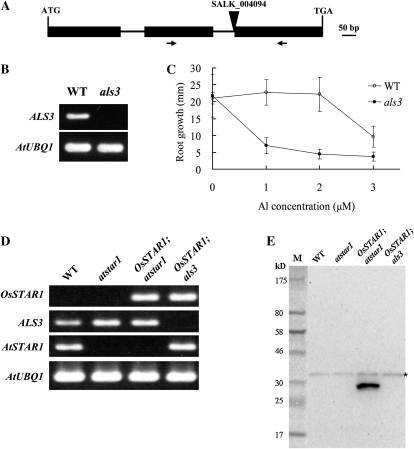
To investigate the possible interaction between AtSTAR1 and ALS3 in Arabidopsis, we first obtained a T-DNA insertion line of ALS3. The T-DNA was inserted in the splicing site between the second intron and the third exon of ALS3 (Fig. 7A), resulting in absence of the full-length transcript of ALS3 (Fig. 7B). Mutation of ALS3 resulted in hypersensitivity to Al (Fig. 7C), whose phenotype was consistent with that of previously reported allelic mutant (Larsen et al., 2005). To investigate the possible interaction between AtSTAR1 (OsSTAR1) and ALS3, we obtained lines carrying OsSTAR1 in als3 mutant background by crossing. Results showed that, in OsSTAR1;atstar1 line, both OsSTAR1 and ALS3 mRNA were expressed (Fig. 7D), while there was no expression of AtSTAR1. By contrast, in the OsSTAR1;als3 line, there was expression of both OsSTAR1 and AtSTAR1 but no expression of ALS3 (Fig. 7D). When western-blot analysis was performed in these lines as well as the wild type and atstar1 by using an antibody against OsSTAR1 (Huang et al., 2009), OsSTAR1 was detected in the OsSTAR1;atstar1 line but not in the wild type and atstar1, indicating the specificity of the antibody for OsSTAR1 in Arabidopsis (Fig. 7E). Interestingly, although OsSTAR1 mRNA was detected in the OsSTAR1;als3 line (Fig. 7D), the protein of OsSTAR1 was not detected. This result indicates that ALS3 might be required for the expression of OsSTAR1 protein in Arabidopsis, indirectly implicating the interaction between AtSTAR1 (OsSTAR1) and ALS3.
Figure 7.
Possible interaction with ALS3. A, A T-DNA insertion line (als3, SALK_004094) of ALS3. Triangle on the top shows insertion position. Arrows show a primer pair for detection of ALS3 expression by RT-PCR. B, mRNA expression of ALS3 in the wild type (WT) and als3. RT-PCR analysis was performed to detect the mRNA expression of ALS3 (30 cycles) and the internal control AtUBQ1 (25 cycles). C, Al sensitivity in the wild type and als3. Seedlings of the wild type and the knockout line were grown in a nutrient solution containing 0, 1, 2, or 3 μm Al at pH 5.0 for 7 d. Data are means ± sd (n = 10–15). D, Expression of OsSTAR1, ALS3, and AtSTAR1 in different lines. RT-PCR analysis was performed to detect mRNA expression of OsSTAR1, ALS3, AtSTAR1, and the internal control AtUBQ1 (28 cycles) in the roots. E, Western-blot analysis of OsSTAR1 protein. Root total protein (40 μg) in each lane was used and detected with an antibody against OsSTAR1. The asterisk indicates a nonspecific band.
DISCUSSION
AtSTAR1 Is Required for Al Tolerance in Arabidopsis
Al is the most abundant metal in the earth's crust, therefore plants have evolved different mechanisms to detoxify this metal (Kochian, 1995; Ma et al., 2001; Kochian et al., 2004; Ma, 2008). One of the well-documented mechanisms is the secretion of organic acid anions, including citrate, malate, and oxalate, from the roots. This mechanism has been observed in a wide variety of plant species, including both dicots and monocots such as wheat (Triticum aestivum), barley (Hordeum vulgare), Arabidopsis, and Cassia tora (Delhaize et al., 1993; Ma et al., 1997; Hoekenga et al., 2003; Zhao et al., 2003). Genes responsible for the Al-induced secretion of malate in wheat (ALMT1; Sasaki et al., 2004), and citrate in barley (HvAACT1; Furukawa et al., 2007) and sorghum (SbMATE; Magalhaes et al., 2007) have been identified. Recently, progress in identification of other Al-tolerance genes has also been made. For example, a gene, ALS1, encoding a tonoplast-localized half-size of an ABC transporter, has been reported to be involved in the Al tolerance in Arabidopsis (Larsen et al., 2007). A bacterial-type ABC transporter OsSTAR1/OsSTAR2 is required for high Al tolerance in rice (Huang et al. 2009). In this study, we characterized the unique homolog of OsSTAR1 in Arabidopsis, AtSTAR1. All results show that like OsSTAR1, AtSTAR1 is also required for Al tolerance in Arabidopsis; knockout of AtSTAR1 resulted in enhanced sensitivity specific to Al (Figs. 2 and 3). These results suggest that dicots like Arabidopsis and monocots like rice share similar mechanism of Al tolerance. This is supported by the finding that OsSTAR1 was able to rescue the hypersensitivity of atstar1 to Al in Arabidopsis (Fig. 6). However, the expression patterns and tissue cell specificity of localization are different between AtSTAR1 and OsSTAR1.
AtSTAR1 was expressed in both the roots and shoots of Arabidopsis (Fig. 4), whereas OsSTAR1 was only expressed in the roots of rice (Huang et al., 2009). Furthermore, the expression of AtSTAR1 was mainly confined to root tip region (Fig. 5), while OsSTAR1 was expressed at the whole root. Within the root tip region, AtSTAR1 was mainly expressed at outer cell layers, including lateral root caps in root meristem region and epidermis in the elongation zone (Fig. 5, E–G). In contrast, rice OsSTAR1 was expressed at all cells of root tips (Huang et al., 2009). Furthermore, the expression of AtSTAR1 was not responsive to Al stress in Arabidopsis, whereas OsSTAR1 is highly induced by Al treatment (Huang et al., 2009). These differences in the expression pattern and localization suggest that in addition to involvement of Al tolerance, AtSTAR1 has other roles different from that of OsSTAR1 in Arabidopsis. Early flowering in the knockout line of AtSTAR1 supports this as described below.
Possible Interaction between AtSTAR1 and ALS3
Work in rice has shown that OsSTAR1 interacts with OsSTAR2 to form a complex and functions as an ABC transporter (Huang et al., 2009). This leads us to hypothesize that AtSTAR1 also requires a protein containing TMDs to be functional. ALS3 was previously reported to be involved in Al tolerance in Arabidopsis (Larsen et al., 2005). It is the unique homolog of OsSTAR2, which encodes only TMDs. Knockout of ALS3 showed similar phenotype of atstar1; enhanced Al sensitivity and early flowering (Figs. 2D and 7C). Furthermore, OsSTAR1 protein, the functional substitute of AtSTAR1, was not detected in the OsSTAR1;als3 line (Fig. 7E). These results suggest that ALS3 interacts with and stabilizes OsSTAR1 to prevent the degradation of OsSTAR1.
However, the tissue specificity of localization is not completely consistent between AtSTAR1 and ALS3. Although in the root tips, both AtSTAR1 and ALS3 are expressed at the outer cell layers, they have different localization in other tissues. ALS3 was primarily expressed at the phloem of all organs, whereas no expression of AtSTAR1 was found in the phloem (Fig. 5; Larsen et al., 2005). Furthermore, the expression of ALS3 was induced by Al (Larsen et al., 2005), but that of AtSTAR1 was not (Fig. 4B). These findings suggest that AtSTAR1 might form a complex with ALS3 in the root tip region to mediate Al tolerance, whereas in other tissues AtSTAR1 interacts with other proteins to fulfill its function. Deeken et al. (2008) isolated a number of potential mobile transcripts from leaf phloem exudates, which also include ALS3. Therefore, there is a possibility that ALS3 is transcribed at the phloem and then moved to AtSTAR1-expressed tissues to perform protein synthesis and interact with AtSTAR1 although further investigation is required in future.
A complex of OsSTAR1/OsSTAR2 transports UDP-Glc, which may be used for modification of cell wall (Huang et al., 2009). Although the transport substrate of AtSTAR1/ALS3 complex was not examined in this study, the complementation of OsSTAR1 in atstar1 suggests a similar function for AtSTAR1/ALS3.
A recent study showed that the expression of OsSTAR1 and OsSTAR2 is regulated by ART1 in rice, a zinc-finger-type transcription factor (Yamaji et al., 2009). Similarly, the expression of ALS3 is regulated by a zinc-finger transcription factor STOP1 in Arabidopsis, but AtSTAR1 was not in the list of downstream genes regulated by STOP1 (Sawaki et al., 2009). Consistently, we found that AtSTAR1 was not induced by Al (Fig. 4), while STOP1 regulates Al-inducible genes (Iuchi et al., 2007); therefore, AtSTAR1 is supposed to be not regulated by STOP1. It would be interesting to compare the different regulation mechanisms between rice and Arabidopsis in the future.
AtSTAR1 and ALS3 Might Also Be Involved in the Regulation of Flowering Time
Knockout of either AtSTAR1 or ALS3 resulted in earlier flowering (Fig. 2D), suggesting that both AtSTAR1 and ALS3 might be also involved in the regulation of flowering time. This phenotype is different from that of osstar1, which showed similar flowering time as wild-type rice (Huang et al., 2009). This difference may be attributed to the expression of AtSTAR1 and ALS3 in the shoots. Flowering time is a complicated trait and many genes have been suggested to be involved (Simpson and Dean, 2002; Komeda, 2004; Roux et al., 2006). Altered mineral nutrient availability is also suggested to affect the flowering time in Arabidopsis (Kolar and Senkova, 2008). However, among nutrients tested, there was no difference in the concentration between wild type and the knockout line of AtSTAR1 (Fig. 3, C and D), suggesting that earlier flowering in the mutant was not related to these mineral nutrient availability. It remains to be elucidated how AtSTAR1/ALS3 is involved in the control of flowering time in the future.
In summary, our results indicate that AtSTAR1 is required for Al tolerance in Arabidopsis, probably functioning as a bacterial-type ABC transporter by forming a complex with ALS3. Furthermore, we found that both AtSTAR1 and ALS3 may be also involved in the regulation of flowering time in Arabidopsis.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana; ecotype Columbia-0) was used for all the control experiments. The T-DNA insertion lines atstar1 (762A06) and als3 (SALK_004094) were obtained from GABI-Kat (http://www.gabi-kat.de) and the Arabidopsis Biological Resource Center, respectively. The homozygous lines were identified through PCR analysis of the genotype of each line.
Evaluation of Sensitivity to Al, Cd, and La
Seeds of each line were soaked in deionized water and kept at 4°C for 2 to 4 d in the dark to synchronize germination. The seeds were then placed on a plastic mesh floating on a low-strength nutrient solution consisting of one-thirtieth-strength Hoagland nutrient solution without NH4H2PO4 and 1 mm CaCl2 in a growth chamber with a 14-h-light/10-h-dark cycle at 22°C. For evaluation of sensitivity to Al, seeds were grown on the low-strength nutrient solution containing 0, 1, 2, 3, or 4 μm AlCl3 at pH 5.0 for 7 d. The solution was renewed every 2 d. After the treatment, root length was measured by a ruler and relative root growth expressed as (root length with Al treatment/root length without Al) × 100 was used to evaluate the Al sensitivity. For evaluation of sensitivity to other metals, seeds were exposed to the same nutrient solution containing 0, 2 μm CdCl2 or 0.1 μm LaCl3 for 7 d as described above.
Determination of Cation Concentrations
To compare the macro- or microcation concentration in the shoots of the wild type and atstar1, plants were cultured in a one-thirtieth-strength Hoagland nutrient solution. After growth for 38 d, the shoots were harvested and dried in an oven at 70°C for a week. The dried tissue was fully digested with concentrated HNO3 (60%) at a temperature up to 140°C. The concentrations of potassium, calcium, magnesium, zinc, iron, and manganese in the digested solution were determined with flame atomic absorption spectrometry (model Z-2000, Hitachi), after appropriate dilution with 0.1 n HNO3.
RNA Isolation and RT-PCR Analysis
To examine the expression pattern of AtSTAR1 in Arabidopsis, different tissues of 40-d-old plants, including roots, leaves, stem, and flowers, were excised and frozen in liquid nitrogen within 5 min of harvest for RNA extraction. For the expression analysis of AtSTAR1 in response to different metals, seedlings were grown on a nutrient solution consisting of one-thirtieth-strength Hoagland nutrient solution (NH4H2PO4 omitted) plus 1 mm CaCl2 at pH 5.0 for 7 d and then exposed to the same nutrient solution containing 0, 3 μm AlCl3, 3 μm CdCl2, or 3 μm LaCl3 at pH 5.0 for 6 h and then the roots were sampled. For the expression analysis of AtSTAR1 expression in response to low pH, 40-d-old plants were exposed to one-tenth-strength Hoagland nutrient solution at pH 6.2 or pH 4.2 for 8 h and then the roots were sampled for RNA isolation. Total RNA was extracted using the RNeasy mini kit (Qiagen). One microgram of DNase I-treated total RNA was used for first-strand cDNA synthesis using a SuperScript II kit (Invitrogen) and an oligo(dT)12-18 primer (Invitrogen) following the manufacturer's instructions. One-tenth of the reaction volume was used as template for PCR of AtSTAR1, and AtUBQ10 was used as an internal control (26 cycles). The primer sequences for RT-PCR of AtSTAR1 were 5′-GTTGAAGAAACCTCTGTGCCATT-3′ and 5′-GTCGTAGAGTTGGAATGCTTTTTC-3′ and the forward and reverse sequences of AtUBQ10 were 5′-GGAGGTGGAGAGTTCTGACA-3′ and 5′-AGACCAAGTGAAGTGTGGAC-3′, respectively. For real-time RT-PCR analysis, 1 μL of 10-fold-dilution cDNA from each sample was used for the quantitative analysis of gene expression performed with SYBR Premix Ex Taq (Takara). AtUBQ1 (forward primer, 5′-TCTACACTTCATCTTGTGTTGAGGC-3′; reverse primer, 5′-CACTGAAACAAGAAAAACAAACCCT-3′) was used as an internal control in this experiment. Data were collected in accordance with the 7500 real-time PCR system (Applied Biosystems).
For RT-PCR analysis of AtSTAR1 and ALS3 expression, 1 mL of 10-fold-dilution cDNA from the wild type and the T-DNA lines was used as templates in a 20 μL solution containing 1× Ex Taq buffer, 0.5 μm of each primer, 0.2 mm deoxynucleotide triphosphates, and 0.1 unit of Ex Taq DNA polymerase. PCR was carried out as follows: 95°C for 3 min, 35 (AtSTAR1) or 30 (ALS3) cycles of 95°C denaturing for 30 s, 58°C annealing for 1 min and 72°C extension for 30 s, and a 5 min extension at 72°C. The primer sequences for AtSTAR1 were 5′-GTTGAAGAAACCTCTGTGCCATT-3′ and 5′-GTCGTAGAGTTGGAATGCTTTTTC-3′ and those of ALS3 were 5′-CAATGTTCTTGCTCGTCCTCCT-3′ and 5′-CCGCCCATTATCATACCAGTCA-3′, respectively. AtUBQ10 or AtUBQ1 was used as internal control. Their primer sequences were 5′-GGAGGTGGAGAGTTCTGACA-3′ and 5′-AGACCAAGTGAAGTGTGGAC-3′ for AtUBQ10, and 5′-TCTACACTTCATCTTGTGTTGAGGC-3′ and 5′-CACTGAAACAAGAAAAACAAACCCT-3′ for AtUBQ1.
Complementation Analysis
For complementation test of atstar1 in Arabidopsis, a 3.8-kb DNA fragment harboring 1.7-kb promoter and AtSTAR1 gene was directly amplified with a pair of primers: 5′-CTCGAGATGCGAAGGTATGAAAAATAAC-3′ and 5′-CACCACAGAAACCCATCCCAAAA-3′. The DNA fragment was inserted into the pPZP2H-lac binary vector (Fuse et al., 2001) and transformed into atstar1 mutant by Agrobacterium tumefaciens-mediated floral-dip method (Clough and Bent, 1998). For the introduction of OsSTAR1 into atstar1 mutant, the open reading frame of OsSTAR1 was amplified from rice (Oryza sativa) root cDNA by PCR using 5′-agaaTTCTCAAGATTACAGGTACACATC-3′ and 5′-aatgtacaaaggGATGCGCATTGCCCGGATTCCC-3′ primer pair. The DNA fragment was cloned into pPZP2Ha3 binary vector that contains a cauliflower mosaic virus 35S promoter (Fuse et al., 2001) and then the construct was transformed into atstar1 mutant.
Cross between OsSTAR1;atstar1 and als3 Mutant
We made a cross between OsSTAR1;atstar1 line and the T-DNA line (als3) to generate OsSTAR1;als3 line. The resultant F1 plants were genotyped to select OsSTAR1-containing plants through direct genomic PCR. After self pollination, a large F2 population from one F1 progeny was subjected for genotyping by genomic PCR and F2 plants with the genotype of OsSTAR1;AtSTAR1;als3 were selected for the collection of F3 seeds. mRNA expression analysis and western-blot analysis was performed in OsSTAR1;atstar1 and OsSTAR1;als3 lines to investigate the possible relationship between OsSTAR1 and ALS3. Since the OsSTAR1 locus was heterozygous in the two lines, we first genotyped each plant by genomic PCR and then mixed the roots of OsSTAR1-positive plants for both mRNA expression analysis and western-blot analysis.
Western-Blot Analysis
Total proteins were extracted from the roots of the wild type, atstar1, OsSTAR1;atstar1, and OsSTAR1;als3 lines. The fresh roots (approximately 0.2 g) of each line were frozen in liquid nitrogen and ground with a homogenizer. The samples were then homogenized in 500 μL lysis buffer consisting of 100 mm Tris-HCl (pH 8.0), 150 mm KCl, 0.5% (w/v) polyvinylpolypyrrolidone, 5 mm EDTA, 3.3 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 10% (v/v) glycerol. After that, the homogenates were centrifuged at 13,000 rpm for 10 min at 4°C. The supernatants were then transferred to new tubes as total protein. Protein concentrations were measured by the Bradford method (Bradford, 1976). Equal amounts of samples (40 μg protein) were mixed with same volume of sample buffer containing 250 mm Tris-HCl, pH 6.8, 8% (w/v) SDS, 40% (v/v) glycerol, 0.01% (w/v) bromphenol blue, and 200 mm β-mercaptoethanol. The mixture was incubated at 65°C for 20 min and then loaded into a SDS-PAGE gel consisting of 11% polyacrylamide and 0.1% SDS for electrophoresis. The gel was then transferred to polyvinylidene difluoride membrane with a semidry blotting system, and the membrane was treated with the primary antibody anti-OsSTAR1 diluted at 1:100. Anti-rabbit IgG HRP conjugate (1:10,000 dilution; GE Healthcare) was used as a secondary antibody, and ECL plus (GE Healthcare) was used for detection via chemiluminescence.
GUS Analysis
For construct of pAtSTAR:AtSTAR1-GUS, a 3.3-kb DNA fragment harboring 2.4-kb promoter region and the AtSTAR1 gene except for the stop codon was amplified by PCR of the Arabidopsis genomic DNA. The primer pair for the amplification and introduction of restriction sites was 5′-ttggtaccGCTATCGCACATAACAAGAGGACA-3′ and 5′-aatctAGAACTGAGTTGAAGAAACCTCTGTGC-3′. The obtained DNA fragment was then fused in frame to the GUS gene and introduced into the pPZP2H-lac binary vector, and finally transformed into Arabidopsis wild-type plants.
For histochemical staining of GUS activity, T2 transgenic plants were used. Roots and leaves were collected from 5-week-old plants grown in one-tenth-strength Hoagland nutrient solution at 22°C. The samples were pretreated with or without cold 90% acetone and immersed into staining buffer consisting of 50 mm sodium phosphate (pH 7.2), 1% Triton X-100, 2 mm potassium ferri- and ferrocyanide, and 1 mm X-Gluc under vacuum for 20 min and then incubated at 37°C overnight. Stained samples were subjected to an ethanol series (20%, 35%, 50%, and 70%) for the removal of chlorophyll or imbedded into 5% agarose for sections. Cross sections (50–100 μm) were made by using a micro-slicer (ZERO1, Dosaka EM). Stained tissues were observed by a microscopy (CKX41, Olympus) and photographed by a camera (DP20, Olympus).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB567722.
Acknowledgments
We would like to thank GABI-Kat (Max Planck Institute for Plant Breeding Research, Cologne, Germany) and the Arabidopsis Biological Resource Center for providing T-DNA insertion lines.
References
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deeken R, Ache P, Kajahn I, Klinkenberg J, Bringmann G, Hedrich R. (2008) Identification of Arabidopsis thaliana phloem RNAs provides a search criterion for phloem-based transcripts hidden in complex datasets of microarray experiments. Plant J 55: 746–759 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Ryan PR, Randall PJ. (1993) Aluminum tolerance in wheat (Triticum aestivum L.): II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103: 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y, Sato K, Katsuhara M, Takeda K, Ma JF. (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48: 1081–1091 [DOI] [PubMed] [Google Scholar]
- Fuse T, Sasaki T, Yano M. (2001) Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol 18: 219–222 [Google Scholar]
- Garcia O, Bouige P, Forestier C, Dassa E. (2004) Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J Mol Biol 343: 249–265 [DOI] [PubMed] [Google Scholar]
- Hoekenga OA, Vision TJ, Shaff JE, Monforte AJ, Lee GP, Howell SH, Kochian LV. (2003) Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta x Columbia) by quantitative trait locus mapping: a physiologically simple but genetically complex trait. Plant Physiol 132: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF. (2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Koyama H, Iuchi A, Kobayashi Y, Kitabayashi S, Kobayashi Y, Ikka T, Hirayama T, Shinozaki K, Kobayashi M. (2007) Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc Natl Acad Sci USA 104: 9900–9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV. (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46: 237–260 [Google Scholar]
- Kochian LV, Hoekenga OA, Pineros MA. (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Kolar J, Senkova J. (2008) Reduction of mineral nutrient availability accelerates flowering of Arabidopsis thaliana. J Plant Physiol 165: 1601–1609 [DOI] [PubMed] [Google Scholar]
- Komeda Y. (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55: 521–535 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Cancel J, Rounds M, Ochoa V. (2007) Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 225: 1447–1458 [DOI] [PubMed] [Google Scholar]
- Larsen PB, Geisler MJB, Jones CA, Williams KM, Cancel JD. (2005) ALS3 encodes a phloem-localized ABC transporter-like protein that is required for aluminum tolerance in Arabidopsis. Plant J 41: 353–363 [DOI] [PubMed] [Google Scholar]
- Ma JF. (2008) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int Rev Cytol 264: 225–253 [DOI] [PubMed] [Google Scholar]
- Ma JF, Nagao S, Huang CF, Nishimura M. (2005) Isolation and characterization of a rice mutant hypersensitive to Al. Plant Cell Physiol 46: 1054–1061 [DOI] [PubMed] [Google Scholar]
- Ma JF, Ryan PR, Delhaize E. (2001) Aluminum tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6: 273–278 [DOI] [PubMed] [Google Scholar]
- Ma JF, Zheng SJ, Matsumoto H. (1997) Specific secretion of citric acid induced by Al stress in Cassia tora L. Plant Cell Physiol 38: 1019–1025 [Google Scholar]
- Magalhaes JV, Liu J, Guimarães CT, Lana UG, Alves VM, Wang YH, Schaffert RE, Hoekenga OA, Piñeros MA, Shaff JE, et al. (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39: 1156–1161 [DOI] [PubMed] [Google Scholar]
- Rea PA. (2007) Plant ATP-binding cassette transporters. Annu Rev Plant Biol 58: 347–375 [DOI] [PubMed] [Google Scholar]
- Roux F, Touzet P, Cuguen J, Corre VL. (2006) How to be early flowering: an evolutionary perspective. Trends Plant Sci 11: 375–381 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H. (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Sawaki Y, Iuchi S, Kobayashi Y, Kobayashi Y, Ikka T, Sakurai N, Fujita M, Shinozaki K, Shibata D, Kobayashi M, et al. (2009) STOP1 regulates multiple genes which protect Arabidopsis from proton and aluminum toxicities. Plant Physiol 150: 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C. (2002) Arabidopsis, the rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Sugiyama A, Shitan N, Yazaki K. (2007) Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-rhizobium symbiosis. Plant Physiol 144: 2000–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, et al. (2008) Plant ABC proteins—a unified nomenclature and updated inventory. Trends Plant Sci 13: 151–159 [DOI] [PubMed] [Google Scholar]
- von Uexkull HR, Mutert E. (1995) Global extent, development and economic impact of acid soils. Plant Soil 171: 1–15 [Google Scholar]
- Yamaji N, Huang CF, Nagao S, Yano M, Sato Y, Nagamura Y, Ma JF. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZQ, Ma JF, Sato K, Takeda K. (2003) Differential Al resistance and citrate secretion in barley (Hordeum vulgare L.). Planta 217: 794–800 [DOI] [PubMed] [Google Scholar]