Abstract
Genetic evidence in Arabidopsis (Arabidopsis thaliana) suggests that the auxin precursor indole-3-butyric acid (IBA) is converted into active indole-3-acetic acid (IAA) by peroxisomal β-oxidation; however, direct evidence that Arabidopsis converts IBA to IAA is lacking, and the role of IBA-derived IAA is not well understood. In this work, we directly demonstrated that Arabidopsis seedlings convert IBA to IAA. Moreover, we found that several IBA-resistant, IAA-sensitive mutants were deficient in IBA-to-IAA conversion, including the indole-3-butyric acid response1 (ibr1) ibr3 ibr10 triple mutant, which is defective in three enzymes likely to be directly involved in peroxisomal IBA β-oxidation. In addition to IBA-to-IAA conversion defects, the ibr1 ibr3 ibr10 triple mutant displayed shorter root hairs and smaller cotyledons than wild type; these cell expansion defects are suggestive of low IAA levels in certain tissues. Consistent with this possibility, we could rescue the ibr1 ibr3 ibr10 short-root-hair phenotype with exogenous auxin. A triple mutant defective in hydrolysis of IAA-amino acid conjugates, a second class of IAA precursor, displayed reduced hypocotyl elongation but normal cotyledon size and only slightly reduced root hair lengths. Our data suggest that IBA β-oxidation and IAA-amino acid conjugate hydrolysis provide auxin for partially distinct developmental processes and that IBA-derived IAA plays a major role in driving root hair and cotyledon cell expansion during seedling development.
The auxin indole-3-acetic acid (IAA) controls both cell division and cell expansion and thereby orchestrates many developmental events and environmental responses. For example, auxin regulates lateral root initiation, root and stem elongation, and leaf expansion (for review, see Davies, 2004). Normal plant morphogenesis and environmental responses require modulation of auxin levels by controlling biosynthesis, regulating transport, and managing storage forms (for review, see Woodward and Bartel, 2005a). In some storage forms, the carboxyl group of IAA is conjugated to amino acids or peptides or to sugars, and free IAA can be released by hydrolases when needed (Bartel et al., 2001; Woodward and Bartel, 2005a). A second potential auxin storage form is the side chain-lengthened compound indole-3-butyric acid (IBA), which can be synthesized from IAA (Epstein and Ludwig-Müller, 1993) and is suggested to be shortened into IAA by peroxisomal β-oxidation (Bartel et al., 2001; Woodward and Bartel, 2005a).
Genetic evidence suggests that the auxin activity of both IAA-amino acid conjugates and IBA requires free IAA to be released from these precursors (Bartel and Fink, 1995; Zolman et al., 2000). Mutation of Arabidopsis (Arabidopsis thaliana) genes encoding IAA-amino acid hydrolases, including ILR1, IAR3, and ILL2, reduces plant sensitivity to the applied IAA-amino acid conjugates that are substrates of these enzymes, including IAA-Leu, IAA-Phe, and IAA-Ala (Bartel and Fink, 1995; Davies et al., 1999; LeClere et al., 2002; Rampey et al., 2004), which are present in Arabidopsis (Tam et al., 2000; Kowalczyk and Sandberg, 2001; Kai et al., 2007).
Unlike the simple one-step release of free IAA from amino acid conjugates, release of IAA from IBA is suggested to require a multistep process (Zolman et al., 2007, 2008). Conversion of IBA to IAA has been demonstrated in a variety of plants (Fawcett et al., 1960; for review, see Epstein and Ludwig-Müller, 1993) and may involve β-oxidation of the four-carbon carboxyl side chain of IBA to the two-carbon side chain of IAA (Fawcett et al., 1960; Zolman et al., 2000, 2007). Mutation of genes encoding the apparent β-oxidation enzymes INDOLE-3-BUTYRIC ACID RESPONSE1 (IBR1), IBR3, or IBR10 results in IBA resistance, but does not alter IAA response or confer a dependence on exogenous carbon sources for growth following germination (Zolman et al., 2000, 2007, 2008), consistent with the possibility that these enzymes function in IBA β-oxidation but not fatty acid β-oxidation.
Both conjugate hydrolysis and IBA β-oxidation appear to be compartmentalized. The IAA-amino acid hydrolases are predicted to be endoplasmic reticulum localized (Bartel and Fink, 1995; Davies et al., 1999) and enzymes required for IBA responses, including IBR1, IBR3, and IBR10, are peroxisomal (Zolman et al., 2007, 2008). Moreover, many peroxisome biogenesis mutants, such as peroxin5 (pex5) and pex7, are resistant to exogenous IBA, but remain IAA sensitive (Zolman et al., 2000; Woodward and Bartel, 2005b).
Although the contributions of auxin transport to environmental and developmental auxin responses are well documented (for review, see Petrášek and Friml, 2009), the roles of various IAA precursors in these processes are less well understood. Expansion of root epidermal cells to control root architecture is an auxin-regulated process in which these roles can be dissected. Root epidermal cells provide soil contact and differentiate into files of either nonhair cells (atrichoblasts) or hair cells (trichoblasts). Root hairs emerge from trichoblasts as tube-shaped outgrowths that increase the root surface area, thus aiding in water and nutrient uptake (for review, see Grierson and Schiefelbein, 2002). Root hair length is determined by the duration of root hair tip growth, which is highly sensitive to auxin levels (for review, see Grierson and Schiefelbein, 2002). Mutants defective in the ABCG36/PDR8/PEN3 ABC transporter display lengthened root hairs and hyperaccumulate [3H]IBA, but not [3H]IAA, in root tip auxin transport assays (Strader and Bartel, 2009), suggesting that ABCG36 functions as an IBA effluxer and that IBA promotes root hair elongation. The related ABCG37/PDR9 transporter also can efflux IBA (Strader et al., 2008b; Růžička et al., 2010) and may have some functional overlap with ABCG36 (Růžička et al., 2010). In addition to lengthened root hairs, abcg36/pdr8/pen3 mutants display enlarged cotyledons, a second high-auxin phenotype. Both of these developmental phenotypes are suppressed by the mildly peroxisome-defective mutant pex5-1 (Strader and Bartel, 2009), suggesting that IBA contributes to cell expansion by serving as a precursor to IAA, which directly drives the increased cell expansion that underlies these phenotypes. However, whether IBA-derived IAA contributes to cell expansion events during development of wild-type plants is not known.
Here, we directly demonstrate that peroxisome-defective mutants are defective in the conversion of IBA to IAA, consistent with previous reports that these genes are necessary for full response to applied IBA. We found that a mutant defective in three suggested IBA-to-IAA conversion enzymes displays low-auxin phenotypes, including decreased root hair expansion and decreased cotyledon size. We further found that these mutants suppress the long-root-hair and enlarged cotyledon phenotypes of an abcg36/pdr8 mutant, suggesting that endogenous IBA-derived IAA drives root hair and cotyledon expansion in wild-type seedlings.
RESULTS
IBA-Resistant Mutants Are Defective in IBA-to-IAA Conversion
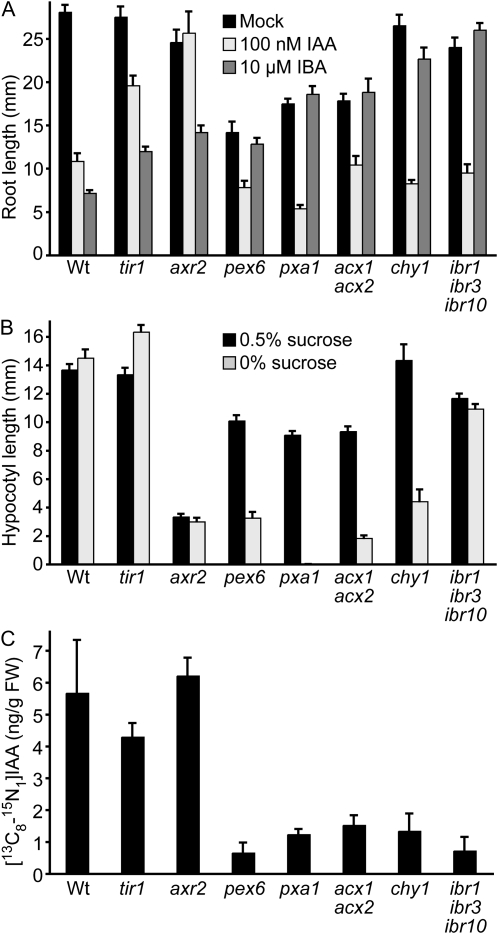
Genetic evidence suggests that the auxin activity of IBA requires conversion to IAA in a peroxisomal process resembling fatty acid β-oxidation (Zolman et al., 2000), but this hypothesis has not been tested directly. Mutants defective in auxin-signaling components, such as tir1-1 (Ruegger et al., 1998) and axr2-1 (Wilson et al., 1990), are resistant to both IBA and IAA in root elongation assays (Fig. 1A; Wilson et al., 1990; Ruegger et al., 1998; Zolman et al., 2000; Strader et al., 2008a). In contrast, mutants defective in peroxins (PEX proteins) required for proper peroxisome function, such as pex6-1, are resistant to IBA but sensitive to IAA in root elongation assays (Fig. 1A; Zolman and Bartel, 2004). Similarly, mutants defective in the peroxisomal ABC transporter PXA1, which may transport substrates into the peroxisome for β-oxidation (Zolman et al., 2001b), and mutants defective in several peroxisome-targeted enzymes, including the acyl-CoA oxidase double mutant acx1-2 acx2-1 (Adham et al., 2005), the branched-chain amino acid catabolism mutant chy1-10 (Zolman et al., 2001a), and the IBA response triple mutant ibr1-2 ibr3-1 ibr10-1 (Zolman et al., 2008), are IBA resistant and IAA sensitive (Fig. 1A; Zolman et al., 2001a, 2001b, 2008; Adham et al., 2005).
Figure 1.
Peroxisomal mutants are defective in IBA-to-IAA conversion. A, Auxin response of Col-0, tir1-1, axr2-1, pex6-1, pxa1-1, acx1-2 acx2-1, chy1-10, and ibr1-2 ibr3-1 ibr10-1. Mean primary root lengths (+se; n ≥ 12) of 8-d-old seedlings grown under yellow-filtered light at 22°C on medium supplemented with ethanol (mock), 100 nm IAA, or 10 μm IBA are shown. B, Suc dependence of Col-0, tir1-1, axr2-1, pex6-1, pxa1-1, acx1-2 acx2-1, chy1-10, and ibr1-2 ibr3-1 ibr10-1. Mean hypocotyl lengths (+se; n = 12) of seedlings grown in the absence or presence of 0.5% Suc were measured 4 d after transfer of 1-d-old seedlings to the dark. C, In planta conversion of IBA to IAA. Eight-day-old Col-0, tir1-1, axr2-1, pex6-1, pxa1-1, acx1-2 acx2-1, chy1-10, and ibr1-2 ibr3-1 ibr10-1 light-grown seedlings were incubated for 1 h in the presence of 10 μm [13C8-15N1]IBA and the resultant [13C8-15N1]IAA levels were measured. Wt, Col-0.
Because genetic evidence suggests that IBA-to-IAA conversion is impaired in mutants defective in peroxisome function or certain peroxisome-targeted enzymes, we examined the mutant biochemical defects in a feeding assay with labeled IBA. We incubated 8-d-old seedlings in a solution containing 10 μm [13C8-15N1]IBA and assayed levels of [13C8-15N1]IAA after 1 h by using gas chromatography-mass spectrometry (GC-MS) with selective ion monitoring (SIM). We found that wild-type seedlings converted some of the provided IBA to IAA during the 1-h labeling period, demonstrating that Arabidopsis, like other plants (Epstein and Lavee, 1984; Epstein et al., 1989; Epstein and Ludwig-Müller, 1993; Ludwig-Müller, 2000), can use IBA as an IAA precursor. Moreover, the tir1-1 and axr2-1 auxin-signaling mutants accumulated similar levels of [13C8-15N1]IAA as wild type (Fig. 1C). However, all of the tested IBA-resistant, IAA-sensitive mutants (pex6-1, pxa1-1, acx1-2 acx2-1, chy1-10, and ibr1-2 ibr3-1 ibr10-1) accumulated only low levels of [13C8-15N1]IAA (Fig. 1C), confirming the hypothesis suggested by genetic evidence that these mutants are defective in IBA-to-IAA conversion.
Because the tested IBA-resistant, IAA-sensitive mutants produced low levels of [13C8-15N1]IAA when supplied with [13C8-15N1]IBA (Fig. 1C), we examined endogenous IAA and IBA levels in wild-type and ibr1 ibr3 ibr10 seedlings. We found that free IAA and IBA levels in ibr1 ibr3 ibr10 seedlings were not notably different from levels found in wild type (Supplemental Fig. S1). Because measuring auxin levels in whole seedlings may mask cell- or tissue-specific variations in auxin levels, we examined ibr1 ibr3 ibr10 for auxin-related phenotypes.
Mutants Deficient in IBA-to-IAA Conversion Fail to Expand Root Hairs
We previously found that abcg36/pdr8/pen3 mutants display defects in IBA efflux and mild developmental phenotypes, including root hairs that were longer than those of wild type (Fig. 2B; Strader and Bartel, 2009), which suggest increased auxin levels in these cells. We therefore examined mutants with decreased IBA-to-IAA conversion for the opposite phenotype. Many of these mutants are defective not only in IBA-to-IAA conversion (Fig. 1C) and IBA responsiveness (Fig. 1A), but also fatty acid β-oxidation, which results in dependence on an external fixed carbon supply such as Suc for growth following germination (Fig. 1B). To avoid complications that might result from the limited fixed carbon available to mutants with defects in fatty acid utilization, we examined root hair lengths of IBA response mutants defective in enzymes suggested to be dedicated to IBA-to-IAA conversion: IBR1, IBR3, and IBR10 (Zolman et al., 2007, 2008). These mutants, unlike many other peroxisome-defective mutants, do not require exogenous carbon sources to fuel early seedling growth (Fig. 1B; Zolman et al., 2000, 2007, 2008), suggesting that fatty acid β-oxidation is functioning normally.
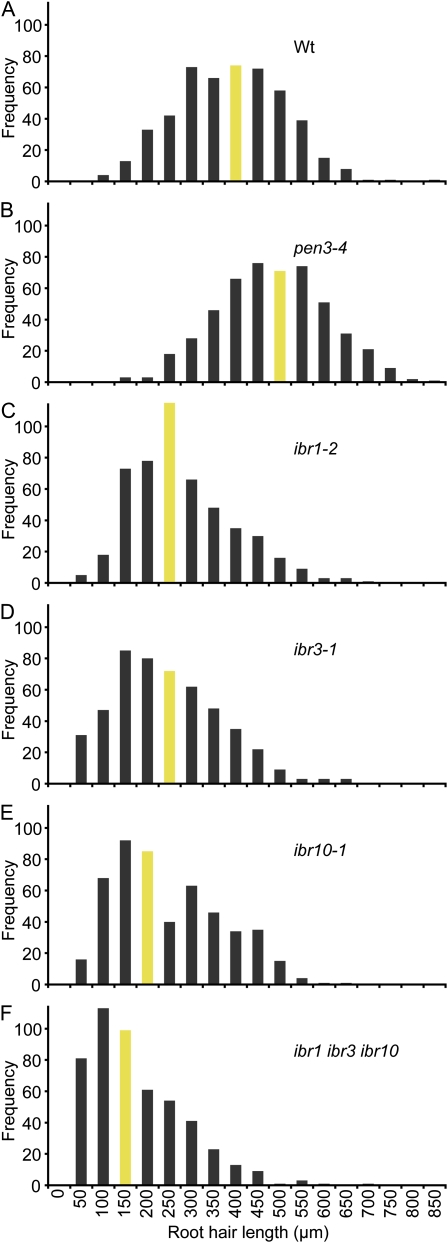
Figure 2.
ibr mutants display root hair elongation defects. A to F, Histograms of root hair lengths of 5-d-old vertically grown Col-0 (Wt; A), pen3-4 (B), ibr1-2 (C), ibr3-1 (D), ibr10-1 (E), and ibr1-2 ibr3-1 ibr10-1 (F) seedlings. Root hairs were measured using NIH Image software (n = 500 total root hairs from at least 10 seedlings). Bars depicting median root hair lengths are shaded. [See online article for color version of this figure.]
Unlike the longer root hairs of pen3 mutants (Fig. 2B), we found that ibr1 (Fig. 2C), ibr3 (Fig. 2D), and ibr10 (Fig. 2E) single mutants each displayed shorter root hairs than wild type (Fig. 2A). Similar to findings in IBA-responsive root elongation assays (Zolman et al., 2008), the ibr1 ibr3 ibr10 triple mutant had shorter root hairs than any of the ibr single mutants (Fig. 2F).
Exogenous Active Auxin Restores ibr1 ibr3 ibr10 Root Hair Elongation
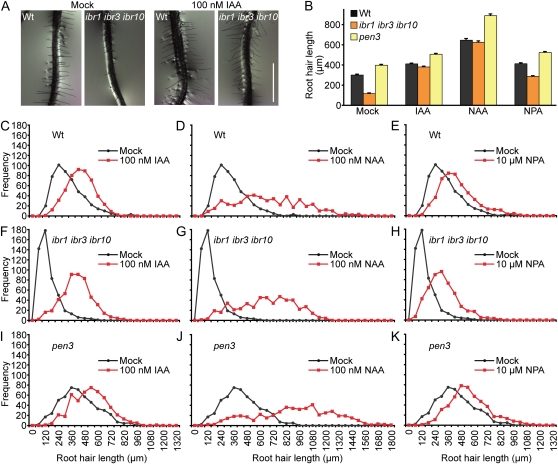
Conversely to the long root hairs of the abcg36/pdr8/pen3 mutants, which suggest increased auxin levels in root hair cells, the shorter root hairs of ibr1 ibr3 ibr10 suggest that auxin levels are low in root hairs of this mutant. We found that supplementation with the naturally occurring active auxin IAA rescued ibr1 ibr3 ibr10 root hairs to wild-type lengths (Fig. 3, A–C and F). Growing pen3 with IAA further lengthened the already long root hairs (Fig. 3, B and I), suggesting that active auxin levels in pen3 can be further increased by exogenous sources. Like IAA, growth on the synthetic auxin 1-naphthalene acetic acid (NAA) lengthened root hairs of wild type (Fig. 3, B and D), ibr1 ibr3 ibr10 (Fig. 3, B and G), and pen3 (Fig. 3, B and J). NAA-treated ibr1 ibr3 ibr10 root hairs resembled NAA-treated wild-type root hairs.
Figure 3.
Auxin application restores root hair elongation to ibr mutants. A, Photographs of 5-d-old Col-0 and ibr1-2 ibr3-1 ibr10-1 seedling roots vertically grown under yellow-filtered light at 22°C on medium supplemented with ethanol (mock) or 100 nm IAA. Scale bar = 1 mm. B, Mean root hair lengths (+se) of 5-d-old Col-0, ibr1-2 ibr3-1 ibr10-1, and pen3-4 seedlings vertically grown under yellow-filtered light at 22°C on medium supplemented with ethanol (mock), 100 nm IAA, 100 nm NAA, or 10 μm NPA. Root hair lengths were measured using NIH Image software (n = 500 total root hairs from at least 12 seedlings). C to K, Histograms of root hair lengths from section B of Col-0 (C–E), ibr1-2 ibr3-1 ibr10-1 (F–H), and pen3-4 (I–K) grown on IAA (C, F, and I), NAA (D, G, and J), or NPA (E, H, and K). Histograms for mock-treated seedlings are shown in every section for comparison. Wt, Col-0. [See online article for color version of this figure.]
Growing seedlings on the auxin efflux inhibitor 1-naphthylphthalamic acid (NPA) resulted in longer root hairs in wild type (Fig. 3, B and E), ibr1 ibr3 ibr10 (Fig. 3, B and H), and pen3 (Fig. 3, B and K). However, unlike IAA and NAA, NPA-treated ibr1 ibr3 ibr10 root hairs remained shorter than NPA-treated wild-type root hairs (Fig. 3, B, E, and H; Supplemental Fig. S2). Similarly, ibr1 ibr3 ibr10 root hairs treated with the auxin efflux inhibitor 2,3,5-triiodobenzoic acid (TIBA) were longer than untreated root hairs but remained shorter than TIBA-treated wild-type root hairs (Supplemental Fig. S2). The ability of natural and synthetic auxins to fully rescue ibr1 ibr3 ibr10 root hair elongation defects and the ability of auxin transport inhibitors to partially rescue ibr1 ibr3 ibr10 root hair elongation defects are consistent with the possibility that endogenous IBA is an important IAA source during normal root hair elongation.
Blocking IBA-to-IAA Conversion Suppresses the Long-Root-Hair Phenotype of abcg36
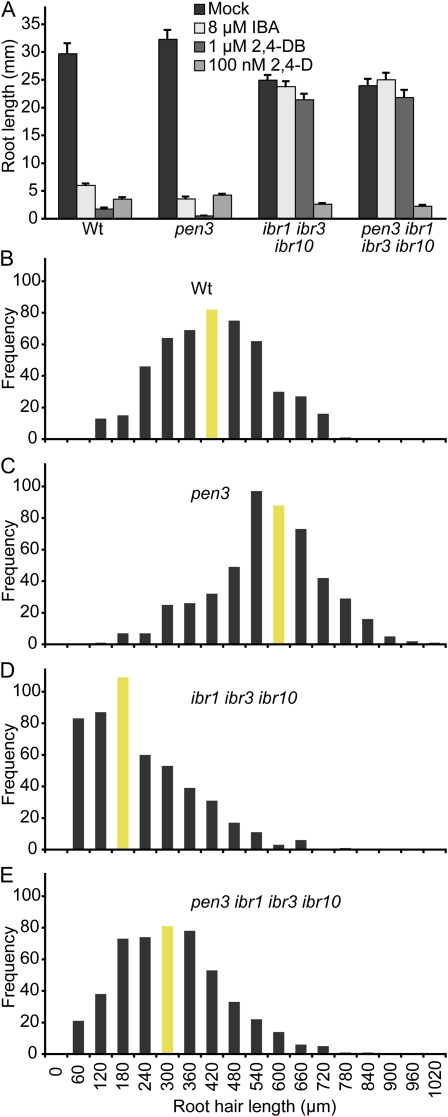
To determine whether the IBR1, IBR3, and IBR10 enzymes act upstream or downstream of the ABCG36 transporter, we crossed ibr1 ibr3 ibr10 to the pen3-4 null allele of abcg36/pdr8/pen3 and compared root hair elongation and auxin-responsive root elongation of wild type, pen3, ibr1 ibr3 ibr10, and the pen3 ibr1 ibr3 ibr10 quadruple mutant. We found that the pen3 ibr1 ibr3 ibr10 quadruple mutant exhibited IBA and 2,4-dichlorophenoxybutyric acid (2,4-DB) resistance similar to the ibr1 ibr3 ibr10 triple mutant parent (Fig. 4A), confirming that IBA-to-IAA and 2,4-DB-to-2,4-dichlorophenoxyacetic acid (2,4-D) conversion are necessary for the heightened sensitivity of pen3 to IBA and 2,4-DB (Strader and Bartel, 2009). In addition, untreated pen3 ibr1 ibr3 ibr10 seedlings (Fig. 4D) displayed shorter root hairs than wild type, like the ibr1 ibr3 ibr10 parent (Fig. 4C). These data are consistent with the hypothesis that IBA-derived IAA causes the elongated root hairs in the pen3 mutant.
Figure 4.
ibr1 ibr3 ibr10 suppresses pen3 root hair hyperelongation. A, Mean primary root lengths (+se) of 8-d-old Col-0, pen3-4, ibr1-2 ibr3-1 ibr1-10, and pen3-4 ibr1-2 ibr3-1 ibr10-1 seedlings grown under yellow-filtered light at 22°C on medium supplemented with ethanol (mock), 8 μm IBA, 1 μm 2,4-DB, or 100 nm 2,4-D (n ≥ 15). B to E, Histograms of root hair lengths of 5-d-old vertically grown Col-0 (B), pen3-4 (C), ibr1-2 ibr3-1 ibr10-1 (D), and pen3-4 ibr1-2 ibr3-1 ibr10-1 (E) seedlings. Root hair lengths were measured using NIH Image software (n = 500 total root hairs from at least 15 seedlings). Bars depicting median root hair lengths are shaded. Wt, Col-0. [See online article for color version of this figure.]
Defects in IAA Conjugate Hydrolases Result in Shortened Root Hairs
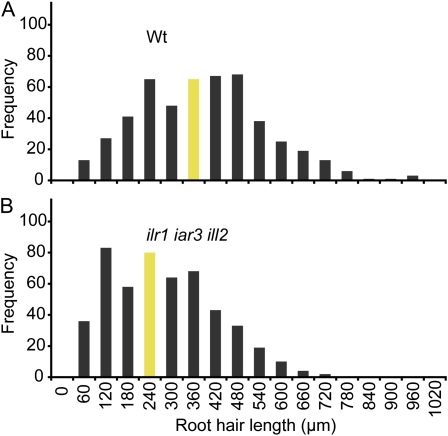
Because IBA-derived IAA appears to play a role in root hair elongation, we assessed whether other auxin storage forms also might contribute to root hair elongation. We examined root hairs in a triple mutant defective in three IAA-amino acid conjugate hydrolases (Rampey et al., 2004): ILR1, an IAA-Leu and IAA-Phe hydrolase (Bartel and Fink, 1995); IAR3, an IAA-Ala hydrolase (Davies et al., 1999); and ILL2, an IAA-Ala hydrolase (LeClere et al., 2002). We found that the ilr1-1 iar3-2 ill2-1 triple mutant root hairs were approximately 30% shorter than wild type (Fig. 5), consistent with a role for conjugate-derived IAA in root hair elongation.
Figure 5.
The ilr1 iar3 ill2 triple auxin-conjugate hydrolase mutant displays defects in root hair elongation. Histograms of root hair lengths of 5-d-old vertically grown Ws-2 (Wt; A) and ilr1-1 iar3-2 ill2-1 (B) seedlings. Root hair lengths were measured using NIH Image software (n = 500 total root hairs from at least 15 seedlings). Bars depicting median root hair lengths are shaded. [See online article for color version of this figure.]
Mutants Deficient in IBA-to-IAA Conversion Have Smaller Cotyledons
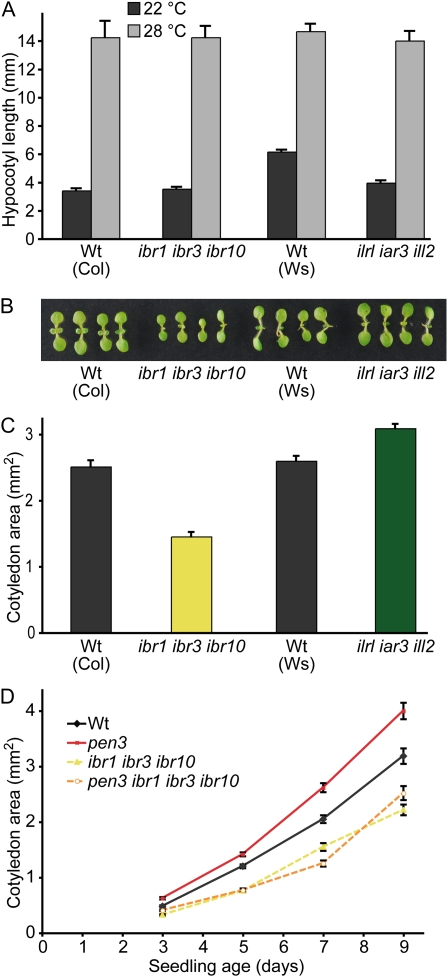
Hypocotyl elongation and cotyledon cell expansion also depend on auxin. The triple hydrolase mutant, which has decreased free IAA levels in whole seedlings (Rampey et al., 2004), displays reduced hypocotyl lengths when grown in the light at 22°C but not at 28°C (Fig. 6A; Rampey et al., 2004), suggesting that conjugates provide some of auxin that drives hypocotyl elongation at normal growth temperatures. When we examined ibr1 ibr3 ibr10 for a similar phenotype, we found that 8-d-old ibr1 ibr3 ibr10 seedlings displayed wild-type hypocotyl lengths at both temperatures (Fig. 6A), suggesting that conjugate-derived IAA has a larger role than IBA-derived IAA in driving hypocotyl elongation in the light and that neither precursor contributes to the increased IAA that drives hypocotyl elongation at high temperature (Gray et al., 1998; Zhao et al., 2002).
Figure 6.
The ibr1 ibr3 ibr10 mutant has small cotyledons but normal hypocotyl elongation. A, Mean hypocotyl lengths (+se) of 8-d-old Col-0 (Wt), ibr1-2 ibr3-1 ibr10-1, Ws-2 (Wt), and ilr1-1 iar3-2 ill2-1 seedlings grown under yellow-filtered light at 22°C or 28°C. B, Photograph of 7-d-old Col-0 (Wt), ibr1-2 ibr3-1 ibr10-1, Ws-2 (Wt), and ilr1-1 iar3-2 ill2-1 light-grown seedlings. C, Mean cotyledon areas (+se) of 7-d-old Col-0 (Wt), ibr1-2 ibr3-1 ibr10-1, Ws-2 (Wt), and ilr1-1 iar3-2 ill2-1 light-grown seedlings (n ≥ 33). D, ibr1 ibr3 ibr10 suppresses pen3 enlarged cotyledons. Col-0 (Wt), pen3-4, ibr1-2 ibr3-1 ibr10-1, and pen3-4 ibr1-2 ibr3-1 ibr10-1 seeds were stratified for 3 d at 4°C prior to growth at 22°C under white light. Mean cotyledon areas (±se) of light-grown seedlings during growth following germination (n ≥ 51). Measurements were made using NIH Image software.
We previously found that abcg36/pdr8/pen3 mutants, in addition to displaying elongated root hairs, display enlarged cotyledons (Fig. 6D; Strader and Bartel, 2009), suggesting that auxin levels are increased not only in root epidermal cells, but also in cotyledon cells. This increase in cotyledon size probably results from increased cotyledon cell expansion, because after germination, Arabidopsis cotyledons grow by cell expansion without cell division (Mansfield and Briarty, 1996). We examined cotyledons of 7-d-old ibr1 ibr3 ibr10 mutants and ilr1 iar3 ill2 mutants and found that ibr1 ibr3 ibr10 displayed smaller cotyledons than wild type, whereas ilr1 iar3 ill2 cotyledons resembled wild-type cotyledons (Fig. 6, B and C). We further found that the pen3 ibr1 ibr3 ibr10 quadruple mutant exhibited cotyledon sizes similar to the ibr1 ibr3 ibr10 triple mutant parent (Fig. 6D). The cotyledons of wild type, pen3, ibr1 ibr3 ibr10, and pen3 ibr1 ibr3 ibr10 appeared to be similarly sized soon after emergence from the seed coat, but pen3 mutant cotyledons expanded more rapidly and ibr1 ibr3 ibr10 and pen3 ibr1 ibr3 ibr10 mutant cotyledons expanded more slowly than those of the wild type (Fig. 6D), suggesting that the small cotyledons of seedlings with impaired IBA-to-IAA conversion result from a cell expansion defect rather than delayed germination.
DISCUSSION
The active auxin pool is tightly controlled through regulation of IAA biosynthesis, regulation of IAA transport, and use of storage forms. One means of controlling free IAA levels is through the formation and hydrolysis of IAA conjugates, wherein IAA is conjugated to amino acids, peptides, or sugars, to be released by hydrolases when free IAA is needed (Bartel et al., 2001; Woodward and Bartel, 2005a). A second potential auxin storage form is the chain-lengthened compound IBA, which requires peroxisomal β-oxidation to IAA for auxin activity (Bartel et al., 2001; Woodward and Bartel, 2005a).
Genetic screens have uncovered a number of genes required for full responsiveness to the protoauxin IBA. These genes encode proteins required for proper peroxisome function, such as PEX4 (Zolman et al., 2005), PEX5 (Zolman et al., 2000), PEX6 (Zolman and Bartel, 2004), and PEX7 (Woodward and Bartel, 2005b; Ramón and Bartel, 2010); the peroxisomal ABC transporter PXA1, which may transport IBA into the peroxisome for β-oxidation (Zolman et al., 2001b); and enzymes that reside in the peroxisome, including ACX enzymes (Adham et al., 2005), CHY1 (Zolman et al., 2001a), IBR1 (Zolman et al., 2008), IBR3 (Zolman et al., 2007), and IBR10 (Zolman et al., 2008). Here, we have provided biochemical evidence that Arabidopsis seedlings can convert IBA into IAA and that multiple mutants identified in genetic screens for IBA resistance are defective in this conversion (Fig. 1C).
Of the identified factors required for full response to IBA, the specificity of indole-3-butyric acid response1 (ibr1), ibr3, and ibr10 defects, along with the identities of the mutated enzymes, are consistent with the possibility that IBR1, IBR3, and IBR10 act directly in the conversion of IBA to IAA (Fig. 1; Zolman et al., 2007, 2008). IBR1, IBR3, and IBR10 encode peroxisomal proteins that resemble β-oxidation enzymes (Zolman et al., 2007, 2008). IBR1 is a member of the dehydrogenase subcategory of the short-chain dehydrogenase/reductase family (Kallberg et al., 2002a, 2002b; Zolman et al., 2008), and is predicted to perform the 3-hydroxyacyl-CoA dehydrogenase step in IBA-to-IAA conversion (Zolman et al., 2008). IBR3 is an acyl-CoA dehydrogenase-like protein that is predicted to oxidize IBA-CoA to the α,β-unsaturated thioester (Zolman et al., 2007). IBR10 resembles enoyl-CoA hydratases and is predicted to produce an IBA hydroxyacyl-CoA thioester intermediate (Zolman et al., 2008). IBR1, IBR3, and IBR10 are peroxisomal proteins, suggesting that these enzymes act along with other identified and unidentified proteins in the peroxisomal conversion of the protoauxin IBA to active IAA.
IBA-derived IAA appears to contribute to several auxin-related developmental processes. We previously found that abcg36/pdr8/pen3 IBA efflux mutants display the high-auxin phenotypes of enlarged cotyledons and lengthened root hairs, both of which are suppressed by the weak peroxisome mutant pex5-1 (Strader and Bartel, 2009), suggesting that IBA-derived IAA drives the increased cell expansion contributing to these phenotypes. However, the pex5-1 single mutant does not display notable auxin-deficient phenotypes (Strader and Bartel, 2009), perhaps because of the mild nature of the PEX5 defect in this allele (Zolman et al., 2000). In contrast, the ibr1 ibr3 ibr10 triple mutant displayed a strong defect in root hair elongation and cotyledon expansion (Figs. 2F and 6C), suggesting that auxin levels were low in these tissues. Additionally, comatose alleles of PXA1, the ABC transporter likely to bring IBA into the peroxisome (Zolman et al., 2001b), display a stamen filament elongation delay that is rescued by NAA application (Footitt et al., 2007), suggesting that the auxin required for filament elongation (Cecchetti et al., 2008) is low in this tissue. Similarly, the ibr1 ibr3 ibr10 short-root-hair phenotype was fully rescued by exogenous IAA and NAA, but only partially rescued by the auxin transport inhibitors NPA or TIBA, consistent with the possibility that active auxin levels are low in root hair cells of this mutant. We also found that ibr1 ibr3 ibr10, like pex5-1 (Strader and Bartel, 2009), suppressed the long-root-hair and enlarged cotyledon phenotypes of the pen3 IBA efflux mutant (Figs. 4E and 6C), confirming that IBA-to-IAA conversion is downstream of ABCG36/PDR8/PEN3.
Because hydrolysis of IAA-amino acid conjugates also can contribute to the active auxin pool, we examined root hairs in the triple auxin-conjugate hydrolase mutant ilr1-1 iar3-2 ill2-1 (Rampey et al., 2004). Light-grown ilr1 iar3 ill2 seedlings have slightly decreased free IAA levels, a longer primary root, and a shorter hypocotyl than wild type (Rampey et al., 2004). We found that ilr1 iar3 ill2 had an approximately 30% reduction in root hair length, compared to the approximately 55% reduction in root hair length in the ibr1 ibr3 ibr10 triple mutant, suggesting that IBA-derived IAA may play a more substantial role in driving root hair expansion than conjugate-derived IAA. The reduced hypocotyl elongation phenotype of light-grown ilr1 iar3 ill2 triple mutant seedlings (Fig. 6A; Rampey et al., 2004) was not observed in ibr1 ibr3 ibr10 (Fig. 6A), suggesting that IBA-derived IAA plays a negligible role in hypocotyl elongation. Conversely, ilr1 iar3 ill2 cotyledon expansion was not impaired, suggesting that conjugate-derived IAA plays a minimal role in this growth process. However, the hydrolase and ibr mutations are present in different Arabidopsis accessions, and a direct comparison of the importance of conjugates versus IBA in providing IAA for root hair elongation, hypocotyl elongation, and cotyledon expansion would be facilitated by the development of a triple hydrolase mutant in the Columbia (Col) accession.
Active auxin pools are controlled by a variety of processes, including regulation of biosynthesis, transport, and storage (for review, see Woodward and Bartel, 2005a). Our data suggest that IBA-derived IAA drives cell expansion in specific tissues, including root hair and cotyledon cells, indicating that IBA is an important IAA precursor in young Arabidopsis seedlings. The reverse reaction, the formation of IBA from IAA, is catalyzed by an IBA synthetase (for review, see Ludwig-Müller 2000) that is induced by drought and abscisic acid in maize (Zea mays) seedlings (Ludwig-Müller et al., 1995). It will be interesting to learn whether and how IBA-to-IAA conversion is regulated and whether IBA β-oxidation contributes to active IAA in other tissue types and developmental stages.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Phenotypic Assays
Arabidopsis (Arabidopsis thaliana) mutants were in the Col-0 or Wassilewskija (Ws-2) accession. PCR analysis of F2 plants was used to identify higher-order mutants as previously described for ibr1-2 (Zolman et al., 2008), ibr3-1(Zolman et al., 2008), and ibr10-1 (Zolman et al., 2008). Amplification of PDR8 with PDR8-1 (5′-GTATCACCCAACTAAATCCTCACG-3′) and PDR8-2 (5′-ATCTGTTACACGGCCAAAGTTAG-3′) yields a 1,450-bp product in wild type and no product in pen3-4 (SALK_000578). Amplification with PDR8-1 and LB1-SALK (5′-CAAACCAGCGTGGACCGCTTGCTGCAACTC-3′) yields an approximately 450-bp product in pen3-4 and no product in wild type.
Surface-sterilized seeds were plated on plant nutrient medium (PN; Haughn and Somerville, 1986) solidified with 0.6% (w/v) agar and supplemented with 0.5% (w/v) Suc (PNS). Seedlings were grown at 22°C under continuous illumination, unless otherwise noted.
To examine Suc-dependent hypocotyl elongation in the dark, seeds were plated on PN or PNS, incubated under white light for 1 d, and then incubated in the dark for an additional 4 d, after which hypocotyl lengths were measured. To examine hypocotyl elongation in the light, seedlings were grown under continuous illumination through yellow long-pass filters for 8 d on PNS at either 22°C or 28°C, and hypocotyl lengths were measured.
To examine auxin-responsive root elongation, seedlings were grown under yellow long-pass filters to slow indolic compound breakdown (Stasinopoulos and Hangarter, 1990) for 8 d on PNS supplemented with the indicated auxin, and the lengths of primary roots were measured.
To examine root hairs, seedlings were vertically grown for 5 d at 22°C under continuous white light (unless otherwise noted), the 4-mm root sections adjacent to the root-shoot junction were imaged using a dissecting microscope, and root hair lengths were measured using National Institutes of Health (NIH) Image software.
To examine cotyledon expansion, cotyledons of seedlings grown under continuous white light at 22°C were removed, mounted, and imaged through a dissecting microscope. Cotyledon areas were measured using NIH Image software.
Synthesis of [13C8-15N1]IBA
Synthesis of [13C8-15N1]IBA was modified from previously described methods (Cohen and Schulze, 1981; Sutter and Cohen, 1992). A total of 0.08 mol of γ-butyrolactone, 4.26 × 10−4 moles (0.05 g) of [U-13C8, 98 atom%+;15N, 96 atom% to 99 atom%]indole (Cambridge Isotope Laboratories Inc.), and 0.08 mol of NaOH were mixed in a 23-mL Teflon insert fitted into a screw-top reaction bomb (Parr Instruments). This mixture was incubated in a heating mantle, temperature was increased 2°C/min, and then the mixture was held at 220°C for 24 h. The reaction was stopped by the addition of 50 mL water, and the mixture stirred in an Erlenmeyer flask until the sample was dissolved. The reaction mixture was extracted with chloroform twice to remove any unreacted indole, and the aqueous layer brought to pH 2.5 with HCl. [13C8-15N1]IBA was then partitioned 3× into ethyl actetate from the aqueous layer. The combined ethyl acetate fractions were evaporated and the [13C8-15N1]IBA was resuspended in 50% isopropanol (v/v). The [13C8-15N1]IBA was purified on a 1.0 × 40 cm column of Sephadex LH-20 (Sigma-Aldrich) with 50% isopropanol (v/v) as the running solvent. The concentration of [13C8-15N1]IBA in the pooled fractions was determined at UV282 (ϵ = 6,060) and confirmed by reverse-isotope dilution GC-MS-SIM with an unlabeled IBA standard.
Feeding Assays with [13C8-15N1]IBA
Eight-day-old seedlings (50–130 mg fresh weight) were incubated in 250 μL uptake buffer (10 mm MES, 10 mm Suc, and 0.5 mm CaSO4, pH 5.6) supplemented with 10 μM [13C8-15N1]IBA. Seedlings were incubated for 1 h at room temperature, rinsed four times in uptake buffer, placed in a 1.5-mL tube, frozen in liquid nitrogen, and stored at −80°C prior to analysis.
Quantification of Auxins
For free [13C8-15N1]IAA analysis, 150 μL of homogenization buffer (65% isopropanol, 35% 0.2 m imidazole buffer, pH 7.0) containing 7.5 ng [13C6]IAA internal standard (99 atom%, Cambridge Isotope Laboratories; Cohen et al., 1986) were added to each tissue sample from the [13C8-15N1]IBA feeding assay. For free IAA and IBA analysis, 150 μL of homogenization buffer containing 10 ng [13C6]IAA and 10 ng [13C8-15N1]IBA were added to each 50 mg fresh weight tissue sample (5-d-old seedlings grown under white light on PNS). Two 3-mm tungsten carbide beads (Qiagen) were added to each tube and samples were homogenized with a Mixer-Mill (Qiagen) for 5 min at 25 Hz, then incubated on ice for 1 h to allow the internal standards to equilibrate with the endogenous auxins in the extracts.
After equilibration, samples were centrifuged for 5 min at 10,000g and 100 μL of supernatant were collected and placed into a deep 96-well plate (Continental Lab Products). IAA extraction was performed according to Barkawi et al. (2008), with the IAA eluted in 600 μL MeOH and transferred to 1.5 mL screw-capped glass vials. For methylation, 900 μL ethereal diazomethane (Cohen, 1984) was added to each sample, and tubes were capped and incubated for 5 min at room temperature, then dried under N2 in a 55°C sand bath and resuspended in 30 μL ethyl acetate. Samples were analyzed by GC-MS-SIM using an Agilent 6890 GC/5973 MS (Agilent Technologies) run in electron impact ionization mode at 70 eV and equipped with a fused silica capillary column (HP-5MS, 30 m × 0.25-mm i.d., 0.25 μm film). The injector temperature was 280°C and the GC oven temperature was programmed to ramp from 70°C to 280°C at 10°C/min. Helium was used as the carrier gas at a flow rate of 1 mL/min.
IAA and IBA levels were calculated by monitoring ions at mass-to-charge ratio (m/z) 130 and 189 for endogenous IAA, m/z 139 and 198 for [13C8-15N1]IAA formed from added [13C8-15N1]IBA, m/z 136 and 195 for the [13C6]IAA standard, m/z 130 and 217 for endogenous IBA, and m/z 139 and 226 for the [13C8-15N1]IBA standard. Quantities were calculated by using standard isotope dilution equations (Cohen et al., 1986).
The Arabidopsis Genome Initiative locus identifiers of genes used in this study are as follows: ACX1, At4g16760; ACX2, At5g65110; AXR2, At3g23050; CHY1, At5g65940; IAR3, At1g51760; IBR1, At4g05530; IBR3, At3g06810; IBR10, At4g14430; ILL2, At5g56660; ILR1, At3g02875; TIR1, At3g62980; PEN3/PDR8/ABCG36, At1g59870; PEX6, At1g03000; and PXA1, At4g39850.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Auxin levels in whole ibr1 ibr3 ibr10 seedlings resemble those in wild type.
Supplemental Figure S2. Auxin transport inhibitors only partially restore root hair elongation to ibr1 ibr3 ibr10.
Supplementary Material
Acknowledgments
We are grateful to Ben Pederson (Macalester College and University of Minnesota) for synthesizing [13C8-15N1]IBA; Xing Liu and Peng Yu (University of Minnesota) for assistance with mass spectrometry; Mark Estelle (University of California, San Diego) for aux1-7; the Arabidopsis Biological Resource Center at The Ohio State University for pen3-4 (SALK_000578); and Sarah Christensen, Wendell Fleming, Naxhiely Ramón, Jerrad Stoddard, and Andrew Woodward (Rice University) for critical comments on the manuscript.
References
- Adham AR, Zolman BK, Millius A, Bartel B. (2005) Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in b-oxidation. Plant J 41: 859–874 [DOI] [PubMed] [Google Scholar]
- Barkawi LS, Tam YY, Tillman JA, Pederson B, Calio J, Al-Amier H, Emerick M, Normanly J, Cohen JD. (2008) A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal Biochem 372: 177–188 [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748 [DOI] [PubMed] [Google Scholar]
- Bartel B, LeClere S, Magidin M, Zolman BK. (2001) Inputs to the active indole-3-acetic acid pool: de novo synthesis, conjugate hydrolysis, and indole-3-butyric acid b-oxidation. J Plant Growth Regul 20: 198–216 [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD. (1984) Convenient apparatus for the generation of small amounts of diazomethane. J Chromatogr A 303: 193–196 [Google Scholar]
- Cohen JD, Baldi BG, Slovin JP. (1986) 13C6-[Benzene ring]-indole-3-acetic acid: a new internal standard for quantitative mass spectral analysis of indole-3-acetic acid in plants. Plant Physiol 80: 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JD, Schulze A. (1981) Double-standard isotope dilution assay. I. Quantitative assay of indole-3-acetic acid. Anal Biochem 112: 249–257 [DOI] [PubMed] [Google Scholar]
- Davies PJ. (2004) Introduction. Davies PJ, , Plant Hormones: Biosynthesis, Signal Transduction, Action! Ed 3. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–35 [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E, Chen KH, Cohen JD. (1989) Identification of indole-3-butyric acid as an endogenous constituent of maize kernels and leaves. Plant Growth Regul 8: 215–223 [Google Scholar]
- Epstein E, Lavee S. (1984) Conversion of indole-3-butyric acid to indole-3-acetic acid by cuttings of grapevine (Vitus vinifera) and olive (Olea europa). Plant Cell Physiol 25: 697–703 [Google Scholar]
- Epstein E, Ludwig-Müller J. (1993) Indole-3-butyric acid in plants: occurrence, synthesis, metabolism, and transport. Physiol Plant 88: 382–389 [Google Scholar]
- Fawcett CH, Wain RL, Wightman F. (1960) The metabolism of 3-indolylalkanecarboxylic acids, and their amides, nitriles and methyl esters in plant tissues. Proc R Soc Lond B Biol Sci 152: 231–254 [DOI] [PubMed] [Google Scholar]
- Footitt S, Dietrich D, Fait A, Fernie AR, Holdsworth MJ, Baker A, Theodoulou FL. (2007) The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol 144: 1467–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. (1998) High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C, Schiefelbein J. (2002) Root Hairs. The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi/, http://www.aspb.org/publications/arabidopsis/ [Google Scholar]
- Haughn GW, Somerville C. (1986) Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet 204: 430–434 [Google Scholar]
- Kai K, Horita J, Wakasa K, Miyagawa H. (2007) Three oxidative metabolites of indole-3-acetic acid from Arabidopsis thaliana. Phytochemistry 68: 1651–1663 [DOI] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jörnvall H, Persson B. (2002a) Short-chain dehydrogenase/reductase (SDR) relationships: a large family with eight clusters common to human, animal, and plant genomes. Protein Sci 11: 636–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jörnvall H, Persson B. (2002b) Short-chain dehydrogenases/reductases (SDRs). Eur J Biochem 269: 4409–4417 [DOI] [PubMed] [Google Scholar]
- Kowalczyk M, Sandberg G. (2001) Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol 127: 1845–1853 [PMC free article] [PubMed] [Google Scholar]
- LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B. (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277: 20446–20452 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. (2000) Indole-3-butyric acid in plant growth and development. Plant Growth Regul 32: 219–230 [Google Scholar]
- Ludwig-Müller J, Schubert B, Pieper K. (1995) Regulation of IBA synthetase from maize (Zea mays L.) by drought stress and ABA. J Exp Bot 46: 423–432 [Google Scholar]
- Mansfield SG, Briarty LG. (1996) The dynamics of seedling and cotyledon cell development in Arabidopsis thaliana during reserve mobilization. Int J Plant Sci 157: 280–295 [Google Scholar]
- Petrášek J, Friml J. (2009) Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Ramón NM, Bartel B. (2010) Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell 21: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol 135: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Strader LC, Bailly A, Yang H, Blakeslee JJ, Łangowski Ł, Nejedlá E, Fujita H, Ito H, Syōno J, et al. (2010) Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc Natl Acad Sci USA 107: 10749–10753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP. (1990) Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol 93: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. (2009) The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Monroe-Augustus M, Bartel B. (2008a) The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Monroe-Augustus M, Rogers KC, Lin GL, Bartel B. (2008b) Arabidopsis iba response5 (ibr5) suppressors separate responses to various hormones. Genetics 180: 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter EG, Cohen JD. (1992) Measurement of indolebutyric acid in plant tissues by isotope dilution gas chromatography-mass spectrometry analysis. Plant Physiol 99: 1719–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam YY, Epstein E, Normanly J. (2000) Characterization of auxin conjugates in Arabidopsis: low steady-state levels of indole-3-acetyl-aspartate, indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol 123: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. (1990) A dominant mutation in Arabidopsis confers resistance to auxin, ethylene, and abscisic acid. Mol Gen Genet 222: 377–383 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005a) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005b) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL. (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B. (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA 101: 1786–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. (2008) Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. (2005) Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell 17: 3422–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SPT, Bartel B. (2001a) chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J Biol Chem 276: 31037–31046 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B. (2007) IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B. (2001b) The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.