Abstract
We used three-dimensional structured illumination microscopy (3D-SIM) to obtain subdiffraction (“super-resolution”) images of plasmodesmata (PD) expressing a green fluorescent protein-tagged viral movement protein (MP) in tobacco (Nicotiana tabacum). In leaf parenchyma cells, we were able to resolve individual components of PD (neck and central cavities) at twice the resolution of a confocal microscope. Within the phloem, MP-green fluorescent protein filaments extended outward from the specialized pore-PD that connect sieve elements (SEs) with their companion cells (CCs) along the tubular sieve element reticulum (SER). The SER was shown to interconnect individual pore-PD at the SE-CC interface. 3D-SIM resolved fine (less than 100 nm) endoplasmic reticulum threads running into individual pore-PD as well as strands that crossed sieve plate pores, structurally linking SEs within a file. Our data reveal that MP entering the SE from the CC may remain associated with the SER. Fluorescence recovery after photobleaching experiments revealed that this MP pool is relatively immobile compared with the membrane probe 3,3’-dihexyloxacarbocyanine iodide, suggesting that MP may become sequestered by the SER once it has entered the SE. The advent of 3D-SIM offers considerable potential in the subdiffraction imaging of plant cells, bridging an important gap between confocal and electron microscopy.
Fluorescence-based imaging has revolutionized cell biology, allowing the localization of proteins to specific cells and organelles (Shaner et al., 2007; Frigault et al., 2009). However, conventional fluorescence microscopy is limited by the diffraction of light to approximately 200 nm in the lateral (x-y) plane and to about 500 nm in the axial (z) plane (Fernandez-Suarez and Ting, 2008; Huang et al., 2009). This is because light traveling through a lens cannot be focused to a point, only to an airy disc with a diameter of about half the wavelength of the visible emitted light (Huang et al., 2009). Confocal laser scanning microscopy has produced improvements in axial resolution due to the removal of out-of-focus flare, but it is also limited by diffraction (Huang et al., 2009). Thus, objects closer than about 200 nm cannot be resolved but appear merged into one. Many subcellular structures of interest to cell biologists lie below this resolution limit and have remained below the diffraction barrier. Such structures can be seen but not resolved.
Recently, major innovations in biological imaging have broken the diffraction barrier. These include photoactivation localization microscopy (PALM) and stimulated emission and depletion (STED; for review, see Fernandez-Suarez and Ting, 2008; Huang et al., 2009). Most subdiffraction or “super-resolution” approaches have improved resolution in either the lateral (x-y) plane or the axial (z) plane, but usually not both (Schermelleh et al., 2008). Many of the structures of interest within plant cells lie some distance from the cell wall, posing problems for some super-resolution approaches (e.g. PALM) where the subject of interest must lie close to the coverslip (Huang et al., 2009). Recently, Schermelleh et al. (2008) described a subdiffraction multicolor imaging protocol using three-dimensional structured illumination microscopy (3D-SIM). In this method, objects beyond the diffraction limit are illuminated with multiple interfering beams of light transmitted through a series of diffraction gratings, producing a resolution of 100 nm in x-y and 200 nm in z (Schermelleh et al., 2008; Huang et al., 2009). These substantial increases in resolution are significant for plant cell imaging. The thickness of the plant cell wall is typically in the region of about 700 nm, allowing limited optical sectioning capacity with a confocal microscope (about 500 nm in z). A further advantage of 3D-SIM is that it permits the imaging of conventional fluorescent reporters and dyes that are compatible with confocal imaging, allowing a direct correlation of 3D-SIM and confocal images (Schermelleh et al., 2008).
The phloem of higher plants is a major conduit for the long-distance transport of solutes (Oparka and Turgeon, 1999) and also functions as a “superhighway” for macromolecular trafficking (Lucas and Lee, 2004; Kehr and Buhtz, 2008; Lee and Cui, 2009). However, the phloem is difficult to image with conventional optical microscopy (Knoblauch and van Bel, 1998; Oparka and Turgeon, 1999; van Bel et al., 2002). Sieve elements (SEs), the conducting cells of the phloem, are enucleate yet contain a plethora of proteins and RNAs associated with long-distance signaling and defense (van Bel and Gaupels, 2004; Lee and Cui, 2009). Many of these macromolecules are synthesized in the companion cell (CC) and passed into the SE via the specialized pore-plasmodesmata (PD) that connect the two cell types (Oparka and Turgeon, 1999; van Bel et al., 2002). Pore-PD have been suggested to be a major “lifeline” from CC to SE (van Bel et al., 2002), but the exact nature of this pathway remains unresolved.
Our current understanding of PD substructure is derived largely from electron microscope studies (Roberts, 2005). Such methods are time-consuming and do not permit facile protein localization within PD. Recent proteomics approaches have been successful in identifying new proteins associated with PD (Maule, 2008). Localization of these proteins with confocal microscopy results in the appearance of discrete punctae at the cell wall, consistent with the location of pit fields (Faulkner et al., 2008), but does not pinpoint specific protein locations within PD. In general, there is a growing gap between proteomics studies of plant organelles, including PD, and the ability to ascribe accurate addresses to these proteins (Millar et al., 2009; Moore and Murphy, 2009). The advent of 3D-SIM prompted us to explore the potential of subdiffraction imaging in plant cells, with a view to obtaining improved florescence resolution of PD. We used 3D-SIM to examine PD in a transgenic tobacco (Nicotiana tabacum) line expressing the viral movement protein (MP) of Tobacco mosaic virus (TMV) fused to GFP. Using a specific antibody to callose, a wall constituent located at the PD collar, we were able to resolve clearly the structure of single, simple PD in epidermal cells at 100-nm resolution, discriminating between the neck region of the pore and the central cavity to which it connects (Roberts and Oparka, 2003; Faulkner et al., 2008). 3D-SIM also revealed details of the central cavities of complex PD seen previously only with the electron microscope (Ding et al., 1992; Ehlers and Kollmann, 2001; Faulkner et al., 2008).
Using 3D-SIM, we were able to image PD sequentially from the epidermis to the phloem within vascular bundles, producing unparalleled images of sieve plate pores and the specialized pore-PD that connect SEs with their CCs. In the SEs, MP was no longer restricted to the central cavities of PD but became distributed along the SE parietal layer, connecting all the pore-PD along the SE-CC interface. We were able to detect fine threads of MP-GFP that extended for up to 40 μm along the SE and also crossed individual sieve plate pores. Fluorescence recovery after photobleaching (FRAP) experiments revealed that this MP-GFP pool was relatively immobile within the SE parietal layer, suggesting that the SE may sequester TMV MP on or within the sieve element reticulum (SER).
Our data reveal that 3D-SIM is especially suited to the subdiffraction imaging of plant cells and yields spatial information not previously possible with conventional fluorescence-based imaging. The unique optical sectioning capacity of 3D-SIM and the ability to produce multicolor imaging with conventional fluorophores offer enormous potential in plant cell biology.
RESULTS
Simple PD
To obtain comparisons of PD using different imaging techniques, we first examined PD using confocal microscopy and subsequently with 3D-SIM. We were unable to locate directly the same PD pores with both techniques but were able to collect images from the same regions of tissue for comparison. In the 3D-SIM microscope, we were able to collect conventional wide-field images in parallel with the 3D-SIM images, providing a direct resolution comparison of the same PD at the same optical depth within the tissue.
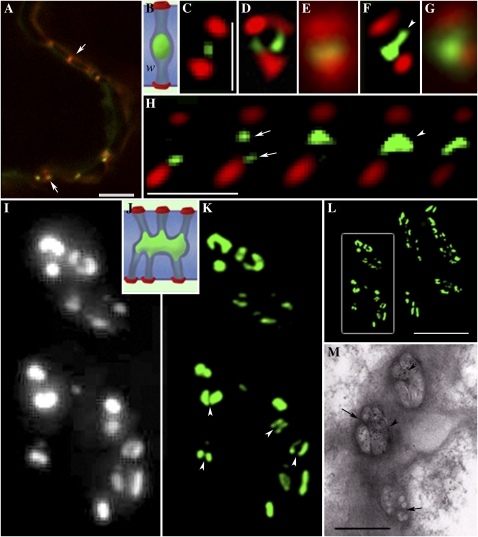
In the leaf epidermis, MP-labeled PD showed the characteristic punctae in the cell wall described previously using confocal imaging (Roberts et al., 2001; Faulkner et al., 2008). When leaf tissue was labeled with callose antibody, we obtained strong signals from both GFP (488 nm) and callose (Alexa 594; Fig. 1A), and these signals overlapped to produce characteristic yellow punctae within the cell wall. However, we were unable to resolve the signals clearly or to identify individual PD pores. Comparisons of the same PD viewed by conventional wide-field fluorescence imaging and 3D-SIM are shown in Figure 1 (C–G). Wide-field imaging revealed a significant overlap of the callose/GFP signals in the cell wall (Fig. 1, E and G). However, using 3D-SIM, we were able to resolve individual simple PD and to separate the GFP and Alexa 594 signals clearly (Fig. 1, C, D, and F). Measurements of individual PD estimated the callose signal to extend laterally in the cell wall for about 187 ± 6 nm (n = 32), slightly larger than the dimension of the raised wall collar that surrounds the PD pore (100 nm; Faulkner et al., 2008). While the dimensions of the callose signal were consistent for each simple pore, those of the central cavities varied considerably. In many pores, the central cavity appeared spherical (Fig. 1C), while in others, it extended laterally in the region of the middle lamella (Fig. 1F). Significantly, 3D-SIM revealed an unlabeled region of about 136 ± 6 nm (n = 15) between the central cavity and the callose collars. This region lacked viral MP and most likely corresponds to the region of pore that connects the PD entrance to the underlying cavity (Fig. 1B). In many cases, we observed two callose collars in one cell face and a single collar in the adjoining cell face (Fig. 1D), consistent with the images of Y-shaped PD often reported in transmission electron microscopy (TEM) studies (Faulkner et al., 2008). Using optical sectioning, we were able to image successive longitudinal “slices” of single PD within the cell wall, locating the two distinct pores that connect to the same central cavity (Fig. 1H).
Figure 1.
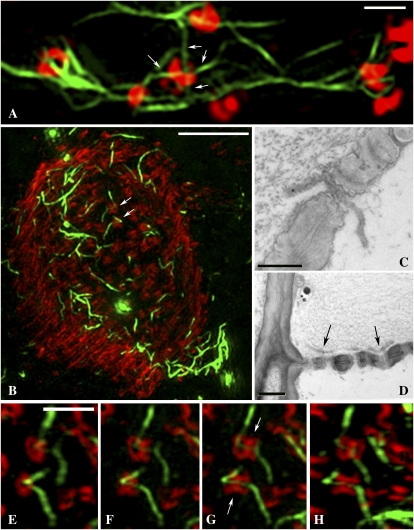
Simple and branched PD viewed with confocal, wide-field, and 3D-SIM imaging. A, Conventional confocal image showing PD containing MP-GFP (green) labeled with callose antibody (Alexa 594; red). Overlapping signals appear yellow (arrows), but individual PD are not resolved. Bar = 5 μm. B to G, 3D-SIM images of simple PD. B, Diagram of a single PD pore, showing callose collars (red) separated from a single central cavity (green). C to F, 3D-SIM spatially resolves the callose collars of individual PD from the central cavity. The corresponding wide-field images of the same pores pictured in D and F are shown in E and G, respectively. A Y-shaped PD configuration is shown in D. An extension of the central cavity is seen in F (arrowhead). W, Cell wall. Bars = 1 μm. H, A z series taken using 3D-SIM of a single PD pore. The individual images are 125 nm apart. Note that two pores can be seen (arrows), each leading to a shared central cavity (arrowhead). Bar = 1 μm. I to M, Comparative images of PD central cavities obtained with wide-field illumination (I), 3D-SIM (K and L), and EM (M). L shows a z stack of an entire pit field, with the boxed region, enlarged in K, revealing fine wall partitions (arrowheads) that separate individual central cavities. Similar partitions are shown in the EM image in M (arrows), in which the viral MP within central cavities is immunolabeled with gold (arrowheads). J shows a diagrammatic representation of a branched PD pore. Bars = 5 μm (L) and 1 μm (M).
Branched PD
While the PD that connect epidermal cells are predominantly simple in architecture, those that connect the epidermis-mesophyll and mesophyll-mesophyll boundaries are branched and arranged in discrete pit fields (Fig. 1, I–L). Using wide-field imaging of the epidermis-mesophyll interface, we could detect characteristic pit fields within which MP-GFP was detected in discrete punctae (Fig. 1I; Faulkner et al., 2008). Imaging these pit fields with 3D-SIM revealed a remarkable complexity in central cavity structure that could not be resolved with wide-field imaging (Fig. 1K) and showed that many of the punctae seen with wide-field imaging could be resolved into two or more closely associated central cavities (Fig. 1K, arrowheads). Some of these central cavities extended for up to 600 nm within the cell wall (Fig. 1K; the complete pit field is shown in Fig. 1L). Many had a horseshoe-shaped cross-section (Fig. 1K), also seen in glancing sections of the wall using comparative EM (Fig. 1M; for interpretation, see Fig. 1J). In the EM, such cavities are separated by narrow wall partitions (Fig. 1M; Faulkner et al., 2008) that could also be resolved with 3D-SIM (Fig. 1K). We were also able to image the complex PD that connect the epidermis with trichomes (Faulkner et al., 2008). By imaging the basal cell wall of the trichome in face view, we were able to optically section in z through all PD at a single wall interface (Fig. 2A). Figure 2B shows a z series, each section 125 nm apart, of an individual complex PD at the trichome-epidermis interface. Note that the channels leading from individual PD pores at the surface become progressively fused in the region of the middle lamella. Following pectinase treatment, we were able to image PD in elongated parenchyma cells surrounding vascular bundles. Here, the central cavities became increasingly elaborate, and optical sections of the cell wall revealed many closely adjacent cavities that had fused to form one extended cavity in the middle lamella region of wall (Fig. 2, C–E). The callose signals revealed that these cavities were shared by numerous overlying PD pores. These central cavities extended for over 1 μm in the middle lamella region of wall (Fig. 2, D and E).
Figure 2.
3D-SIM images of branched PD. A, A z stack of all the branched PD (MP-GFP) at the trichome-epidermis interface. A single branched pore is shown (arrow). Bar = 5 μm. B, Single images (125 nm apart) of a single branched PD. Note that six branches can be detected entering from the trichome and only three entering from the epidermis. Single pores entering from the epidermis and trichome become fused in the middle lamella region of the wall. Bar = 1 μm. C to E, 3D-SIM images of pit fields in a phloem parenchyma cell. Callose is labeled with Alexa 594 (red; C), and central cavities are labeled with MP-GFP (D). Note that the central cavities extend for over 1 μm from the collars that lead into them (arrow; merged image in E). Bar = 1 μm.
Phloem PD
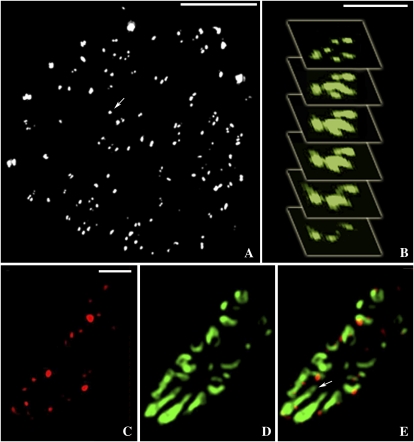
In pectinase-treated tissues, we were able to separate out phloem strands from surrounding tissues (van Bel and Koops, 1985) and to examine the PD that connect different phloem cell types. As above, we compared wide-field fluorescence and 3D-SIM images of the same cells using the same oil-immersion objective. Callose labeling of SEs with Alexa 594 revealed an exceptionally strong signal from sieve plates (Fig. 3). Using conventional imaging, we could detect small red spots on the sieve plates, consistent with the location of callose around the sieve plate pores, but were unable to resolve clearly the individual pores (Fig. 3A). By contrast, 3D-SIM resolved the individual sieve plate pores clearly (Fig. 3B). The average diameter of the pores was 276 ± 16 nm (n = 21), and the callose collars extended this diameter to 568 ± 22 nm (n = 21).
Figure 3.
3D-SIM images of SEs. A, A sieve plate (SP) imaged using confocal microscopy (callose, red; cellulose, blue). Sieve plate pores are seen as red dots but cannot be resolved. Bar = 5 μm. B, Comparative 3D-SIM image. Note that the individual sieve plate pores, and the callose collars lining them, are resolved clearly. Bar = 5 μm. C, SE-CC complex. The image is a stack of 67 images taken with 3D-SIM. Callose (red) is associated with sieve plate pores and with pore-PD (arrows). Bar = 5 μm. D, SE showing lateral sieve area pores (arrows) near the sieve plate. Bar = 5 μm.
Using 3D-SIM, we were also able to generate optical stacks through entire SE-CC complexes. For example, Figure 3C shows an optical stack of 67 separate sections, each 125 nm apart. The rotation of this image can be viewed in Supplemental Movie S1. Using optical stacking, we were also able to resolve the individual lateral sieve area pores connecting adjacent SEs (Fig. 3D; Supplemental Movie S2) and the pore-PD that connect SEs and CCs at their shared interface (Figs. 3C and 4, C–E).
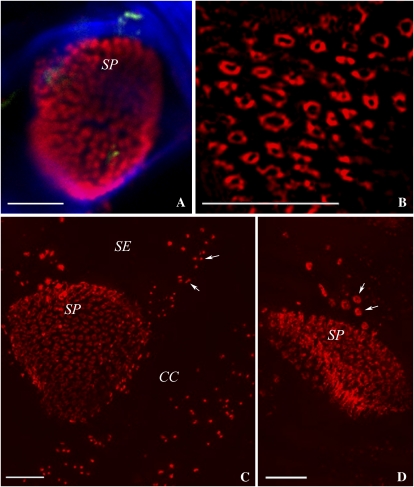
Figure 4.
Viral MP decorates the SER. A, Confocal image showing MP-GFP associated with the SE-CC interface (arrows). Bar = 5 μm. B, 3D-SIM image of the surface of the SE parietal layer revealing strands of MP-decorated SER (green) associated with pore-PD (red). Bar = 5 μm. C to E, 3D-SIM images (125 nm apart) showing individual strands of MP-decorated SER associated with pore-PD. Note that the callose collars of the pore-PD are resolved (arrowheads). Bar = 5 μm. F, The corresponding wide-field image to E fails to resolve the structure of the pore-PD.
Fine Strands of MP Interconnect Pore-PD at the SE-CC Interface
In confocal images of the phloem, we noticed that MP-GFP was no longer located exclusively in the central cavities of PD but extended longitudinally within the SEs (Fig. 4A). Using 3D-SIM, we could resolve MP-decorated strands of about 144 ± 6 nm (n = 30) diameter that could be traced for up to 40 μm along the SE parietal layer, specifically at the SE-CC junction, and noticed that these strands connected individual pore-PD (Fig. 4B). In surface views of the SE-CC interface, we were able to resolve the callose collars of the individual pore-PD leading into the SE (Figs. 4, C–E, and 5A). Measurements showed the pores to have a diameter of 188 ± 8 nm (n = 26), significantly smaller than the sieve plate pores. The callose collars extended the pore diameter to 510 ± 19 nm (n = 26). The intricate association of the MP strands with the pores is shown in Fig. 4 (C–E) and in Supplemental Movie S3. Optical sectioning of the SE-CC interface revealed that the strands were linked into a network, often running into the center or around the perimeters, of the pore-PD (Figs. 4, C–E, and 5A). Such fine strands could not be resolved with wide-field fluorescence imaging (Fig. 4F).
Figure 5.
High-magnification views of pore-PD and sieve plate pores. A, High-magnification view of the pore-PD shown in Figure 4E. Four individual SER strands (less than 100 nm diameter; arrows) can be seen to enter a single pore. Bar = 1 μm. B, A z stack of a sieve plate (callose, red) showing fine strands of MP-decorated SER (green) traversing the sieve plate pores (arrows). Bar = 5 μm. C and D, EM images of petiole phloem fixed using the same protocol as for 3D-SIM. C depicts a single ER profile traversing a sieve plate pore, and D shows ER elements (arrows) lying adjacent to a sieve plate. Bars = 0.5 μm (C) and 1 μm (D). E to H, Details of the sieve plate pores shown in B. The images are from a rotated z series of the sieve plate. Note that the SER strands (arrows in G) can be traced from one SE to another through the pores. Bar = 1 μm.
In many SEs, we observed up to four fine strands extending from each pore (Fig. 5A). The average diameter of these strands was 89 ± 6 nm (n = 6), at the limit of resolution of 3D-SIM. However, we were unable to detect equivalent strands of MP in the adjoining CC (data not shown). The SER is composed of an elaborate system of tubular elements and stacked cisternae that, together with the plasma membrane, mitochondria, and plastids, form the SE parietal layer (Evert, 1990; van Bel et al., 2002). The cytoskeleton is absent from enucleate SEs (Evert, 1990), making the tubular SER a likely candidate for MP targeting. To confirm this, we stained SEs with the red vital ER stain, hexyl rhodamine B (Fig. 6, A and B). As this dye photobleaches rapidly and has an emission of 574 nm, we were unable to use the 594-nm line of the 3D-SIM microscope. Using confocal microscopy, we found that MP colocalized with the tubular component of the SER (Fig. 6, A and B) but not with the stacked parietal cisternae. Also, while MP labeled the ER at the SE-CC interface only (Fig. 6B), hexyl rhodamine B stained the entire SER (data not shown). To obtain comparative TEM images, tissue was postfixed in osmium and embedded in resin. The TEM images showed good preservation of SEs, as judged by a lack of plasmolysis and the intact appearance of SE plastids, organelles acutely sensitive to SE turgor loss (Supplemental Fig. S1, A and B; Barclay et al., 1976; Ehlers et al., 2000). Using the MP-GFP tobacco line, we were able to image the SER using TEM for comparison with the 3D-SIM images. In glancing sections of the SE wall, the SER was visible as a fine, tubular network identical to that seen with 3D-SIM (Supplemental Fig. S1, B and C). Measurements of the tubular SER revealed an average diameter of 77 ± 3 nm (n = 11).
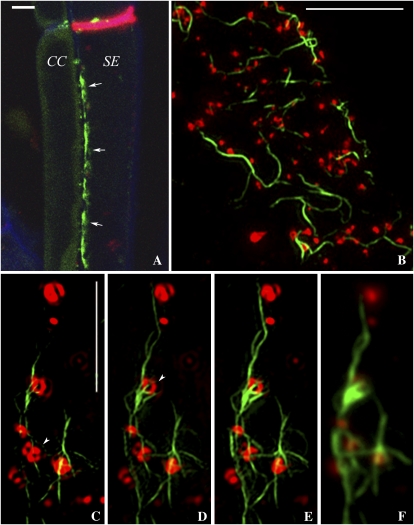
Figure 6.
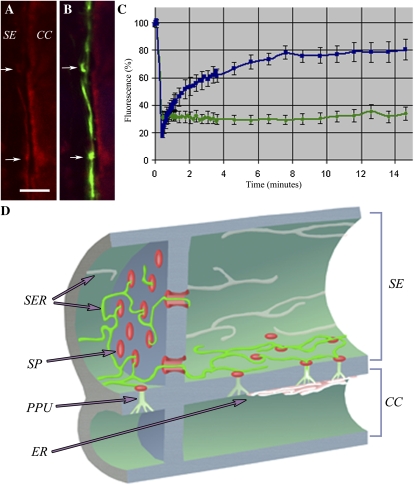
A and B, Confocal images of the SE-CC interface stained with the red ER-specific dye, hexyl rhodamine B (arrows; A). The corresponding MP-GFP image (B) shows that MP decorates the SER specifically within the SE (arrows). Bar = 5 μm. C, FRAP data in which the GFP-labeled SER was photobleached with high-intensity illumination at 488 nm. The recovery profiles for DiOC6 (blue) and GFP (green) are shown for the same bleach conditions. D, Diagrammatic representation of the relationship of the SER with sieve plate pores (SP) and pore-PD (PPU) as revealed by 3D-SIM. MP-GFP (green) highlights the SER at the SE-CC interface only and labels the SER strands that connect individual pore-PD. The SER forms a continuum between SE and CC. Callose associated with pore-PD and sieve plate pores is shown in red.
100-nm Strands of ER Traverse Sieve Plate Pores
Next, we examined the surfaces of sieve plates connecting individual SEs. Here, we also found tubular elements of MP-decorated SER (Fig. 5B). In optical stacks of sieve plates, the sieve plate pores had the appearance of “cotton reels” due to the callose cylinders lining them (Fig. 5, B and E–H; Supplemental Movie S4). In many of these pores, we noticed that the tubular SER became thinned down into fine strands of about 103 ± 3 nm (n = 30) in diameter that appeared to traverse the sieve plates (Fig. 5B). To examine this further, we rotated the optical stack to obtain 3D images of single sieve plate pores. This confirmed that the fine strands were running longitudinally across individual pores from one SE to the next (Fig. 5, E–H). By TEM, we were able to detect similar ER strands of about 80 nm traversing sieve plate pores (Fig. 5C) and lying against sieve plates (Fig. 5D).
FRAP of Enucleate SEs
To determine if the MP-GFP located on the SER was mobile, we imaged phloem strands from intact petioles and photobleached the SER using high-intensity illumination at 488 nm with the confocal microscope. MP-GFP showed little recovery into the photobleached area (Fig. 6C). To determine if tissue isolation had disrupted the SER, we also performed FRAP on SEs stained with 3,3’-dihexyloxacarbocyanine iodide (DiOC6), a membrane potential probe that selectively stains the ER (Martens et al., 2006). In contrast to MP-GFP, DiOC6 showed a rapid recovery into the photobleached area (Fig. 6C), indicating that the SER was continuous between the bleached and unbleached regions of the SE.
DISCUSSION
Despite a historical acceptance that resolution is limited by diffraction (for review, see Huang et al., 2009), there has been a rapid expansion of imaging techniques that break the diffraction barrier. Many of these have achieved improved resolution in either the x-y or z axes, but 3D-SIM is currently the only method that achieves 2-fold increases in resolution in both dimensions (Schermelleh et al., 2008). The relative ease of 3D-SIM and its improved capacity for optical z stacking over confocal microscopy made it an ideal choice for imaging PD, structures below the limit of resolution of conventional fluorescence imaging. At present, 3D-SIM, in common with many of the other super-resolution approaches, does not achieve live-cell imaging and is restricted to samples that must remain static during imaging. TEM remains the highest resolution technique for imaging plant cells, although the major advantages of 3D-SIM over TEM are the ability to localize a wide range of proteins rapidly using conventional fluorophores and its ability to rapidly produce 3D information. Unlike confocal imaging, 3D-SIM cannot yet be used to image protein dynamics using photoactivatable probes, although recent reports suggest that super-resolution techniques will enter the domain of live-cell imaging in the near future (Huang et al., 2009).
Although the membrane-lined pore of PD is about 50 nm (Roberts, 2005), the width of the raised wall collars extends the structural diameter of tobacco PD to about 100 nm (Faulkner et al., 2008), within the resolution limit of 3D-SIM (Schermelleh et al., 2008). We have shown that different components of PD (collar and central cavity) can be spatially resolved with accuracy. Within the phloem, a tissue recalcitrant to conventional imaging, 3D-SIM resolved sieve plate pores, lateral sieve areas, and the pore-PD that connect CC and SE. In the last of these, we were able to image individual strands of ER entering the pore from the SE side of the cell wall. Recently, Truernit et al. (2008) described a clearing method for measuring a range of phloem parameters in three dimensions using confocal imaging. This method is still limited by diffraction, but used in combination with 3D-SIM it may offer an approach to studying phloem architecture at super-resolution.
Previously, we showed that the MP of Cucumber mosaic virus (CMV) entered SEs during virus infection, and we noted a fibrillar structure within SEs that was below the limit of resolution (Blackman et al., 1998). We show here that this structure is likely to be the tubular SER and that the TMV MP also enters SEs via PD to become distributed throughout the SER (shown diagrammatically in Fig. 6D). Although MP-GFP must enter the enucleate SE from the CC (Oparka and Turgeon, 1999), our FRAP data suggest that MP is relatively immobile in the SE parietal layer compared with the ER membrane probe (DiOC6). TMV MP has been shown previously to accumulate as a fibrillar material in the central cavities of PD, possibly within the extended desmotubule lumen (Ding et al., 1992). However, we have been unable to determine the exact location (membrane or lumen) of MP-GFP that decorates the SER. The lack of mobility of MP in the SER may reflect the fact that it is sequestered on this unique structure. Electron micrographs depicting SER in surface views are extremely rare, but those that do (Esau, 1978; Warmbrodt, 1986) reveal an intricate strand-like network identical to that reported here using 3D-SIM (Supplemental Fig. S1C). This fine SER network is quite unlike that of nucleate parenchyma cells (Brandizzi et al., 2002; Runions et al., 2006). The SER may be involved in a range of functions (van Bel et al., 2002) and has recently been suggested to be a Ca2+ reservoir essential for the maintenance of long-distance electrical signals in the phloem (Furch et al., 2007). Only a limited number of organelles remain in the mature SE (Evert, 1990; Oparka and Turgeon, 1999; van Bel et al., 2002), but despite the selective autolysis of the cell, the SE parietal layer contains several biosynthetic enzymes (Bird et al., 2003; Wang et al., 2003), transporters (Meyer et al., 2004), protein signals (Khan et al., 2007), and transcripts (for review, see Lee and Cui, 2009). These raw materials are supplied by the CC and passed selectively into the SE (Oparka and Turgeon, 1999; van Bel et al., 2002). To achieve this feat, the SE must anchor these components in the face of mass flow occurring through the sieve tube lumen (Sjolund, 1997; Ehlers et al., 2000).
Many viral ribonucleoprotein complexes enter SEs for long-distance transport and exit the phloem at distant sites in the plant (Kehr and Buhtz, 2008; Lee and Cui, 2009). We showed recently that infectious TMV RNA becomes anchored to the cortical ER within seconds of its injection into a single trichome cell (Christensen et al., 2009). The TMV MP remodels the cortical ER during infection (Heinlein et al., 1998; Gillespie et al., 2002) and transfers the viral genome through PD (Guenoune-Gelbart et al., 2008). The continuous ER network through PD in the form of the central desmotubule (Roberts, 2005), as well as the maintained continuity of ER within and between SEs (Evert, 1990), suggest that some viruses may require to locate the ER only once during infection, subsequently depending on the cortical ER to “hitch a ride” to the SE (Christensen et al., 2009). At present, we have been unable to determine why MP is restricted to the SE-CC interface, as the SER encircles the entire SE parietal layer (Evert, 1990; Martens et al., 2006). The radial distances for MP diffusion on the SER within the SE are small compared with the axial dimensions, so there appears to be no a priori reason why MP should not associate with the entire SER. One possibility is that a functionally unique form of SER occurs at the SE-CC interface, making it possible for selective protein transport to occur between these two cell types.
Previously, we showed using FRAP that ER coupling between CCs and SEs is extensive compared with other cell types (Martens et al., 2006). It has been suggested that the TMV MP may also diffuse between infected and noninfected cells via the membrane of the desmotubule (Guenoune-Gelbart et al., 2008). Our data here suggest that small membrane-bound molecules (e.g. DiOC6), in addition to moving between CC and SE, may also diffuse along the SER, although the lateral diffusion of the TMV MP appears to be restricted. During natural TMV infection, it is thus possible that the SER represents the “end of the line” for viral MP once invasion of the SE has occurred. The appearance of MP on the SER begs the question of why MP is not normally retained on the ER of nonvascular parenchyma cells during TMV infection. During early TMV infection, MP decorates the ER but is subsequently removed by the host cell, later appearing on microtubules (Gillespie et al., 2002). The most likely explanation for the removal of MP from the ER is that nucleate parenchyma cells have a cytoskeleton-based degradation pathway (Gillespie et al., 2002; Reichel et al., 2006) that is absent from the enucleate SEs.
Over 40 years ago, using conventional light microscopy, Thaine and colleagues (Thaine, 1961; Thaine et al., 1967) described fine “transcellular strands” in enucleate SEs and suggested that these were involved in phloem translocation. These membranous strands were later dismissed as diffraction artifacts (Esau, 1963) or bundles of P-protein filaments created by wounding (Johnson et al., 1976). Thaine and colleagues would have been unable to resolve the fine strands that we observed here using subdiffraction imaging. However, their images may have represented larger bundles of SER strands running longitudinally along the SE-CC interface.
Using 3D-SIM, we have been able to resolve several structures seen previously only with the electron microscope. Significantly, we were able to use standard cytological methods and fluorophores compatible with confocal imaging. The fields of cell biology and molecular plant pathology are likely to benefit enormously from the application of 3D-SIM.
MATERIALS AND METHODS
Plant Material
Tobacco (Nicotiana tabacum ‘Xanthi’) plants expressing TMV MP-GFP (Roberts et al., 2001) and cv Samsun nn plants were grown from seed in a heated glasshouse and used for experiments between 30 and 55 d old.
Confocal Microscopy
For imaging GFP, longitudinal sections of petiole were prepared using a Vibroslice Tissue Slicer HA752 (Camden Instruments). Petioles were embedded in 5% Phytoagar (Duchefa Biochemie) to stabilize before cutting into approximately 100-μm sections. The excess Phytoagar was trimmed before the tissue was briefly counterstained with aniline blue to visualize callose at sieve plates. The tissue was mounted in water and viewed through a coverslip.
To image the ER in SEs, tissue sections were prepared as above and then stained with hexyl rhodamine B (Invitrogen) at 0.1 μg mL−1.
The tissues were imaged using a Leica SP2 confocal laser scanning microscope (Leica Microsystems) with a ×63 water-immersion lens (HCX PLAPO CS; Leica Microsystems). Aniline blue was excited using a 405-nm laser, GFP with a 488-nm laser, and hexyl rhodamine B with 561-nm laser source.
Immunolocalization
Longitudinal petiole tissue sections were prepared as above and then fixed in 4% paraformaldehyde/0.25% glutaraldehyde in PME buffer (50 mm PIPES, 2 mm EGTA, and 2 mm MgSO4, pH 6.8). The tissue was washed with PME before the vasculature was separated following a 15-min pectinase treatment at 37°C: pectinase (2%; Invitrogen) in 1% (w/v) phosphate-buffered saline (PBS) with 1% (w/v) bovine serum albumin and 5 mm EDTA. The sections were then rinsed in PME with 10% (v/v) glycerol and 0.02% (v/v) Triton X-100 before washing further with 1% (w/v) PBS. Nonspecific binding sites were blocked using 3% (w/v) PBS and 50 mm Gly in 1% (w/v) PBS for 20 min. The tissue was then incubated overnight in mouse anti-callose (Biosupplies) antibody at a 1:400 dilution. Sections were rinsed briefly in the blocking solution before incubating with secondary antibody, anti-mouse Alexa 594 (Invitrogen) diluted 1:500, for 3 h. All antibodies were diluted in 1% (w/v) bovine serum albumin, 0.02% (v/v) Tween in 1% (w/v) PBS and incubated at 37°C. The petiole sections were mounted in Citifluor AF1 antifade agent (Agar Scientific) under a coverslip and then sealed with nail varnish. Tissues were then imaged with a Leica SP2 confocal scanning microscope, as described above using a 591-nm laser source for the Alexa 594, or with 3D-SIM as described below.
TEM
For TEM, tissue was fixed as described above in 3% glutaraldehyde in 0.1 m sodium cacodylate buffer followed by postfixation in 1% osmium tetroxide in 0.1 m sodium cacodylate buffer prior to embedding in Araldite (Oparka and Turgeon, 1999). EM immunolocalization of viral MP within PD was performed exactly as described by Faulkner et al. (2008).
Structured Illumination
The protocol applied was based on that described by Schermelleh et al. (2008). Images were acquired using a UPlanSApochromat 100×, numerical aperture 1.4, oil-immersion objective lens (Olympus) and back-illuminated Cascade II 512 × 512 EMCCD camera (Photometrics) on the OMX version 2 system (Applied Precision) equipped with 405, 488, and 593 solid-state lasers.
Samples were illuminated by a coherent scrambled laser light source that had passed through a diffraction grating to generate the structured illumination by interference of light orders in the image plane to create a 3D sinusoidal pattern, with lateral stripes approximately 0.2 μm apart. The pattern was shifted laterally through five phases and through three angular rotations of 60′ for each z section, separated by 0.125 μm.
Exposure times were typically between 50 and 250 ms, and the power of each laser was adjusted to achieve optimal intensities of between 2,000 and 4,000 counts in a raw image of 16-bit dynamic range, at the lowest possible laser power to minimize photobleaching. Where required, the cameras were run in EMCCD mode and gain was applied. Each frame acquisition was separated by a 300-ms pause.
Image Processing
Raw images were processed and reconstructed to reveal structures with greater resolution (Gustafsson et al., 2008). The channels were then aligned in x, y, and rotationally using predetermined shifts as measured using a target lens and 100-nm Tetraspeck fluorescent microspheres (Invitrogen) in the Softworx alignment tool (Applied Precision).
FRAP
Large petiole pieces were immersed in PBS and cut into fine longitudinal sections with a scalpel. Sections from MP-GFP plants were incubated in 0.25 mg mL−1 aniline blue solution, while sections from wild-type tobacco (cv Samsun nn) plants were incubated in a 0.25 mg mL−1 aniline blue/2 μg mL−1 DiOC6 solution for 10 min. Sections were then immersed in clean PBS and washed for 5 min with agitation before being mounted on slides with coverslips.
FRAP was performed using the Leica Confocal Suite FRAP Wizard. Five prebleach images were collected with a 488-nm laser at 20% intensity (×63 water objective, unidirectional scan, zoom ×3, resolution 512 × 512), and then a 10-μm region was specifically bleached using 10 scans with the 488-nm laser at 100% intensity. Postbleach images were then collected using the same settings as the prebleach images for approximately 15 min in total. Scans were taken every 1 s for the first 30 s, then every 10 s for the next 150 s, and then every 1 min for the subsequent 12 min.
The background fluorescence was subtracted from all values, and each series was corrected for bleaching during the postbleach acquisition phase using an area of fluorescence outside of the cell containing the region of interest for reference. Fluorescence readings were then normalized to percentage values of the prebleach fluorescence, and the normalized measurements were averaged.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. SEs and CCs.
Supplemental Movie S1. Movie showing the rotation of a single SE (from Fig. 3C).
Supplemental Movie S2. Movie showing the rotation of two adjacent sieve plates (one shown in Fig. 3D).
Supplemental Movie S3. Movie of Figure 4B, showing the MP-decorated SER and its relationship with pore-PD
Supplemental Movie S4. Movie of Figure 5E, showing the rotation of sieve plate pores.
Supplementary Material
Acknowledgments
We are grateful to Danae Paultre for technical assistance and to the Scottish Universities Life Sciences Alliance and the Light Microscopy Facility, University of Dundee, for access to the 3D-SIM microscope.
References
- Barclay G, Oparka KJ, Johnson RPC. (1976) Induced disruption of sieve-element plastids in Heracleum mantegazzianum L. J Exp Bot 28: 709–717 [Google Scholar]
- Bird DA, Franceschi VR, Facchini PJ. (2003) A tale of three cell types: alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15: 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman LM, Boevink P, Santa Cruz S, Palukaitis P, Oparka KJ. (1998) The movement protein of cucumber mosaic virus traffics into sieve elements in minor veins of Nicotiana clevelandii. Plant Cell 10: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. (2002) Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen NM, Faulkner C, Oparka KJ. (2009) Evidence for unidirectional flow through plasmodesmata. Plant Physiol 150: 96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Haudenshield JS, Hull RJ, Wolf S, Beachy RN, Lucas WJ. (1992) Secondary plasmodesmata are specific sites of localization of the tobacco mosaic virus movement protein in transgenic tobacco plants. Plant Cell 4: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers K, Knoblauch M, van Bel AJE. (2000) Ultrastructural features of well-preserved and injured sieve elements: minute clamps keep the phloem conduits free for mass flow. Protoplasma 214: 80–92 [Google Scholar]
- Ehlers K, Kollmann R. (2001) Primary and secondary plasmodesmata: structure, origin, and functioning. Protoplasma 216: 1–30 [DOI] [PubMed] [Google Scholar]
- Esau K. (1963) Ultrastructure of differentiated cells in higher plants. Am J Bot 50: 495–506 [Google Scholar]
- Esau K. (1978) Developmental features of the primary phloem in Phaseolus vulgaris L. Ann Bot (Lond) 42: 1–13 [Google Scholar]
- Evert RF. (1990) Dicotyledons. In H-D Behnke, RD Sjolund, eds, Sieve Elements: Comparative Structure, Induction and Development. Springer-Verlag, Berlin, pp 103–137 [Google Scholar]
- Faulkner C, Akman OE, Bell K, Jeffree C, Oparka K. (2008) Peeking into pit fields: a multiple twinning model of secondary plasmodesmata formation in tobacco. Plant Cell 20: 1504–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Ting AY. (2008) Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol 9: 929–943 [DOI] [PubMed] [Google Scholar]
- Frigault MM, Lacoste J, Swift JL, Brown CM. (2009) Live-cell microscopy: tips and tools. J Cell Sci 122: 753–767 [DOI] [PubMed] [Google Scholar]
- Furch ACU, Hafke JB, Schulz A, van Bel AJE. (2007) Ca2+-mediated remote control of reversible sieve tube occlusion in Vicia faba. J Exp Bot 58: 2827–2838 [DOI] [PubMed] [Google Scholar]
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ. (2002) Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the cell-to-cell movement of tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenoune-Gelbart D, Elbaum M, Sagi G, Levy A, Epel BL. (2008) Tobacco mosaic virus (TMV) replicase and movement protein function synergistically in facilitating TMV spread by lateral diffusion in the plasmodesmal desmotubule of Nicotiana benthamiana. Mol Plant Microbe Interact 21: 335–345 [DOI] [PubMed] [Google Scholar]
- Gustafsson MG, Shao L, Carlton PM, Wang CJ, Golubovskaya IN, Cande WZ, Agard DA, Sedat JW. (2008) Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J 94: 4957–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel B, Beachy RN. (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang XW. (2009) Super-resolution fluorescence microscopy. Annu Rev Biochem 78: 993–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RPC, Freundlich A, Barclay GF. (1976) Transcellular strands in sieve tubes: what are they? J Exp Bot 27: 1117–1136 [Google Scholar]
- Kehr J, Buhtz A. (2008) Long distance transport and movement of RNA through the phloem. J Exp Bot 59: 85–92 [DOI] [PubMed] [Google Scholar]
- Khan JA, Wang Q, Sjolund RD, Schulz A, Thompson GA. (2007) An early nodulin-like protein accumulates in the sieve element plasma membrane of Arabidopsis. Plant Physiol 143: 1576–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblauch M, van Bel AJE. (1998) Sieve tubes in action. Plant Cell 10: 35–50 [Google Scholar]
- Lee JY, Cui WE. (2009) Non-cell autonomous RNA trafficking and long-distance signaling. J Plant Biol 52: 10–18 [Google Scholar]
- Lucas WJ, Lee JY. (2004) Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol 5: 712–726 [DOI] [PubMed] [Google Scholar]
- Martens HJ, Roberts AG, Oparka KJ, Schulz A. (2006) Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiol 142: 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ. (2008) Plasmodesmata: structure, function and biogenesis. Curr Opin Plant Biol 11: 1–7 [DOI] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N. (2004) Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol 134: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Carrie C, Pogson B, Whelan J. (2009) Exploring the function-location nexus: using multiple lines of evidence in defining the subcellular location of plant proteins. Plant Cell 21: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Murphy A. (2009) Validating the location of fluorescent protein fusions in the endomembrane system. Plant Cell 21: 1632–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oparka KJ, Turgeon R. (1999) Sieve elements and companion cells: traffic control centers of the phloem. Plant Cell 11: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel C, Ruehland C, Reiser COA, Hess J. (2006) Protein-based nanosystems: virus-like-particles in modern vaccine development. Journal of Biomedical Nanotechnology 2: 186–200 [Google Scholar]
- Roberts AG. (2005) Plasmodesmal structure and development. In KJ Oparka, ed, Plasmodesmata: Annual Plant Reviews, Vol 18. Blackwell Publishing, Oxford, pp 1–32 [Google Scholar]
- Roberts AG, Oparka KJ. (2003) Plasmodesmata and the control of symplasmic transport. Plant Cell Environ 26: 103–124 [Google Scholar]
- Roberts IM, Boevink P, Roberts AG, Sauer N, Reichel C, Oparka KJ. (2001) Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma 218: 31–44 [DOI] [PubMed] [Google Scholar]
- Runions J, Brach T, Kuhner S, Hawes C. (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot 57: 43–50 [DOI] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MGL.et al (2008) Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320: 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Patterson GH, Davidson MW. (2007) Advances in fluorescent protein technology. J Cell Sci 120: 4247–4260 [DOI] [PubMed] [Google Scholar]
- Sjolund RD. (1997) The phloem sieve element: a river runs through it. Plant Cell 9: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaine R. (1961) Transcellular strands and particle movement in mature sieve tubes. Nature 192: 772–773 [Google Scholar]
- Thaine R, Probine MC, Dyer PY. (1967) Existence of transcellular strands in mature sieve elements. J Exp Bot 18: 110–127 [Google Scholar]
- Truernit E, Bauby H, Dubreucq B, Grandjean O, Runions J, Barthelemy J, Palauqui JC. (2008) High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bel AJE, Ehlers K, Knoblauch M. (2002) Sieve elements caught in the act. Trends Plant Sci 7: 126–132 [DOI] [PubMed] [Google Scholar]
- van Bel AJE, Gaupels F. (2004) Pathogen-induced resistance and alarm signals in the phloem. Mol Plant Pathol 5: 495–504 [DOI] [PubMed] [Google Scholar]
- van Bel AJE, Koops AJ. (1985) Uptake of [14C]sucrose in isolated minor-vein networks of Commelina benghalensis L. Planta 164: 362–369 [DOI] [PubMed] [Google Scholar]
- Wang W, Hause B, Peumans WJ, Smagghe G, Mackie A, Fraser R, Van Damme EJM. (2003) The Tn antigen-specific lectin from ground ivy is an insecticidal protein with an unusual physiology. Plant Physiol 132: 1322–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmbrodt RD. (1986) Structural aspects of the primary tissues of the Cucurbita pepo L. root with special reference to the phloem. New Phytol 102: 175–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.