Abstract
Objective
To determine whether or not there are any significant differences in the effects of wound dressings on bacterial bioburden.
Method
A selection of non-occlusive, non-adhesive dressings was tested for their effect on bacterial bioburden. The dressings selected included two dressings with antimicrobial properties (one containing silver and one containing PHMB), a cotton-based dressing enclosed in a perforated sleeve of poly(ethylene terephthalate), a carboxymethyl cellulose-based dressing, a fibre-free alginate dressing, and a 12-ply 100% cotton gauze. Using the colony-drip flow reactor (DFR) model, a meticillin-resistant Staphylococcus aureus (MRSA) or Pseudomonas aeruginosa biofilm was grown underneath a dressing sample. Biofilm growth was examined via plate counts, fluorescent microscopy and scanning electron microscopy.
Results
The dressings containing antimicrobial agents had the greatest effect on bacterial load. In the MRSA experiments, both antimicrobial dressings produced lower bacteria counts than the other dressings (p≤0.001), while in the P. aeruginosa experiments, only the silver-containing sample had fewer bacteria (p≤0.0001). However, neither antimicrobial dressing was able to completely eradicate the bacteria when testing with either microorganism.
Conclusion
The results presented herein illustrate that bacteria can grow unchallenged within the dressing environment and that an antimicrobial dressing can limit this bacterial growth.
Keywords: antimicrobial dressings, biofilms, plate counts, fluorescent microscopy, scanning electron microscopy
Wounds are an ideal environment for bacterial colonisation and biofilm formation. The wound bed provides both a surface on which to grow and an ample supply of nutrients. Recent studies have shown that bacteria inoculated into wounds form biofilms.1,2 Another study found that as many as 60% of chronic wound specimens contained a biofilm.3 Current research indicates that biofilms are present in chronic wounds.3–6 Furthermore, recent studies suggest that biofilms should be targeted when developing and testing therapies for these wounds.7–11
A literature review of culture data from 62 published studies dating from 1969 to 1997 revealed that the predominant wound isolate in both chronic and acute wounds was Staphylococcus aureus (reported in 63% of the studies), followed by coliforms (45%), Bacteroides spp. (39%), Peptostreptococcus spp. (36%), Pseudomonas aeruginosa (29%), Enterococcus spp. (26%) and Streptococcus pyogenes (13%).12
A principal component of wound care is the choice of dressing. Many modern wound dressings have a variety of different attributes, with the aim of creating a supportive wound-healing environment. Such dressings are designed to absorb exudate, provide an optimum moisture balance at the wound surface, prevent maceration of surrounding tissue, and control bacterial colonisation.
This study aimed to evaluate the effect of wound dressings on bacterial bioburden. Specifically, six different wound dressings were selected and their effects on the growth of two pathogens commonly found in biofilms, meticillin-resistant S. aureus (MRSA) and P. aeruginosa biofilms, was examined.
Materials and method
This study used a method of growing in vitro MRSA and P. aeruginosa biofilms under conditions that parallel those of a wound biofilm, using the colony-drip flow reactor (DFR) model.
This model is based on characteristics of both the colony biofilm model13 and the drip-flow reactor (DFR) model.14
In the colony model, biofilms are grown on a semipermeable membrane that sits on an agar plate. The bacteria are given a new supply of nutrients by moving the membrane to a new plate.
In the DFR model, biofilms are grown on an inclined microscope slide that sits inside a testing channel and is continuously supplied with fresh medium.
In the colony-DFR model, biofilms are grown on a membrane similar to the colony model, but the membrane is used in the DFR apparatus.
Wound dressings
A selection of commonly used non-adhesive and non-occlusive dressings was used in this investigation. The wound dressings tested included two containing an antimicrobial agent:
Excilon AMD (Covidien, Mansfield, MA), a cotton dressing containing 0.2% polyhexamethylene biguanide (PHMB)
Silvercel (Systagenix Wound Management, UK), a non-woven dressing composed of alginate, carboxymethylcellulose, and silver-coated nylon fibres, and which contains 8% elemental silver.
The remaining dressings did not contain an antimicrobial agent:
Sterile gauze (Fisher Scientific, Pittsburg, PA); this was a 12-ply, 100% cotton gauze, and was used to represent a conventional wound dressing
Aquacel (ConvaTec, Skillman, NJ), a non-woven dressing comprised mainly of sodium carboxymethyl cellulose
Telfa (Covidien, Mansfield, MA), which is composed of a thin layer of cotton fibres enclosed in a perforated sleeve of poly(ethylene terephthalate)
SeaSorb (Coloplast, Minneapolis, MN) an alginate dressing with a nylon mesh layer to provide strength and one-piece removal.
All dressings were aseptically divided into 2.5 × 2.5cm test squares and stored until use. The thickness of dressing was not altered.
Bacterial strains
Clinical isolates of MRSA and P. aeruginosa were obtained from chronic wounds using methods previously described.3 Experiments with MRSA and P. aeruginosa were conducted separately, but used identical methods.
The microorganisms were cultured in 100% tryptic soy broth (TSB) at 37°C for 24 hours. The culture was then diluted to an optical density of 0.05 at 600nm and used in the colony-DFR as described below.
Colony-DFR experiments
The model system used in this investigation combines attributes of both the colony model13 and the DFR model.15
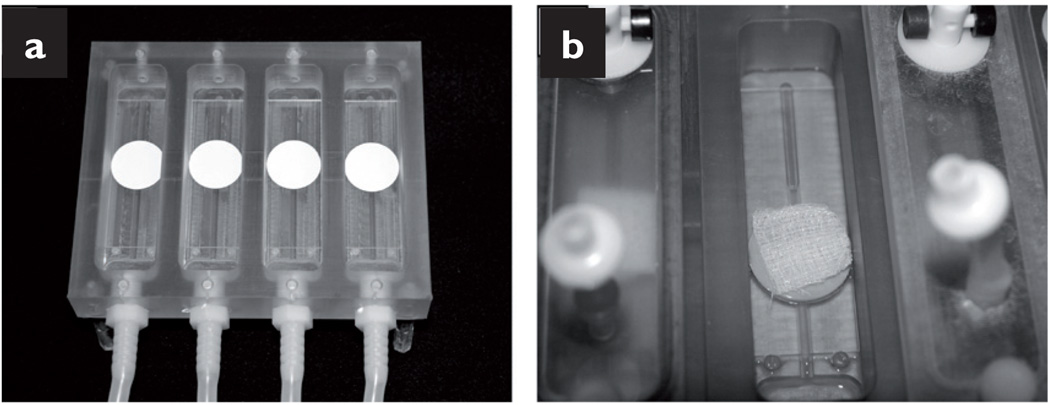
To prepare the reactor apparatus, 25mm (diameter) absorbent pads (Millipore, Billerica, MA) were glued with silicon-based aquarium sealant to clean glass microscope slides and placed in the channels of the DFR (Biosurface Technology, Bozeman, MT) (Fig 1a). The entire set-up was autoclaved and maintained sterile until use.
Fig 1.
Images of a colony-DFR reactor: absorbent pads glued onto glass microscope slides (a);gauze dressing placed on top of the inoculated membrane (b)
Experiments began by hydrating the absorbent pads with 0.5ml of 10% TSB and then ultraviolet (UV)-sterilised 0.22µm porous polycarbonate membranes (GE Water & Process Technologies, Trevose, PA) were placed on these absorbent pads. Next, the membranes were inoculated with 10µl of a TSB-diluted overnight culture (optical density of 0.05 at 600nm). The system was left undisturbed for 30 minutes while the inoculum was allowed to dry. Afterwards, a sterile dressing sample was placed directly on top of the inoculated membrane (Fig 1b).
The reactor was then attached to a medium reservoir and the medium (10% TSB) was pumped through the system at 5ml/h/channel. This reactor and set-up allowed for the medium to drip down the microscope slide and absorb into the pad, which then supplied nutrients to the bacteria growing on the top side of membrane.
The bacteria were allowed to grow for 72 hours at room temperature (average day time temperature of 21.5°C, range 19.9–23.2°C).
Afterwards, the samples were examined with plate counts, fluorescent microscopy and scanning electron microscopy (SEM).
Experimental runs were also conducted with non-inoculated dressings to examine the dressings in their native state.
Plate counting
After the 72-hour growth period, the samples were removed from the chambers and the dressing/membrane pairs were separated and processed separately, to determine whether or not there were any differences in bacterial growth on the dressing compared with the membrane.
Each piece was placed in 10ml of sterile phosphate buffered saline (PBS). The samples were then subjected to 30 seconds of vortexing and two minutes of sonication, followed by an additional 30 seconds of vortexing. Serial 10-fold dilutions were made using sterile PBS, and the dilutions were plated on tryptic soy agar (TSA).
After 24 hours of incubation at 37°C, the plates were counted and the number of colony forming units (CFU) per membrane or per dressing was calculated. It should be noted that serial dilution was used to neutralise any antimicrobial elements, and that this neutralisation was verified (data not shown).
A minimum of five samples of each dressing type were analysed via plate counting. Mean averages and standard deviations (SD) were then generated. For two-group comparisons, statistical analysis for significance was determined using as a two-tailed t-test assuming unequal variances, with α=0.5 and p≤0.05 considered to be significant. For comparisons involving three or more groups, statistical analysis for significance was determined using an ANOVA with a Tukey’s HSD post-hoc test, with α=0.5 and p≤0.05 considered to be significant.
Scanning electron microscopy
After the growth period, both the dressing and the membrane were prepared for SEM. High resolution SEM analysis was used to investigate bacterial growth on individual dressing fibres. The dressing was separated from the membrane and each was placed (biofilm side up) on a formalin-saturated (0.5ml) UV-sterilised absorbent pad. The samples were fixed for two hours. This method allowed the formalin to wick upwards into the sample without disrupting the integrity of the biofilm.
Afterwards, the samples were dehydrated in an ethanol gradient consisting of 50%, 70% and 95% ethanol for 30 minutes each, followed by 100% ethanol for an hour and another change to 100% ethanol overnight.
These steps were performed in a manner similar to the fixation step; the sample was placed on top of an absorbent pad saturated with 0.5ml of the ethanol solution. The samples were air-dried and mounted to a half-inch specimen mount with either double-sided carbon tape or colloidal carbon.
The samples were then coated with iridium for conductivity and imaged using a Zeiss Supra 55VP field emission scanning electron microscope with 1kV accelerating voltage. A minimum of two samples of each dressing type was analysed by SEM.
Fluorescence microscopy
After 72 hours of growth, the samples were prepared for microscopy. Analysis by fluorescence microscopy involved staining the dressing samples for microbial DNA to investigate bacterial presence throughout each dressing. The dressing/membrane pairs were removed from the reactor and placed in a sterile Petri dish on top of an absorbent pad saturated with 0.75ml of SYBR green (Lonza, Rockland, ME) staining solution (5µl of stock SYBR green/ml of filtered water), to identify bacterial nucleic acids. Another saturated absorbent pad was placed on top of the dressing/membrane pair, sandwiching the sample. The sample was allowed to absorb the stain for 25 minutes at room temperature.
Next, the sample was embedded in OCT (Saukura Finetek, Torrance, CA) and frozen on dry ice and stored at −80°C in preparation for cryosectioning. The samples were then sectioned into 10µm cross-sections using a Leica CM 1850 Cryostat (Leica, Wetzlar, Germany) and examined using a Nikon Eclipse E800 microscope with a 20× dry objective (Nikon, Melveill, NY). Images were taken with an Olympus Q-color 5 camera (Olympus, Center Valley, PA).
A minimum of two samples of each dressing type was analysed by fluoresence microscopy.
Results
Plate counting
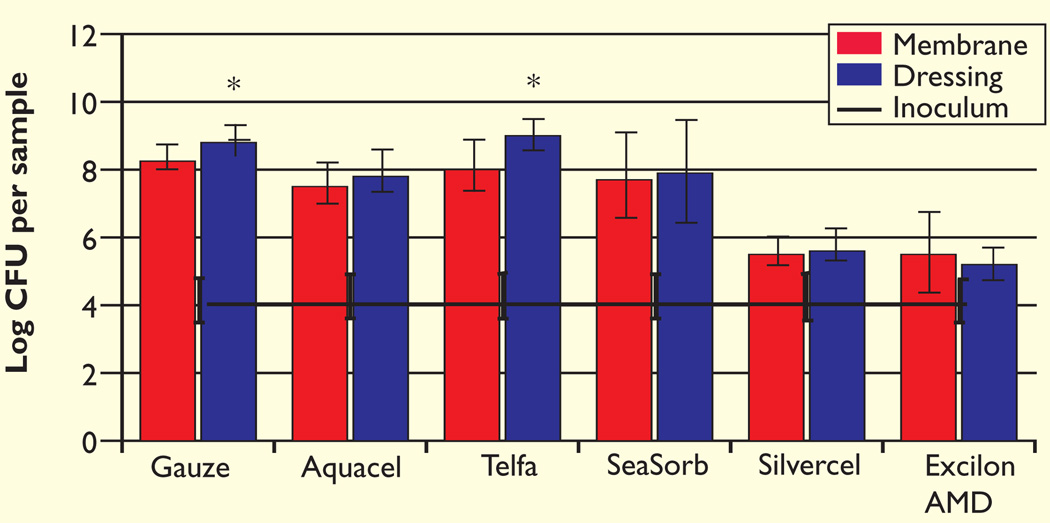
In the MRSA studies, both gauze and Telfa had a greater number of bacteria on the dressing than the membrane (p=0.04 and p=0.04, respectively, Fig 2). MRSA growth was detected on each sample, including the antimicrobial dressings. Combining both the membrane and dressing CFU values resulted in the total CFU for the dressing/membrane pair. The total log CFU/pair for the antimicrobial dressings was significantly lower than for the non-antimicrobial dressings (p≤0.001). When compared with gauze, Silvercel displayed a 3.11 ± 0.22 log difference in MRSA growth and Excilon AMD a 3.20 ± 0.60 log difference.
Fig 2.
The initial inoculum and log CFU per sample of MRSA after three days growth in the colony-DFR model
The number of bacteria remaining on the membrane and contained within the dressing were enumerated separately
*Significantly different from corresponding membrane at p=0.04
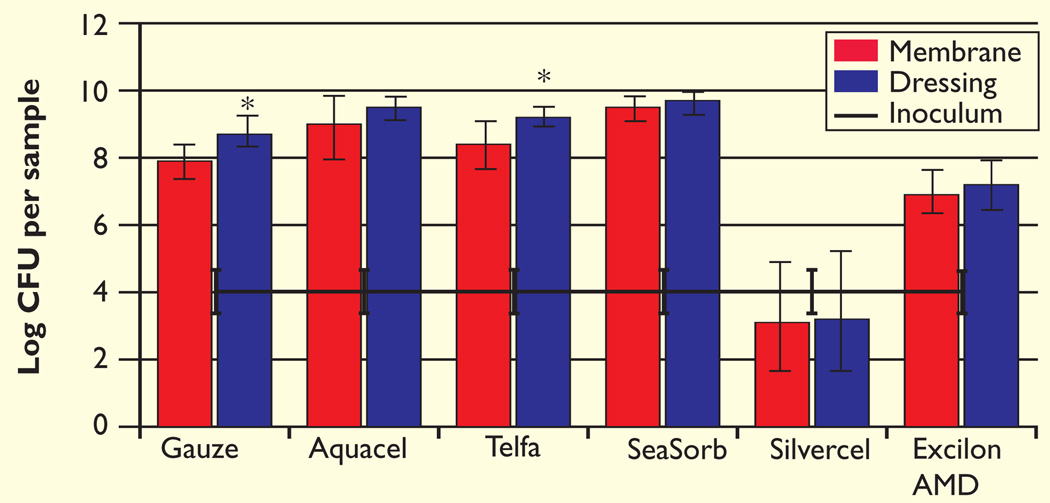
In the P. aeruginosa investigations, the gauze and Telfa dressings had a greater number of bacteria on the dressing than the membrane (p=0.02 and p=0.05, respectively, Fig 3). Overall, the Silvercel membrane/dressing pair had fewer bacteria than any of the other sets (p≤0.0001). Furthermore, the total bacterial count for the Silvercel samples was not statistically different from the initial inoculum (p=0.97). Total bacterial growth on the Excilon AMD was statistically lower than on all the other non-antimicrobial dressings (p≤0.01) except for gauze (p=0.07). When compared with gauze, Silvercel and Excilon AMD displayed 5.37 ± 0.87 and 1.48 ± 0.61 log difference respectively. The non-antimicroial dressings (Aquacel, Telfa and SeaSorb) all exhibited higher bacterial populations compared with gauze.
Fig 3.
The initial inoculum log CFU per sample of P. aeruginosa after three days growth in the colony-DFR model
The number of bacteria remaining on the membrane and contained within the dressing were enumerated separately
*Significantly different from corresponding membrane at p≤0.05
Scanning electron microscopy
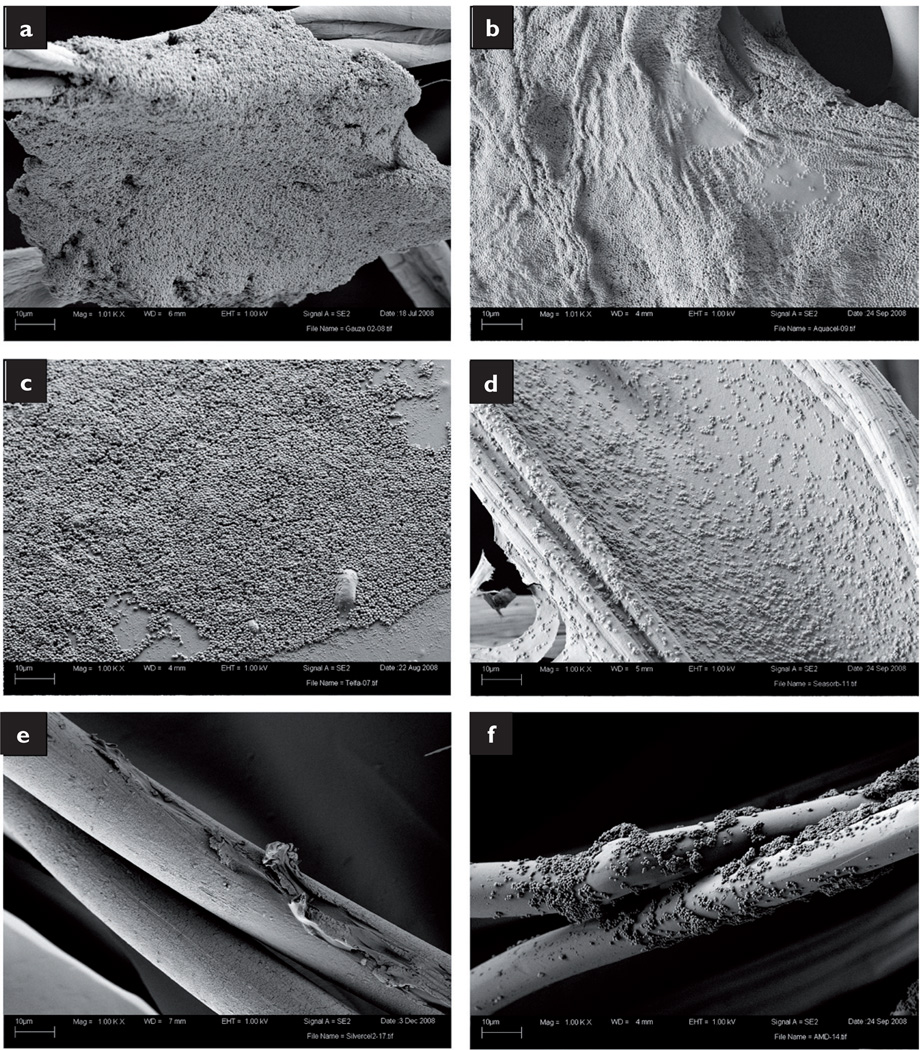
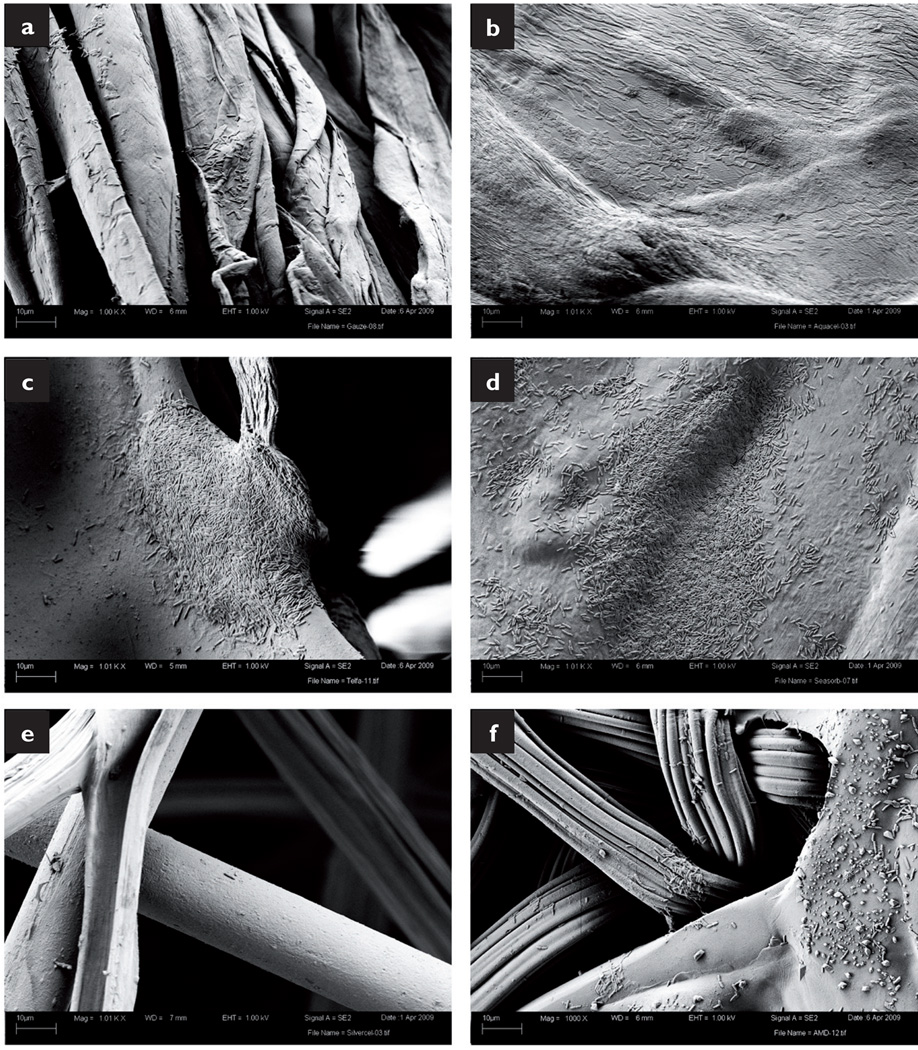
SEM analysis revealed heavy microorganism infiltration. MRSA grew along the cotton fibres of the gauze and formed biofilms large enough to connect neighbouring strands (Fig 4a). The SEM preparation collapsed and dehydrated the Aquacel dressing, but still numerous MRSA cells covered the surface (Fig 4b). MRSA also grew on the surface of the Telfa dressing, covering the majority of the sample in a biofilm several cells thick (Fig 4c). While the bacteria were not as closely packed in the SeaSorb sample, a uniform coating of bacteria was present (Fig 4d). However, only a few, isolated cells were found on the strands of the Slivercel dressing (Fig 4e). Colonies of MRSA were also found growing along the Excilon AMD surface, although they were not as large and homogeneous as those found in the non-antimicrobial samples (Fig 4f).
Fig 4.
SEM images of MRSA within the dressings: gauze (a); Aquacel (b); Telfa (c); SeaSorb (d); Silvercel (e); Excilion AMD (f)
Similar results were obtained when the dressings were exposed to P. aeruginosa. Bacterial cells were found on the gauze fibres and almost entirely covered the surface of the Aquacel dressing (Figs 5a and b respectively). P. aeruginosa formed large colonies in areas of irregularity (junction points and ridges) on the Telfa and SeaSorb dressings (Figs 5c and 5d respectively). In contrast, only a few individual bacterial cells were observed on the Silvercel dressing (Fig 5e). Finally, small microcolonies of P. aeruginosa were detected on the Excilon AMD fibres (Fig 5f).
Fig 5.
SEM images of P. aeruginosa within dressings: gauze (a); Aquacel (b); Telfa (c); SeaSorb (d); Silvercel (e); Excilion AMD (f). All images taken at 3000× magnification
Evaluation of the non-inoculated dressings did not reveal any structures resembling bacteria or bacterial biofilms (data not shown).
Fluorescence microscopy
The membrane/dressing pair was also stained and examined under both brightfield and fluorescence microscopy. The samples were stained with SYBR green, to identify bacterial nucleic acids. The stain, however, was also taken up by many dressing materials. By overlaying the brightfield and fluorescence images, the spatial relationship of the stained bacteria within the dressing was observed. the results were similar to the SEM evaluation.
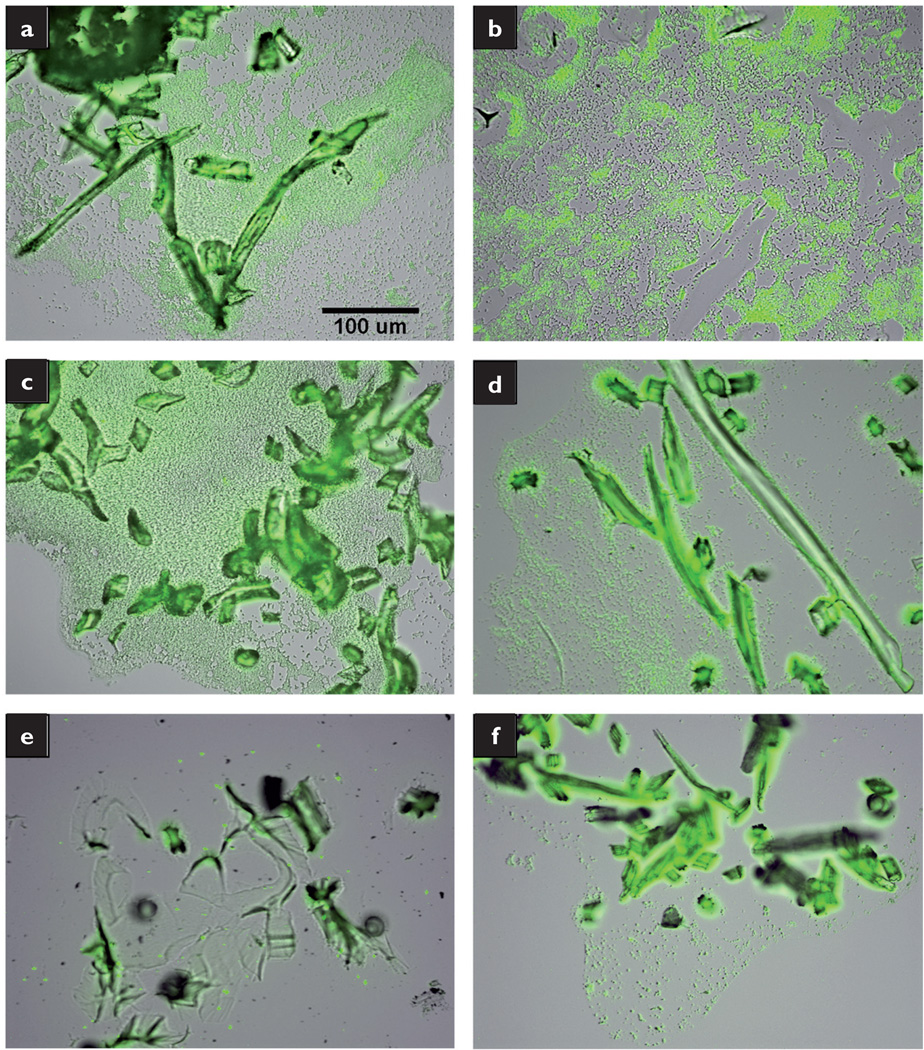
Large MRSA biofilms were observed around the gauze fibres (Fig 6a), and expansive colonies of MRSA were detected throughout the Aquacel, Telfa and SeaSorb dressings (Figs 6b–d). Comparatively, there was minimal bacterial presence in the Silvercel and Excilon AMD dressings (Figs 6e and 6f respectively).
Fig 6.
Fluorescent overlay images of MRSA grown in membrane/dressing pairs: gauze (a); Aquacel (b); Telfa (c); SeaSorb (d); Silvercel (e); Excilion AMD (f). All images taken at 20× magnification
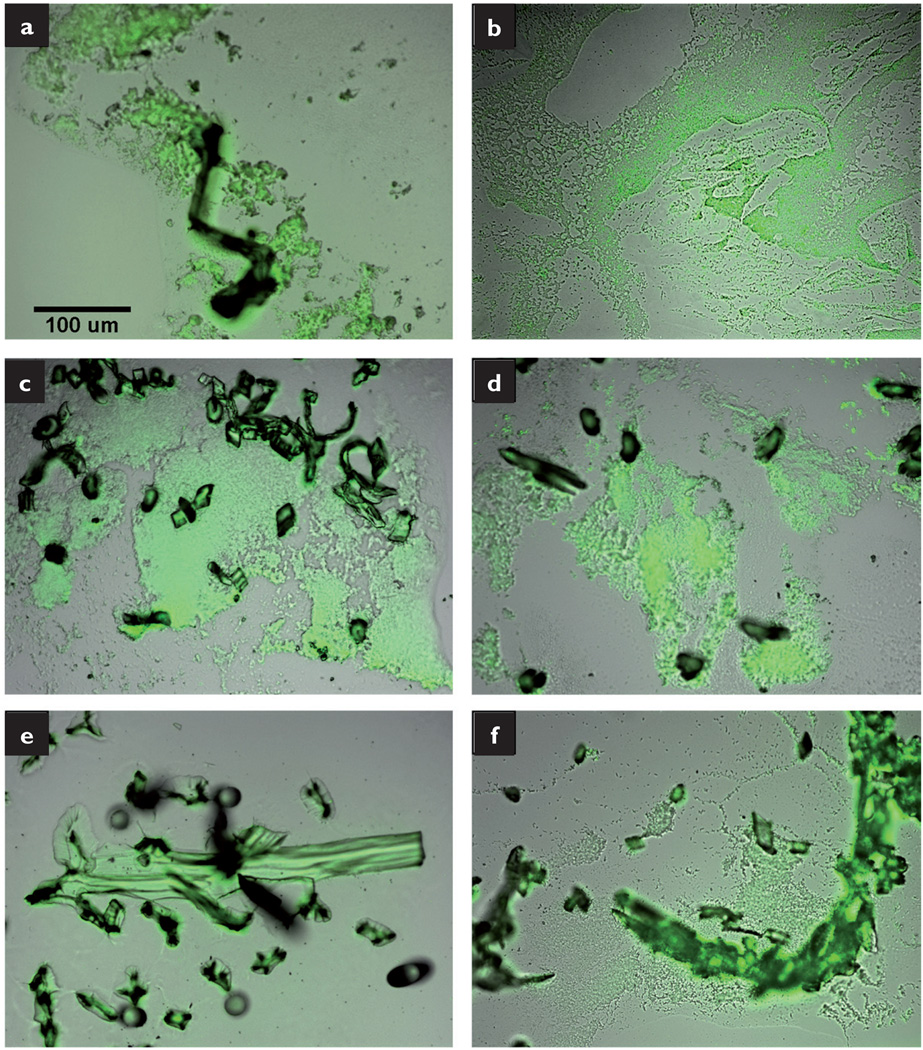
The P. aeruginosa experiments produced similar results. Clusters of bacteria were located around the gauze fibres (Fig 7a), while extensive bacteria staining was observed throughout the cryosection of the Aquacel, Telfa and SeaSorb dressings (Figs 7b–d). While the Silvercel fibres stained intensely, there was minimal bacterial staining (Fig 7e). The Excilon AMD displayed diffuse clusters of bacteria (Fig 7f).
Fig 7.
Fluorescent overlay images of P. aeruginosa grown on membrane/dressing pairs: gauze (a); Aquacel (b); Telfa (c); SeaSorb (d); Silvercel (e); Excilion AMD (f). All images taken at 20× magnification
The staining of non-inoculated dressings demonstrated the background staining of materials, but there was no evidence of any additional staining (data not shown).
Discussion
The antimicrobial properties of Silvercel16,17 and several AMD products18–20 against Gram-positive and Gram-negative bacteria, as well as yeasts and fungi, have been demonstrated in several in vitro and in vivo studies. Furthermore, the infection-controlling properties of Aquacel have been demonstrated by its ability to sequester and retain bacteria.21–23 However, the majority of these studies investigated the dressing’s ability to kill16–20 or sequester21–23 planktonic bacteria, not bacterial biofilms.
Recently, two in vitro techniques that assess the effect of antimicrobial wound dressings on biofilms were described.24,25 One method uses confocal microscopy to evaluate of the effects of silver-containing dressings on established biofilms.24 While this technique obtains real-time images of cell viability, it does so using a closed system (that is, the biofilms are grown in a batch culture with no nutrient replenishment or flow). The dynamic condition of a wound biofilm is not represented.25 The second method uses a novel perfusion chamber that allows for a dynamic growth environment.25 However, the instrumentation is intricate and custom-made, and only one sample can be tested at a time.
The colony-DFR model is a practical method of growing in vitro biofilms in a manner that mimics a chronic wound environment. It is a variation of an American Society for Testing and Materials (ASTM) method.26 The components are commercially available and multiple samples or time points can be evaluated in one experimental run. Furthermore, the bacteria are seeded on a semiporous surface and are continuously fed from below with fresh, flowing nutrients, growing a biofilm under dynamic conditions. Furthermore, the wicking of nutrients through the pad and membrane to the biofilm mimics biofilm growth in the wound environment. Therefore, the method of biofilm growth in the colony-DFR model can be considered more similar to wound biofilm growth than previous models.27
However, the colony-DFR model has limitations, including the lack of host immune factors. Furthermore, the experimental conditions chosen for this study do not ideally mimic the chronic wound environment. For instance, trauma wounds have been reported to range in temperature from 25.3 to 37.3°C28 and the wound bed temperature of chronic leg ulcers has been reported as ranging from 24°C to 26°C.29 Rather than run the experiments at 37°C, which would reflect core body temperature, our experiments were conducted at room temperature (range 19.9–23.2°C), which was closer, but not identical to, the chronic wound bed temperature.
The medium selected for use in our studies was 10% TSB. TSB is a proteinaceous nutrient medium that supports the growth of a wide variety of microorganisms, but does not contain many of the components of wound exudate (such as fibrin, cytokines and proteinases). However, at present there is no standardised nutrient medium to replicate wound exudate in laboratory studies. Furthermore, the biochemical components of wound fluid vary depending on the individual,30 the type of wound,31 and the healing stage of the wound.30,31 Thus, for this investigation a standard bacterial growth medium was selected.
Finally, most chronic wounds are contaminated with multiple species of bacteria.3 This model can be used to grow multispecies biofilms, and this is currently being investigated. However, the addition of a second species increases the complexity of the model by introducing interspecies interactions. The complexity of a multispecies model makes it more difficult to elucidate the effect of the dressing. Thus, in this first study, we examined the effect of dressings on single-species biofilms.
Each dressing used was subjected to uniform conditions and analysed for biofilm formation via plate counts and microscopy. The experiments began with the inoculation of a membrane with either MRSA or P. aeruginosa. The initial inocula were approximately 104 CFU for both microorganisms (Figs 2 and 3). Clinically, wounds presenting with a level greater than or equal to 105 CFU per gram of tissue are considered to be infected32 and wounds with 106 CFU per gram of tissue are associated with impaired healing.1 Thus, these experiments were modelled to represent a clinical situation wherein the wound would not necessarily present as infected and would be treated with a standard dressing.
After the 72-hour growth period, the dressing/membrane pairs were processed separately to explore the bacterial load associated with the dressing. In both the MRSA and P. aeruginosa studies, the gauze and Telfa dressings had more bacteria than the membrane (Figs 2 and 3, respectively). Both dressings are made of cotton, while Telfa also has a plastic covering that is perforated to reveal the cotton fibres. MRSA and P. aeruginosa have been shown to thrive on cotton and synthetic materials.33–35 Overall, however, there was no difference in total bacterial load between any of the non-antimicrobial samples. During the experimental period, the bacteria grew to approximately 108 CFU and microscopic analysis revealed large bacterial biofilms.
Bacterial biofilms are composed of both microorganisms and extracellular polymeric substance (EPS). While EPS is a major constituent of bacterial biofilms,36 the target of this study was the overall bacterial viability associated with the dressings. Therefore, EPS imaging was not performed.
The dressings shown to have the greatest effect on bacterial bioburden were those containing antimicrobial agents: Silvercel and Excilon AMD. In the MRSA experiments, both samples had statistically fewer total bacteria then the non-antimicrobial samples (p≤0.001). In the P. aeruginosa experiments, Silvercel had statistically fewer bacteria than the other dressings (p≤0.0001).
However, neither of the antimicrobial dressings was able to completely eradicate the bacteria and prevent biofilm formation, as evident from the microscopic analyses. Bacteria within biofilms are hundreds to thousands of times more resistant to antibiotics and biocides than planktonic bacteria,5 and our experiments demonstrated that both silver and PHMB-containing dressings cannot prevent MRSA or P. aeruginosa biofilm formation.
These studies also demonstrated that dressings can provide an environment in which microbes can grow unchallenged, leading to an increase in bioburden. Furthermore, none of the dressings tested decreased the initial bioburden. Only in one condition, the P. aeruginosa membrane treated with Silvercel, was the ultimate bioburden controlled to the level of the initial inoculum. In the MRSA experiments, both silver and PHMB-containing dressings allowed for bacterial growth, although the bacterial load was significantly lower than that found when non-antimicrobial dressings were used.
Conclusion
Our results illustrate that bacteria can grow unchallenged within a dressing environment and that an antimicrobial dressing can limit this bacterial growth. Limiting microbial growth within the wound environment may improve wound healing outcomes. In one example, the institution-wide replacement of gauze dressings to antimicrobial gauze dressings was associated with reduced surgical site infections, morbidity rates, patient hospital stays, and post-surgical care costs.37
Footnotes
Declaration of interest: None.
References
- 1.Falanga V. The chronic wound: impaired healing and solutions in the context of wound bed preparation. Blood Cells Mol Dis. 2004;32(1):88–94. doi: 10.1016/j.bcmd.2003.09.020. [DOI] [PubMed] [Google Scholar]
- 2.Serralta VW, Harrison-Balestra C, Cazzaniga AL, et al. Lifestyles of bacteria in wounds: Pr absorbent wound dressings. Int Wound J. 2004;1(3):177–181. doi: 10.1111/j.1742-4801.2004.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James GA, Swogger E, Wolcott R, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16(1):37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 4.Davis SC, Ricotti C, Cazzaniga A, et al. Microscopic and physiologic evidence for biofilm-associated wound colonization in vivo. Wound Repair Regen. 2008;16(1):23–29. doi: 10.1111/j.1524-475X.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 5.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 7.Bjarnsholt T, Kirketerp-Moller K, Jensen PO, et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 2008;16(1):2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 8.Bjarnsholt T, Kirketerp-Moller K, Kristiansen S, et al. Silver against Pseudomonas aeruginosa biofilms. APMIS. 2007;115(8):921–928. doi: 10.1111/j.1600-0463.2007.apm_646.x. [DOI] [PubMed] [Google Scholar]
- 9.Dowd SE, Sun Y, Secor PR, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Percival SL, Bowler PG. Biofilms and their potential role in wound healing. Wounds. 2004;16(7):234–240. [Google Scholar]
- 11.Sun Y, Dowd SE, Smith E, et al. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008;16(6):805–813. doi: 10.1111/j.1524-475X.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 12.Bowler PG. The anaerobic and aerobic microbiology of wounds: a review. Wounds. 1998;10(6):170–178. [Google Scholar]
- 13.Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44(7):1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckingham-Meyer K, Goeres DM, Hamilton MA. Comparative evaluation of biofilm disinfectant efficacy tests. J Microbiol Methods. 2007;70(2):236–244. doi: 10.1016/j.mimet.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Goeres DM, Hamilton MA, Beck NA, et al. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc. 2009;4(5):783–788. doi: 10.1038/nprot.2009.59. [DOI] [PubMed] [Google Scholar]
- 16.Castellano JJ, Shafii SM, Ko F, et al. Comparative evaluation of silver-containing antimicrobial dressings and drugs. Int Wound J. 2007;4(2):114–122. doi: 10.1111/j.1742-481X.2007.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsons D, Bowler PG, Myles V, Jones S. Silver antimicrobial dressings in wound management: a comparison of antibacterial, physical, and chemical characteristics. Wounds. 2005;17(8):222–232. [Google Scholar]
- 18.Cazzaniga A, Serralta V, Davis S, et al. The Effect of an Antimicrobial Gauze dressing impregnated with 0.2-percent polyhexamethylene biquanide as a barrier to prevent Pseudomonas aeruginosa wound invastion. Wounds. 2002;14(5):169–176. [Google Scholar]
- 19.Gallant-Behm CL, Yin HQ, Liu S, et al. Comparison of in vitro disc diffusion and time kill-kinetic assays for the evaluation of antimicrobial wound dressing efficacy. Wound Repair Regen. 2005;13(4):412–421. doi: 10.1111/j.1067-1927.2005.130409.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee WR, Tobias KM, Bemis DA, Rohrbach BW. In vitro efficacy of a polyhexamethylene biguanide-impregnated gauze dressing against bacteria found in veterinary patients. Vet Surg. 2004;33(4):404–411. doi: 10.1111/j.1532-950X.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 21.Bowler PG, Jones SA, Davies BJ, Coyle E. Infection control properties of some wound dressings. J Wound Care. 1999;8(10):499–502. doi: 10.12968/jowc.1999.8.10.26356. [DOI] [PubMed] [Google Scholar]
- 22.Tachi M, Hirabayashi S, Yonehara Y, et al. Comparison of bacteria-retaining ability of absorbent wound dressings. Int Wound J. 2004;1(3):177–181. doi: 10.1111/j.1742-4801.2004.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker M, Hobot JA, Newman GR, Bowler PG. Scanning electron microscopic examination of bacterial immobilisation in a carboxymethyl cellulose (AQUACEL) and alginate dressings. Biomaterials. 2003;24(5):883–890. doi: 10.1016/s0142-9612(02)00414-3. [DOI] [PubMed] [Google Scholar]; 23 Walker M, Hobot JA, Newman GR, Bowler PG. Scanning electron microscopic examination of bacterial immobilisation in a carboxymethyl cellulose (AQUACEL) and alginate dressings. Biomaterials. 2003;24(5):883–890. doi: 10.1016/s0142-9612(02)00414-3. [DOI] [PubMed] [Google Scholar]
- 24.Percival SL, Bowler P, Woods EJ. Assessing the effect of an antimicrobial wound dressing on biofilms. Wound Repair Regen. 2008;16(1):52–57. doi: 10.1111/j.1524-475X.2007.00350.x. [DOI] [PubMed] [Google Scholar]
- 25.Thorn RM, Greenman J. A novel in vitro flat-bed perfusion biofilm model for determining the potential antimicrobial efficacy of topical wound treatments. J Appl Microbiol. 2009;107(6):2070–2079. doi: 10.1111/j.1365-2672.2009.04398.x. [DOI] [PubMed] [Google Scholar]
- 26.Standard A. ASTM E2647 - 08 Standard Test Method for Quantification of a Pseudomonas aeruginosa Biofilm Grown Using a Drip Flow Biofilm Reactor with Low Shear and Continuous Flow. ASTM International. 2008 [Google Scholar]
- 27.James GA, Agostinho AM, Pulcini E. In vitro models for the growth and analysis of chronic wound biofilms. In: Sen CK, editor. Wound Healing Society Yearbook: Advances in wound care. Mary Ann Leibert; 2010. [Google Scholar]
- 28.McGuiness W, Vella E, Harrison D. Influence of dressing changes on wound temperature. J Wound Care. 2004;13(9):383–385. doi: 10.12968/jowc.2004.13.9.26702. [DOI] [PubMed] [Google Scholar]
- 29.Romanelli M, Gaggio G, Coluccia M, et al. Technological advances in wound bed measurements. Wounds. 2002;14(2):58–66. [Google Scholar]
- 30.Trengove NJ, Langton SR, Stacey MC. Biochemical analysis of wound fluid from nonhealing and healing chronic leg ulcers. Wound Repair Regen. 1996;4(2):234–239. doi: 10.1046/j.1524-475X.1996.40211.x. [DOI] [PubMed] [Google Scholar]
- 31.Trengove NJ, Stacey MC, MacAuley S, et al. Analysis of the acute and chronic wound environments: the role of proteases and their inhibitors. Wound Repair Regen. 1999;7(6):442–452. doi: 10.1046/j.1524-475x.1999.00442.x. [DOI] [PubMed] [Google Scholar]
- 32.Bowler PG. The 10(5) bacterial growth guideline: reassessing its clinical relevance in wound healing. Ostomy Wound Manage. 2003;49(1):44–53. [PubMed] [Google Scholar]
- 33.Cohen HA, Amir J, Matalon A, et al. Stethoscopes and otoscopes: a potential vector of infection? Fam Pract. 1997;14(6):446–449. doi: 10.1093/fampra/14.6.446. [DOI] [PubMed] [Google Scholar]
- 34.Neely AN. A survey of Gram-negative bacteria survival on hospital fabrics and plastics. J Burn Care Rehabil. 2000;21(6):523–527. doi: 10.1097/00004630-200021060-00009. [DOI] [PubMed] [Google Scholar]
- 35.Neely AN, Maley MP. Survival of enterococci and staphylococci on hospital fabrics and plastic. J Clin Microbiol. 2000;38(2):724–726. doi: 10.1128/jcm.38.2.724-726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flemming HC, Neu TR, Wozniak DJ. The EPS matrix: the ‘house of biofilm cells’. J Bacteriol. 2007;189(22):7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller SW, Krebsbach LE. Impact of an antimicrobial-impregnated gauze dressing on surgical site infections including methicillin-resistant Staphylococcus aureus infections. Am J Infect Control. 2008;36(9):651–655. doi: 10.1016/j.ajic.2007.12.005. [DOI] [PubMed] [Google Scholar]