Abstract
The ETS transcription factor Fifth Ewing Variant (FEV) mRNA, a homologue of the rodent Pet-1 gene that is exclusively expressed in serotonin-containing neurons and is a critical determinant of serotonin neuronal differentiation and development, was examined in human postmortem brain tissue using in situ hybridization histochemistry. Studies revealed that FEV mRNA is robustly and exclusively expressed in the major serotonin-containing cell groups of the dorsal and median raphe nuclei located in the midbrain and pons of the human brainstem. The localization of transcription factor, FEV, mRNA in serotonin-containing neurons of the human brain raises questions regarding the functional significance of this transcription factor in regulating serotonin-related genes and its potential role in psychiatric illness.
Keywords: dorsal raphe, human, gene expression, midbrain, mood disorders, neuron, postmortem, serotonin, transcription factor
Introduction
The central serotonin (5-HT) system in the brain of mammals consists of discrete neuronal populations located in the raphe nuclei of the midbrain, pons, and caudal medulla (Dahlstrom and Fuxe, 1964). Serotonin neurons of the dorsal and median raphe nuclei provide the major ascending 5-HT axonal projections to the forebrain innervating various cortical regions and numerous subcortical structures (Azmitia and Gannon, 1986; Steinbusch, 1981; Tork, 1990). Given its widespread distribution, the serotonin system has been implicated in a variety of physiological functions as well as mediating various behaviors (Murphy et al., 1998). Aberrant serotonin function has also been implicated in a number of psychiatric disorders such as depression, suicide, impulsive and aggressive behaviors, and anxiety disorders (Davidson et al., 2000; Silverstone, 2004; Stockmeier, 2003; van Heeringen, 2003). Despite the significant role of the serotonin system in a wide variety of biological processes, only recently has information regarding the genetic mechanisms that control the development, specification, and differentiation of serotonin neurons begun to emerge (reviewed in Scott and Deneris (2005)).
Recent studies have identified specific transcription factors involved in controlling the differentiation and development of serotonin neurons. The ETS domain family of transcription factors has been shown to play important roles in the development, differentiation, cell proliferation, and apoptosis (Fyodorov et al., 1998; Lelievre et al., 2001; Sementchenko and Watson, 2000; Wasylyk et al., 1993). Several ETS genes are expressed in the central nervous system and one in particular, Pet-1, has recently been reported to be specifically expressed in serotonin neurons in the rodent brainstem (Hendricks et al., 1999). Pet-1 expression is present at all developmental stages from embryonic to adult brain, it precedes the expression of serotonin and it is restricted to the serotonin-containing neurons of the raphe nuclei in the midbrain, pons, and caudal medulla (Hendricks et al., 1999). Hendricks and colleagues identified conserved Pet-1 binding sites within 2.5 kb of transcription start sites of both the human and mouse 5-HT1A receptor, serotonin transporter (SERT), and tryptophan hydroxylase genes (Hendricks et al., 1999). In a subsequent study, these investigators developed Pet-1 knockout mice that showed a dramatically defective development of the 5-HT system that was associated with increased anxiety and aggressive behaviors in adult mice (Hendricks et al., 2003). Taken together, these findings suggest that Pet-1 is an essential element of a transcriptional program involved in the differentiation of serotonin neurons and of 5-HT-associated behaviors.
A human homolog of Pet-1 is the Fifth Ewing Variant (FEV) gene (Maurer et al., 2003; Peter et al., 1997). The predicted human FEV protein shares ∼96% homology with the predicted mouse Pet-1 protein (Peter et al., 1997). Although a previous RT-PCR study (Maurer et al., 2004) has indicated that FEV mRNA is expressed in the human raphe nuclei, no study to date has determined the cellular and neuroanatomical localization of this gene in the normal human brain. Therefore, the present study was designed to determine the regional distribution and cellular expression of FEV mRNA in normal postmortem human brain, using in situ hybridization histochemistry.
Materials and Methods
Tissue collection and preparation
All procedures in our study were approved by the University of Pittsburgh's Institutional Review Board for Biomedical Research. Human brain specimens were obtained in the course of routine autopsies conducted at the Allegheny County Coroner's Office, Pittsburgh, PA, after obtaining consent from a surviving family member. Seven psychiatrically healthy control subjects were used for this study. All subjects were male with an average age of 56.6 ± 2.65 years and mean postmortem interval of 15.0 ± 2.1 h.
Upon removal of the brain from the cranium, the cerebellum was removed and the brainstem was separated by a transverse cut at the rostral border of the superior colliculi. The brainstems were then cut into midbrain/pons and caudal medulla blocks, immediately frozen in isopentane and stored at −80°C. The frozen midbrain/pontine blocks were sectioned transversely at 20 μm, sections were thaw-mounted onto gelatin-coated microscope slides and every 10th tissue section stained for Nissl substance with cresyl violet. The slides were stored at −80°C until processed. Slide-mounted tissue sections were selected from the dorsal raphe and the median raphe for assay.
Construction of templates for run-off-transcription of cRNA probes
A human bacterial artificial chromosome was used to amplify FEV 3′-untranslated sequences 5′-1409–1832-3′ (Peter et al., 1997). This fragment was subcloned into the EcoR1 and Xho1 sites of Bluescript SK (Stratagene, La Jolla, CA). The resulting plasmid was used to prepare FEV antisense/sense probes using a nested-PCR approach. Two pairs of primers were synthesized; the first or outer pair was gene specific and consisted the following primer sequences: 5′-GCTCCCTCAATCCTTGTCTG-3′ (1455–1474) and 5′-GCAGTTCCTTGGGGAAGAG-3′ (1687–1706), and the second primer (inner pair) contained T7 and SP6 RNA polymerase promoter binding sites (underlined) in addition to gene-specific sequences (T7 5′-CTGTAA TACGACTCACTATAGGGGCTCCCTCAATCCTTGTCTG-3′ and SP6 5′-GGATTTAGGTGACACTATAGAAAGCAGTTCCTTGGGGAAGAG-3′). Amplification conditions with outer primers were composed of the following parameters: an initial 2 min at 94°C followed by 30 s at 94°C, 1 min at 55°C, and 1 min at 72°C. Following this there was a final extension at 72°C for 10 min. Because of the presence of multiple PCR products obtained with the inner primer pair, annealing and elongation steps were combined at a higher temperature. The conditions here are as follows: initial denaturation at 94°C for 5min followed by 1 min at 94°C, and 2 min at 72°C for a total of 35 cycles with a final extension at 72°C for 10 min. Final PCR products were visualized by electrophoresis in 1% TAE agarose gels, and purified by the Qiaquick PCR purification kit (Qiagen, Inc., Valencia, CA). Purified products were quantified and stored at −20°C until further use.
In situ hybridization
In situ hybridization was essentially carried out as previously described (Austin and O'Donnell, 1999) with a few modifications to adapt it to the FEV-specific riboprobes. Briefly, slides were removed from − 80°C freezer and fixed in 2% paraformaldehyde in 0.1 M phosphate-buffered saline (PBS) for 15 min, rinsed once in PBS and followed by treatment in 0.25% acetic anhydride in 0.1 M triethanolamine/0.9% NaCl, pH 8.0 for 10 min. This was followed by a series of 1-min rinses in the following solutions: 2XSSC, 50%, 70%, 95%, and 100% ethanol after which tissue sections were placed in chloroform for an additional 10 min. A final 1-min placement in 95% and 100% ethanol was made and the sections were allowed to dry at room temperature before proceeding to hybridization. Labeled sense and antisense RNA probes were generated using the MAXIscript T7/SP6 in vitro transcription kit (Ambion, Austin, TX) with 35S-CTP following the manufacturers' directions. Briefly, the reaction mix contained 600 ng of purified DNA templates and 0.0625 μCi 35S-CTP including 0.5 mM each of ATP, GTP, and UTP respectively with either T7 or SP6 RNA polymerase. Reactions were incubated at 37°C for 2 h, following which labeled probes were purified using the RNAeasy mini kit from Qiagen, Inc.
Labeled RNA probes were applied at a concentration of 100 ng/ml in hybridization buffer consisting of 50% formamide, 750 mM NaCl, 20 mM PIPES, 10% (wt/vol) dextran sulfate, 5× Denhardt's solution, 50 mM DTT, 0.2% SDS (wt/vol), and 100 mg/ml yeast tRNA. Before hybridization, probe solutions were heated to 72°C for 10 min. For each section ∼100 μl of probe solution was applied, followed by placing a cut glass coverslip to cover the entire section. Tissue sections were incubated for 16–20 h. Immediately after hybridization, slides were placed in 4XSSC (room temperature) containing 1.25% (vol/vol) β-mercaptoethanol for 15 min and coverslips were gently pulled off afterwards. Slides were then washed in 4XSSC 15 min, 50% formamide buffer (0.3 M NaCl, 20 mM Tris, 1 mM EDTA, and 0.08% HCl (vol/vol)) at 68°C 20 min, 2XSSC 5 min, RNase A buffer (500 mM NaCl, 10 mM Tris, and 1 mM EDTA) at 37°C 30 min, 2XSSC 5 min, 1XSSC 5 min, 0.5XSSC 5 min, 0.1XSSC at 68°C 30 min, 0.1XSSC containing 0.5% triton 5 min, followed by three quick dips in 60% ethanol containing 330 mM NH4OAc. After the washes, slides were dried for at least 3 h, after which they were exposed to Kodak Biomax film for 4–7 days depending on the intensity of the signals. The film exposure slides were dipped in photographic emulsion (NTB2, Kodak) for 4 weeks and then developed, fixed and counterstained with hematoxylin and eosin.
Results
The results of the in situ hybridization experiments using 35S-CTP labeled antisense ribonucleotide probes generated from a fragment of the human FEV gene revealed that FEV mRNA is robustly expressed in the major serotonin-containing cell groups located in the midbrain and pons. The dorsal raphe nuclei revealed dense hybridization signal for FEV mRNA at multiple anatomical levels from rostral to caudal (Figs. 1A–1D). The median raphe also exhibited hybridization signal for FEV mRNA (Figs. 1D and 1E). It is evident from the autoradiographs that FEV mRNA is present in the majority of cells in these raphe nuclei. Control experiments to confirm the specificity of the FEV riboprobes showed that tissue sections of the dorsal raphe hybridized with 35S-labeled sense FEV riboprobes exhibited no specific labeling (Fig. 1F). Furthermore, tissue sections of the human hippocampus, amygdala, and prefrontal cortex revealed no specific hybridization signal after hybridization with 35S-labeled antisense FEV riboprobes, nor was there any specific hybridization signal in the adjacent noradrenergic cells of the locus coeruleus in the pontine tissue sections (Figs. 1D and 1E).
Fig. 1.
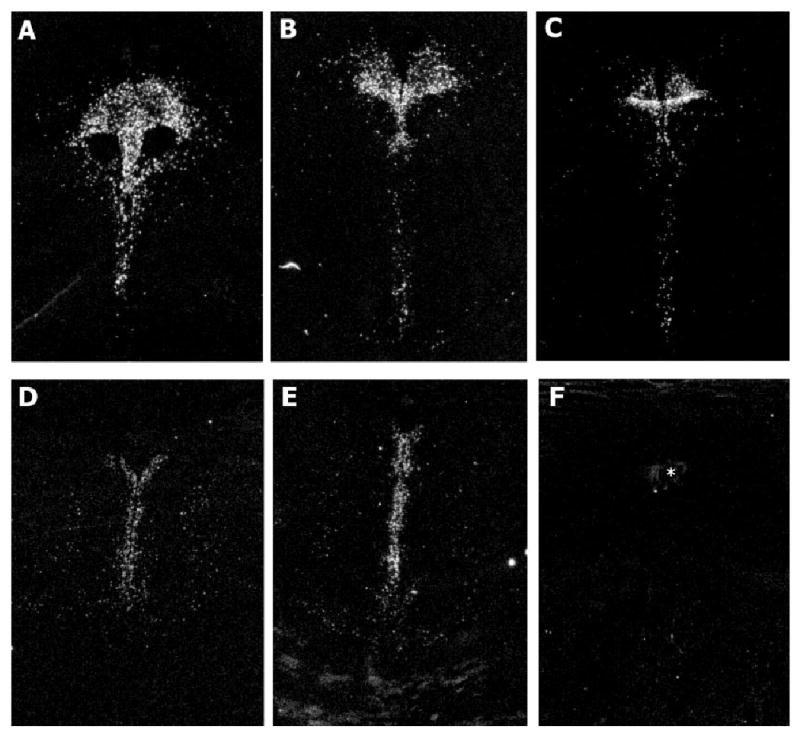
Film autoradiographic images of FEV mRNA in the dorsal raphe nucleus proceeding from rostral to caudal anatomical levels (A–D), and in the median raphe nucleus (D and E). F: Represents a tissue section adjacent to the section in Figure 1A containing the dorsal raphe that was hybridized with labeled sense FEV riboprobes. No specific hybridization signal is evident in the dorsal raphe. Asterisk (*) identifies the cerebral aqueduct.
The cellular localization of FEV mRNA in the brainstem is illustrated in Figure 2. Dense clustering of silver grains are evident, overlying neurons located in the subnuclear divisions of the ventral (Fig. 2A) and ventrolateral (Fig. 2B) nuclei of the dorsal raphe, as well as in the median raphe (Fig. 2C). FEV mRNA-positive neurons were also observed in the caudal linear nucleus, oral pontine, and supralemniscal serotonergic cell groups.
Fig. 2.
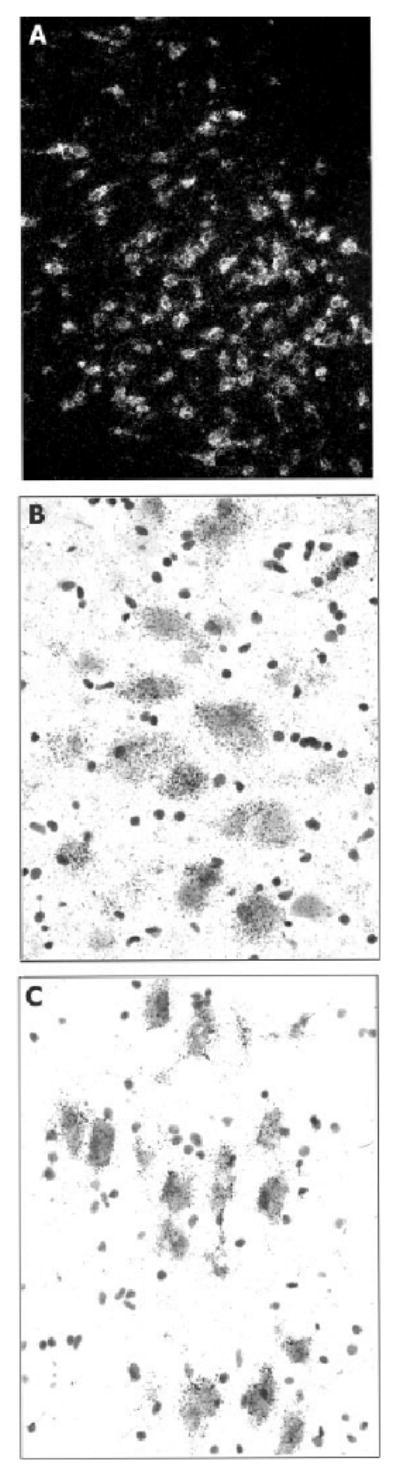
A: Darkfield photomicrograph illustrating FEV mRNA in neurons of the ventral subnucleus of the dorsal raphe. B: Bright-field photomicrograph of neurons in the ventrolateral subnucleus of the dorsal raphe containing dense silver grains corresponding to FEV mRNA. C: Brightfield photomicrograph of FEV mRNA-positive neurons in the median raphe.
Discussion
Previous studies have examined the brain localization and function of the rodent Pet-1 gene. Pet-1 mRNA is exclusively expressed in the serotonin neurons of the raphe nuclei in the mouse and rat brain (Hendricks et al., 1999; Pfaar et al., 2002). Furthermore, in Pet-1 knockout mice the embryonic development of 5-HT neurons was disrupted and was associated with increased anxiety-like and aggressive behaviors in adult mice (Hendricks et al., 2003). These findings suggest that Pet-1 is a critical component of a transcriptional program that governs the development of 5-HT modulated behaviors in adults.
Earlier attempts to characterize the precise cellular location of FEV mRNA in the human brain by in situ hybridization were unsuccessful because of the inability to generate an efficient in situ hybridization probe (Maurer et al., 2003). As an alternative approach, this group used a more sensitive RT-PCR technique that revealed that FEV mRNA is expressed in human raphe tissue homogenates. Although this study confirms the presence of the transcription factor, FEV in the human brain, it does not answer questions about the precise cellular localization or the regional distribution of FEV in the human brainstem.
Our investigation, using in situ hybridization histochemistry, revealed a specific and exclusive localization of FEV mRNA to the serotonergic neuronal populations of the dorsal and median raphe as well as the other serotonin-containing cell populations in the midbrain and pons. Furthermore, the cellular expression of FEV mRNA in serotonin-containing neurons was quite robust. Given the previous rodent studies demonstrating expression of Pet-1 at early embryonic developmental stages and its role in the differentiation of serotonin neurons, the observations that robust expression of FEV mRNA is maintained in the brain of mid- to late-aged human adults suggest a continued role of this transcription factor in the maintenance and transcriptional regulation of serotonin neurons throughout adulthood.
The nearly identical primary structures of Pet-1 and FEV raise the possibility that these proteins perform similar functions in the developing and adult brain. The similar neuroanatomical and cellular expression of the FEV gene in the adult human brain, when compared with Pet-1 in the rodent brain, provides additional support for this idea and suggests that transcriptional mechanisms controlling FEV and Pet-1 expression are conserved. Enhancer sequences directing mouse Pet-1 expression to 5-HT neurons have recently been identified in a conserved region immediately upstream of the Pet-1 coding region and therefore homologous sequences in the FEV upstream region may control its expression in the human brain (Scott et al., 2005). However, it remains to be determined whether the temporal pattern of FEV expression in the human brain is similar to that of Pet-1 and whether these proteins perform similar functions at the transcriptional level.
Considerable evidence has accumulated supporting the notion that serotonin neurotransmission is reduced in the brain of depressed subjects and individuals exhibiting suicidal behavior (for reviews see Stockmeier (2003) and van Heeringen (2003)). Specifically, alterations have been reported in the SERT and 5-HT1A receptors in the brain of depressed subjects (Austin et al., 2002; Sargent et al., 2000; Stockmeier et al., 1998). Based upon the transcriptional role of Pet-1 in regulating specific serotonergic phenotype, it is interesting to speculate that the transcriptional program controlled by FEV may be dysfunctional in the brain of depressed patients and may contribute to the deficiency in serotonin neurotransmission in mood disorders. The integrity of FEV expression in the brain of depressed subjects is an area that warrants further research.
Acknowledgments
The authors thank Drs. Gretchen Haas, Carol Sue Johnston, Cameron Carter, and Matcheri Keshavan for their participation in the diagnostic conferences, and Heather Murphy for assisting with the tissue processing.
Contract grant sponsor: USPHS; Contract grant numbers: MH57011, MH45156 (MCA), MH62723 (ESD); Contract grant sponsor: the Institutional Development Award (IDeA) Program of the National Center for Research Resources; Contract grant number: NIH P20 RR 17701.
References
- Austin MC, O'Donnell SM. Regional distribution and cellular expression of tryptophan hydroxylase messenger RNA in postmortem human brainstem and pineal gland. J Neurochem. 1999;72:2065–2073. doi: 10.1046/j.1471-4159.1999.0722065.x. [DOI] [PubMed] [Google Scholar]
- Austin MC, Whitehead RE, Edgar CL, Janosky JE, Lewis DA. Localized decrease in serotonin transporter-immunoreactive axons in the prefrontal cortex of depressed subjects committing suicide. Neuroscience. 2002;114:807–815. doi: 10.1016/s0306-4522(02)00289-0. [DOI] [PubMed] [Google Scholar]
- Azmitia EC, Gannon PJ. The primate serotonergic system: a review of human and animal studies and a report on Macaca fascicularis. Adv Neurol. 1986;43:407–468. [PubMed] [Google Scholar]
- Dahlstrom A, Fuxe K. Evidence for the existence of monoamine-containing neurons in the central nervous system. I. Demonstration of monoamines in the cell bodies of brainstem neurons. Acta Physiol Scand Suppl. 1964;232(Suppl. 62):1–55. [PubMed] [Google Scholar]
- Davidson RJ, Putnam KM, Larson CL. Dysfunction in the neural circuitry of emotion regulation—a possible prelude to violence. Science. 2000;289:591–594. doi: 10.1126/science.289.5479.591. [DOI] [PubMed] [Google Scholar]
- Fyodorov D, Nelson T, Deneris E. Pet-1, a novel ETS domain factor that can activate neuronal nAchR gene transcription. J Neurobiol. 1998;34:151–163. [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Lelievre E, Lionneton F, Soncin F, Vandenbunder B. The ETS family contains transcriptional activators and repressors involved in angiogenesis. Int J Biochem Cell Biol. 2001;33:391–407. doi: 10.1016/s1357-2725(01)00025-5. [DOI] [PubMed] [Google Scholar]
- Maurer P, T'Sas F, Coutte L, Callens N, Brenner C, Van Lint C, de Launoit Y, Baert JL. FEV acts as a transcriptional repressor through its DNA-binding ETS domain and alanine-rich domain. Oncogene. 2003;22:3319–3329. doi: 10.1038/sj.onc.1206572. [DOI] [PubMed] [Google Scholar]
- Maurer P, Rorive S, Kerchove d'Exaerde A, Schiffmann SN, Salmon I, Launoit Y. The ETS transcription factor FEV is specifically expressed in the human central serotonergic neurons. Neurosci Lett. 2004;357:215–218. doi: 10.1016/j.neulet.2003.12.086. [DOI] [PubMed] [Google Scholar]
- Murphy DL, Andrews AM, Wichems CH, Li Q, Tohda M, Greenberg B. Brain serotonin neurotransmission: an overview and update with emphasis on serotonin subsystem heterogeneity, multiple receptors, interactions with other neurotransmitter systems, and consequent implications for understanding the action of serotonergic drugs. J Clin Psychiatry. 1998;15:4–12. [PubMed] [Google Scholar]
- Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O. A new member of the ETS family fused to EWS in Ewing tumors. Oncogene. 1997;14:1159–1164. doi: 10.1038/sj.onc.1200933. [DOI] [PubMed] [Google Scholar]
- Pfaar H, von Holst A, Vogt DM, Weisenhorn C, Brodski J, Guimera J, Wurst W. mPet-1, a mouse ETS-domain transcription factor, is expressed in central serotonergic neurons. Dev Genes Evol. 2002;212:43–46. doi: 10.1007/s00427-001-0208-x. [DOI] [PubMed] [Google Scholar]
- Sargent PA, Kjaer KH, Bench CJ, Rabiner EA, Messa C, Meyer J, Gunn RN, Grasby PM, Cowen PJ. Brain serotonin1A receptor binding measured by positron emission tomography with [11C]WAY-100635: effects of depression and antidepressant treatment. Arch Gen Psychiatry. 2000;57:174–180. doi: 10.1001/archpsyc.57.2.174. [DOI] [PubMed] [Google Scholar]
- Scott MS, Deneris ES. Making and breaking serotonin neurons and autism. In: DiCicco-Bloom E, editor. Special issue on autism: modeling human brain abnormalities in developing animal systems. Int J Dev Neurosci. 2–3. Vol. 23. 2005. pp. 277–285. [DOI] [PubMed] [Google Scholar]
- Scott MS, Krueger K, Deneris ES. A differentially autoregulated Pet-1 enhancer region is a critical target of the transcriptional cascade that governs serotonin neuron development. J Neurosci. 2005;25:2628–2636. doi: 10.1523/JNEUROSCI.4979-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sementchenko VI, Watson DK. ETS target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- Silverstone PH. Qualitative review of SNRIs in anxiety. J Clin Psychiatry. 2004;65(Suppl. 17):19–28. [PubMed] [Google Scholar]
- Steinbusch W. Distribution of serotonin-immunoreactivity in the central nervous system of the rat-cell bodies and terminals. Neuroscience. 1981;6:557–618. doi: 10.1016/0306-4522(81)90146-9. [DOI] [PubMed] [Google Scholar]
- Stockmeier CA. Involvement of serotonin in depression: evidence from postmortem and imaging studies of serotonin receptors and the serotonin transporter. J Psychiatric Res. 2003;37:357–373. doi: 10.1016/s0022-3956(03)00050-5. [DOI] [PubMed] [Google Scholar]
- Stockmeier CA, Shapiro LA, Dilley GE, Kolli TN, Friedman L, Rajkowska G. Increase in serotonin-1A autoreceptors in the midbrain of suicide victims with major depression postmortem evidence for decreased serotonin activity. J Neurosci. 1998;18:7394–7401. doi: 10.1523/JNEUROSCI.18-18-07394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tork I. Anatomy of the serotonergic system. In: Whitaker-Azmitia P, Peroutka SJ, editors. The Neuropharmacology of Serotonin. Vol. 600. Ann NY Acad Sci; 1990. pp. 9–35. [DOI] [PubMed] [Google Scholar]
- van Heeringen K. The neurobiology of suicide and suicidality. Can J Psychiatry. 2003;48:292–300. doi: 10.1177/070674370304800504. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Hahn SL, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]


