Abstract
In the Single Protein Production (SPP) method, all E. coli cellular mRNAs are eliminated by the induction of MazF, an ACA-specific mRNA interferase. When an mRNA for a membrane protein, engineered to have no ACA sequences without altering its amino acid sequence, is induced in the MazF-induced cells, E. coli is converted into a bioreactor producing only the targeted membrane protein. Here we demonstrate that three prokaryotic inner membrane proteins, two prokaryotic outer membrane proteins, and one human virus membrane protein can be produced at very high levels, and assembled in appropriate membrane fractions. The condensed SPP (cSPP) system was used to selectively produce isotope-enriched membrane proteins for NMR studies in up to 150-fold condensed culture without affecting protein yields, providing more than 99% cost saving for isotopes. As a novel application of the cSPP system for studies of membrane proteins prior to purification we also demonstrate, for the first time, fast detergent screening by microcoil NMR and well-resolved NMR spectra of several targeted integral membrane proteins obtained without purification.
Keywords: cSPP, membrane protein, NMR
Introduction
A major bottleneck in structural studies of membrane proteins is their expression and subsequently purification to homogeneity. To circumvent this problem, we have developed a novel technology to express in E. coli cells a single membrane protein of interest in high yield. We have previously demonstrated that a single soluble protein can be produced in E. coli by the method termed the single-protein-production (SPP) (Suzuki et al., 2005). In this system, MazF, an mRNA interferase, is induced, which functions as an ACA-specific endoribonuclease to eliminate almost all cellular mRNAs. Subsequent induction of an ACA-less RNA for a target protein results in the production of only the target protein in the cells, without producing any other cellular proteins. Importantly, any mRNAs can be engineered to have no ACA sequences, without altering the amino acid sequences of the proteins encoded by the mRNAs.
Here we apply the condensed SPP (cSPP) technology to a number of integral membrane proteins, demonstrating that they are produced at very high levels and properly assembled in the membrane. We further demonstrate that these cell cultures can be condensed up to 150 fold without affecting protein yields, resulting in more than 99% cost saving. This cSPP system was used to produce isotope-enriched membrane proteins for NMR studies. The membrane fraction, simply dissolved in an appropriate detergent, was then used directly for NMR studies of the membrane proteins without purification (Scheme 1). Well resolved 2D TROSY [HN-15N]-HSQC (Pervushin et al., 1997) spectra of these membrane proteins have been obtained, demonstrating that the production of membrane proteins by the SPP method is highly cost effective for isotope enrichment and NMR structural studies.
Scheme 1. Schematic representation of the concept of SPP production of a membrane protein.
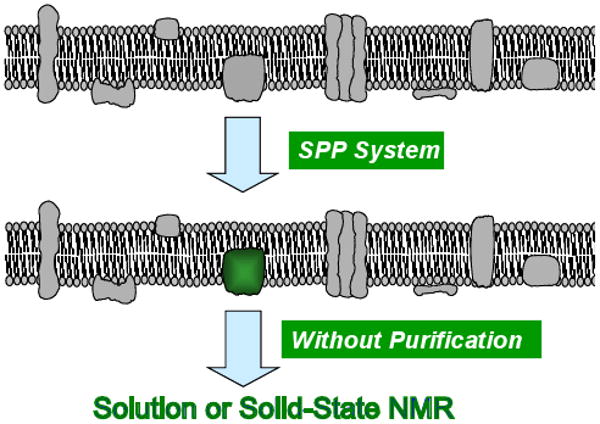
Only a single targeted membrane protein is produced, allowing specific enrichment with isotopes (e.g., 13C, 15N, etc.), as indicated in green.
Since the yields of membrane proteins by the cSPP method are quite high, membrane proteins produced with the cSPP system can be also purified to homogeneity in a few steps. Such samples may be used for NMR and crystallization experiments, as well as for other biophysical and biochemical characterizations. The application of the cSPP method for membrane proteins opens a new exciting avenue for structural and functional studies of membrane proteins.
Materials and Methods
Protein expression
E. coli BL21 (DE3) cells, transformed with pACYCmazF (Suzuki et al., 2005) and pColdI(SP4) (Suzuki et al., 2007) plasmids harboring the target gene, were grown in M9-glucose medium at 37°C. When OD600 reached 0.5 - 0.6 units, the culture was chilled on ice for 5 min and shifted to 15 °C for 45 min in order to acclimate the cells to cold temperature. After the cold-shock treatment, the expression of both MazF and the target gene were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG).
Membrane preparation
The cell pellet from a 1L non-condensed culture was suspended in 10 ml of 50 mM Tris buffer (pH 7.4). Cells were then lysed by a French press at 15,000 psi. The membrane fraction was collected by centrifugation at 100,000 × g for 1 hr at 4 °C. The membrane pellet was resuspended in 1 ml of 50 mM Tris buffer (pH 7.4) by sonication. Fifty micro liter of the membrane suspension was used for further separation of the inner and outer membranes. The remaining suspension was divided into two equal 500-μl fractions. These fractions were centrifuged at 100,000 × g for 1 hr at 4°C and the membrane pellets were stored at -80°C.
Separation of inner and outer membranes
Two hundred micro liter of 50 mM Tris buffer (pH 7.4) was added to the 50-μl membrane suspension. Subsequently, 250 μl of 1% Sarkosyl was added and the mixture was incubated at room temperature for 20 min to dissolve the inner membrane proteins. The inner membrane fraction was then separated as the supernatant from the pellet of the outer membrane fraction by centrifugation at 135,000 × g for 30 min at 4 °C. These membrane fractions were analyzed by SDS-PAGE to determine the localization of a target membrane protein. For NMR study of outer membrane proteins (OmpA and OmpX), the membrane pellet prepared as described in the previous section was used to isolate the outer membrane fraction.
Protein purification by Ni-NTA column
Membrane pellet from 500 ml non-condensed culture was suspended in 500 μl of 25 mM MES (pH 6.0) buffer containing 10% LOPG, incubated at room temperature for 1 hr, and centrifuged at 135,000 × g for 30 min at 4 °C. The supernatant was then mixed with 1 ml of Ni-agarose slurry (50% w/v) and incubated at 4 °C for 1 hr with gentle shaking. The mixture was poured into an empty column, which was washed two times with 4 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0). The slurry of the column was resuspended in 1 ml of wash buffer, transferred to a microcentrifuge tube, and centrifuged (1000 × g) to remove the supernatant. The target protein was purified by suspending the slurry in 500 μl of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0), containing 10% LOPG and incubated at 4 °C for 1 hr. The mixture was transferred to a centrifugal filter tube (Millipore) and the target protein was obtained in the supernatant by centrifugation (12,000 × g).
NMR measurements
NMR measurements of uniformly 15N-labeled YaiZ and YnaJ in 10 % LOPG were obtained using an 800 MHz Bruker AVANCE spectrometer with a cryoprobe at 40 °C. TROSY [HN -15N] HSQC(Pervushin et al., 1997) NMR data were acquired using spectral widths of 14 ppm in 1H dimension and 32 ppm in 15N dimension. The matrix size of collected spectra was 1024 × 256 total data points.
In all NMR experiments, data processing involved zero-filling and weighting by sine squared window function in acquisition and evolution dimensions. All NMR spectra were processed and examined using the program NMR-Pipe/NMR-Draw software package(Delaglio et al., 1995). The program SPARKY(Goddard and Kneller) was used for data visualization and analysis. Chemical shifts were referenced to external 2, 2-dimethyl-2-silapentane-5-sulfonic acid (DSS).
Results
Expression of two inner membrane proteins from E. coli
E. coli YaiZ and YnaJ, predicted to be inner membrane proteins, were chosen for development of the cSPP method for preparing membrane proteins and for membrane protein NMR studies without purification. YaiZ and YnaJ consist of 70 and 85 amino-acid residues, respectively. All the ACA sequences in the corresponding gene DNA sequences were altered to non-MazF cleavable sequences without changing the amino acid sequences. The ACA-less genes were then inserted into pCold vectors, including pColdI(SP4), pColdIII(SP4) and pColdIV(SP4). These three pCold vectors are similar to each other, except in the multiple cloning region. pColdI (SP4) has a translational enhancing element (TEE), 6-His tag, and a Factor Xa cleavage site. pColdIII(SP4) has only the TEE element, while pColdIV(SP4) has none of the three elements, producing a target protein without any extra amino acids(Suzuki et al., 2007).
pColdI(SP4)yaiZ was transformed into E. coli BL21(DE3) cells which contain the pACYCmazF plasmid, harboring a lac-inducible mazF gene(Suzuki et al., 2005). A 1-liter exponentially growing culture at 37 °C (OD600 of 0.5) was cold-shocked at 15° C for 1 hr, then 1 mM IPTG was added to induce both of YaiZ and MazF endoribonuclease. The culture was further incubated at 15 °C for 3 hr, followed by centrifuging the culture (5, 000 × g, 15min, at 4°C) to collect the cells. The cells were washed once with 50 ml of 1 × M9 (without NH4Cl) and suspended in 50 ml of M9 minimal medium containing 15NH4Cl to make the final culture 20-fold condensed. The condensed culture was then incubated at 15 °C for 24 additional hours to produce uniformly 15N-enriched YaiZ (Suzuki et al., 2007). The total membrane fraction was prepared from the condensed culture, and the inner membrane and outer membrane fractions were separated as described in Experimental Section. SDS polyacrylamide gel electrophoresis (PAGE) analyses are shown in Figure 1a. YaiZ was well expressed by the cSPP system [compare lane 2 (induced) with lane 1 (0 hr)], which remained in the supernatant fraction after low speed centrifugation (lane 3), indicating that YaiZ was not in inclusion bodies. This fraction was then ultra-centrifuged to separate the cytoplasmic soluble fraction (lane 4) and the membrane fraction (lane 5). YaiZ was found to be exclusively localized in the membrane fraction, and furthermore, exclusively in the inner membrane fraction (lane 6), but not in the outer membrane fraction (lane 7). Notably, following induction YaiZ is the most abundant protein in the inner membrane.
Figure. 1. Expression of YaiZ and YnaJ by the SPP system.
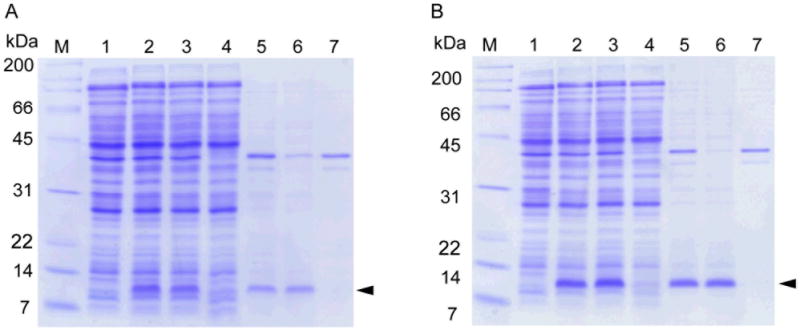
(A), An ACA-less gene for YaiZ was expressed at 15 °C using pColdI(SP4), in the cells expressing MazF from pACYCmazF. M, molecular weight markers; lane 1, at 0 hr when 1 mM IPTG was added. Protein induction was continued overnight. Lane 2, whole cell; lane 3, cell lysate after low speed centrifugation; lane 4, soluble fraction; lane 5, whole membrane fraction; lane 6, inner membrane; and lane 7, outer membrane. (B), The ACA-less gene for YnaJ was expressed from pColdI(SP4) in the same manner as described for YaiZ above. M, molecular weight markers; lane 1, at 0 hr when 1 mM IPTG was added. Protein induction was continued overnight. Lane 2, whole cell; lane 3, cell lysate after low speed centrifugation; lane 4, soluble fraction; lane 5, whole membrane fraction; lane 6, inner membrane; and lane 7, outer membrane. M: molecular weight marker. Positions of YaiZ or YnaJ are designated by arrowheads.
The cSPP expression of YnaJ was carried out in the same manner as described for YaiZ, with 20-fold condensation. SDS polyacrylamide gel electrophoresis (PAGE) analysis (Figure 1b) demonstrates that YnaJ was expressed at high levels (lane 2), and remains in the low speed centrifugation supernatant (lane 3) after removing cell debris and inclusion bodies. Further fractionation demonstrates its exclusive localization in the total membrane fraction (lane 5), but not in the cytoplasmic fraction (lane 4). YnaJ is further exclusively localized in the inner membrane (lane 6) but not in the outer membrane fractions (lane 7).
Expression of outer membrane proteins, OmpA and OmpX
ACA-less genes for two E. coli outer membrane proteins, OmpA and OmpX, were generated and cloned into pColdIV(SP4). E. coli strain BL21(DE3) was engineered to be ompA- and ompF- for two major outer membrane proteins, a strategy aimed at enhancing target outer membrane protein expression. Both OmpA and OmpX were expressed at high levels (compare lane 2 with lane 1, in each of Figure 2a and 2b) and assembled into the outer membrane, rather than as inclusion bodies (Arora et al., 2001; Fernandez et al., 2001). In these production systems, OmpA and OmpX are the major proteins in the outer membrane fraction (lane 7, Figure 2a and 2b).
Figure. 2. Expression of E. coli OmpA(1-176) and OmpX by the cSPP system in E. coli strain ompA-/ompF- BL21 (DE3).
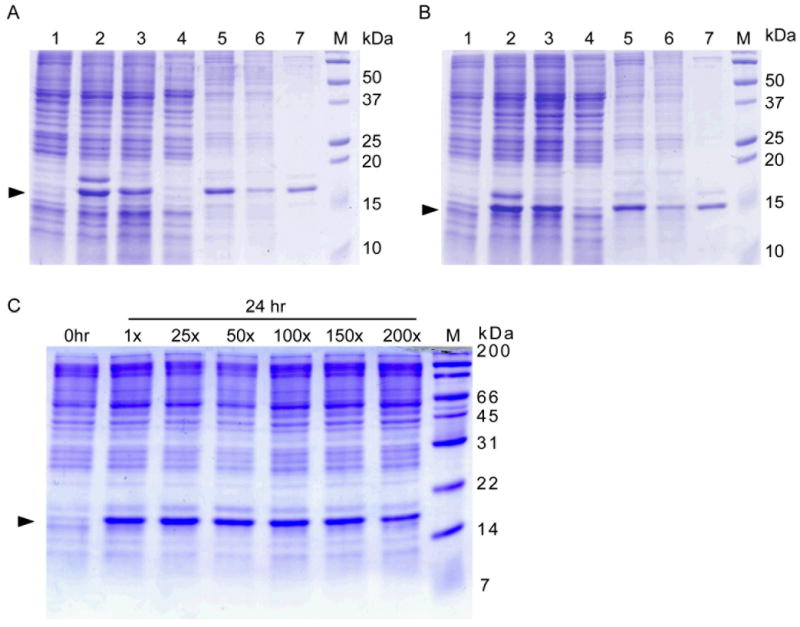
(A), ACA-less gene for OmpA(1-176) was expressed from pColdIV(SP4) along with MazF from pACYCmazF in the same manner as described above. Lane 1, at 0 hr when 1 mM IPTG was added. The proteins were induced overnight. Lane 2, whole cell; lane 3, cell lysate after low speed centrifugation; lane 4, soluble fraction; lane 5, whole membrane fraction; lane 6, inner membrane; and lane 7, outer membrane. M: molecular weight marker. The position of OmpA (1-176) is indicated by an arrowhead. (B), ACA-less gene for OmpX was expressed from pColdIV(SP4) along with MazF from pACYCmazF. Lane 1, at 0 hr when 1 mM IPTG was added. The proteins were induced overnight. Lane 2, whole cell; lane 3, cell lysate after low speed centrifugation; lane 4, soluble fraction; lane 5, whole membrane fraction; lane 6, inner membrane; and lane 7, outer membrane. M: molecular weight marker. The position of OmpX is indicated by an arrowhead. (C), Expression cultures of OmpX can be condensed up to 150 times without sacrificing protein yield. OmpX was expressed from pColdIV(SP4) containing the ACA-less OmpX gene together with MazF from pACYCmazF. The cell culture was grown at 37 °C to an OD600 of 0.5, shifted to 15 °C for 45 min, and condensed to the different levels as shown on top of the figure. Expression of OmpX was then induced by 1 mM IPTG for 24 hr at 15 °C. M: molecular weight marker. The position of OmpX are is shown by an arrowhead.
Additionally, since it is very important to maintain the M9 medium at pH 7.4, we further explored the cSPP system by using 2 × M9 salt to examine how much culture condensation could be achieved without affecting protein yields using OmpX as a model protein. As shown in Figure 2c, the cell culture can be condensed by up to 150 fold (1L culture is reduced to 6.7 ml) without reducing protein yield. This in turn reduces the cost for isotope labeling to only about 0.7% of the cost by the conventional method (> 99% cost saving for isotopes used for the culture).
Detergent screening with microcoil NMR
YaiZ and YnaJ were expressed at high levels as described above, with exclusive localization in the inner membrane of E. coli. Detergent screening (Krueger-Koplin et al., 2004; Sorgen et al., 2002) was carried with < 10 microliter volumes of detergent-extracted samples using a 1-mm microcoil NMR probe at 600 MHz(Aramini et al., 2007; Zhang et al., 2008). Membrane pellets, equal to 500 ml non-condensed culture, were resuspended in 500 μl of 50 mM Tris.HCl (pH 7.4), divided into 50 μl each, and the centrifuged at 135,000 × g for 30 min at 4 °C. Each cell pellet was then suspended in 50 μl of 25 mM MES buffer (pH 6.0), containing 5% of 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine (LMPC), 1-myristoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] (LMPG), 1-oleoyl-2-hydroxysn-glycero-3- phosphocholine (LOPC), 1-oleoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] (LOPG), 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol) (LPPG), n-octyl-β-D-glucopyranoside (β-OG) and Zwittergent 3-16, respectively(Krueger-Koplin et al., 2004). The membrane suspensions were incubated at room temperature for 1 hr, followed by centrifugation at 135, 000 × g for 30 min at 4 °C to collect the supernatants. 5% 2H2O was added to these samples, which were then loaded into 1-mm microcoil NMR tubes using a robotic liquid handling robot. Preliminary detergent screening was carried out on unpurified protein by collecting 1D [1H-15N]-HSQC NMR spectra of each sample using a 1-mm microcoil NMR probe(Aramini et al., 2007; Zhang et al., 2008). Among the screened detergents, samples extracted by LMPG, LOPG and LPPG give the most promising spectra for both YaiZ and YnaJ (Figure 3). After the 1D detergent screening, further optimization of the detergent concentration and the sample temperature was carried out by collecting 2D [1H-15N]-TROSY-HSQC spectra on 5-mm NMR tubes with sample volume of about 300 μl (data not shown). Using these data, 10% LOPG was found to be the optimal detergent condition for purification and further NMR structural studies of both YaiZ and YnaJ.
Figure. 3. 1D [1H-15N]-HSQC NMR screening of YaiZ and YnaJ extracted by seven detergents at 20 °C.
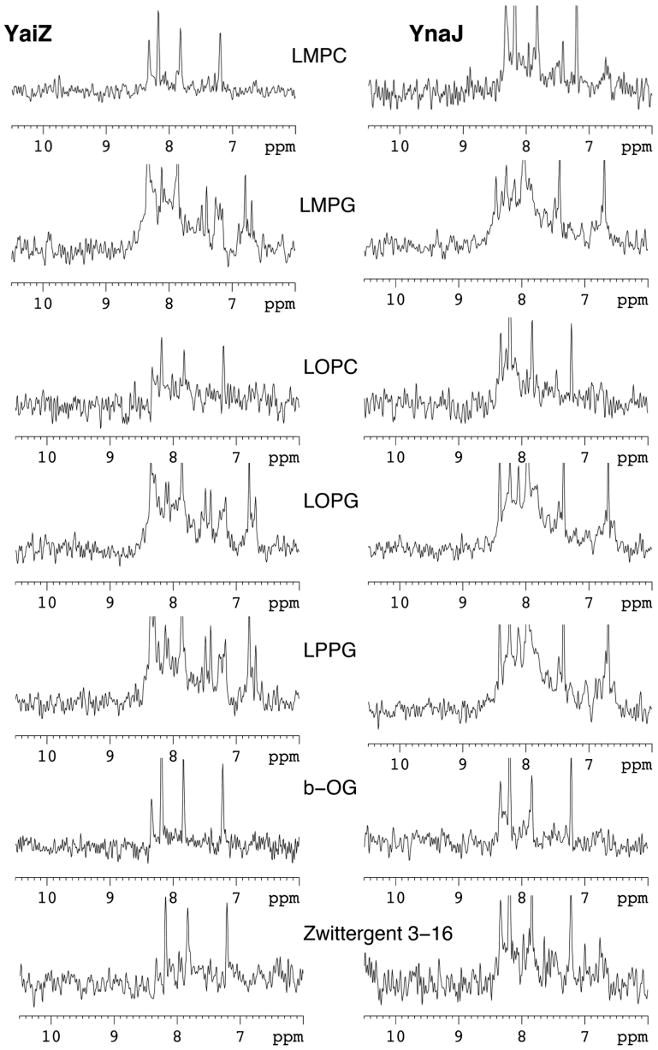
The detergents screened include: LMPC (5%), LMPG (5%), LOPC (5%), LOPG (5%), LPPG (5%), β-OG (5%) and Zwittergent 3-16 (5%). Since the target proteins are selectively isotope-enriched by the SPP system, these data were obtained without any purification of the target protein aside from the simple detergent extraction.
NMR spectroscopy without purification
NMR spectra were recorded for uniformly 15N-enriched YaiZ. [HN -15N] TROSY NMR spectra at high magnetic fields, which have sharper line shapes for relatively slowly tumbling systems(Fernandez et al., 2001; Pervushin et al., 1997), are generally required for studies of such micelle-solubilized membrane proteins. Since the cSPP system allows isotopic-labeling of target membrane proteins with little or no labeling of other endogenous membrane proteins, it can be used to study membrane proteins directly in membrane fractions using solid state NMR(Schneider et al., 2009), or by simply extracting membrane fractions with appropriate detergents using solution-state NMR, without any further purification steps (Scheme 1).
YaiZ was also cloned into pColdIII(SP4) and a uniformly 15N-enriched sample was prepared as described below. A 500-ml exponentially growing culture at 37°C (OD600 of 0.5 units) was cold-shocked at 15° C for 1 hr, then 1 mM IPTG was added to induce MazF mRNA interferase. The culture was further incubated at 15° C for 3 hr, followed by 20-fold condensation in M9 minimal medium (to 25 ml) containing 15NH4Cl. The condensed culture was incubated overnight at 15 °C to produce YaiZ with uniform 15N enrichment. The membrane fraction from the condensed culture expressing YaiZ was resuspended in 500 μl of 25 mM MES buffer (pH 6.0) containing 95% H2O / 5% 2H2O and 10% LOPG, followed by ultracentrifugation at 135,000 × g for 30 mins at 4 °C. The supernatant was then used for obtaining NMR spectra. The concentration of YaiZ in this sample was ∼ 0.2 mM. YnaJ was also cloned into pColdIII(SP4) and a uniformly 15N-enriched sample was prepared in the same procedure as described for YaiZ, with the final concentration of ∼ 0.2 mM.
TROSY [HN -15N]]-HSQC spectra of these 15N-enriched YaiZ and YnaJ samples were collected on 800 MHz spectrometer at 40 °C (Figures 4a and 4b). These well-resolved spectra demonstrate the feasibility of obtaining NMR data for membrane proteins without purification beyond a simple membrane extraction. Notably, there are four tryptophan residues in YaiZ and one in YnaJ, and all of the expected indole Nε1H resonances of tryptophan residues are observed in these 2D TROSY-[HN-15N] -HSQC spectra.
Figure. 4. NMR spectroscopy and purification of YaiZ and YnaJ.
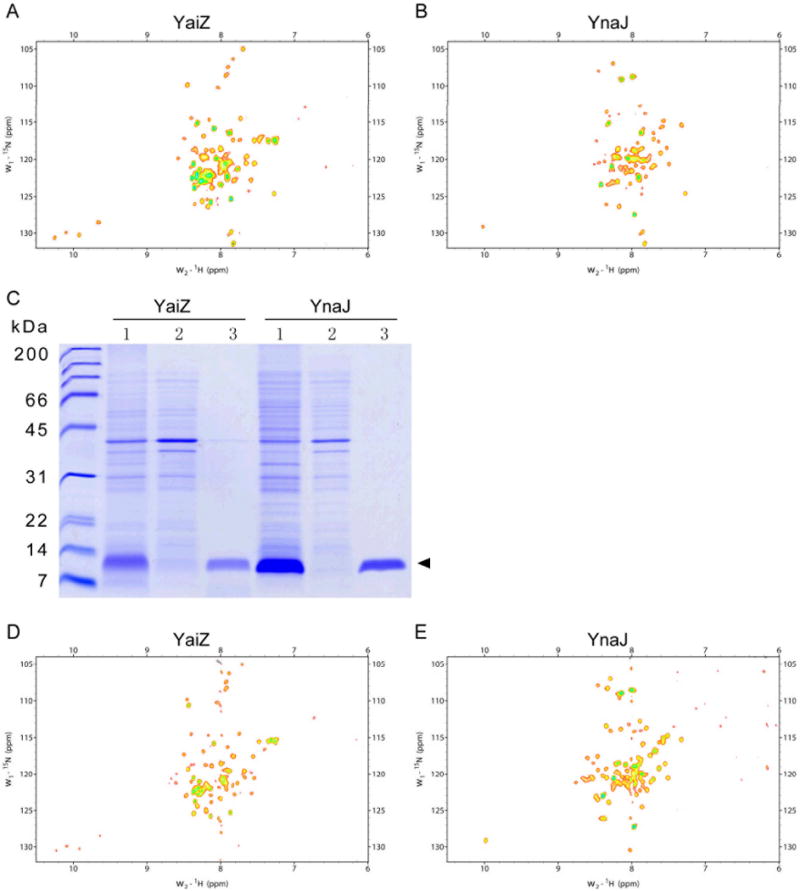
(A), 800 MHz TROSY-[HN-15N]-HSQC NMR spectrum obtained without substantial purification for uniformly 15N-enriched YaiZ. (B), 800 MHz TROSY-[HN-15N]-HSQC NMR spectrum obtained without substantial purification for uniformly 15N-enriched YnaJ. Target protein (YaiZ or YnaJ) was produced using the cSPP system and simply extracted from the membrane fraction by 25 mM MES buffer, pH 6.0, containing 10% LOPG in 95 % H2O / 5 % 2H2O. (C), One step purification of uniformly 15N-enriched YaiZ and YnaJ in 10% LOPG by a Ni-NTA column. YaiZ or YaiJ produced by the cSPP system was extracted from the membrane fraction by 10% LOPG (1-oleoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)]) in 25 mM MES buffer, pH 6.0 (lane 1), and then applied to a Ni-NTA column; lane 2, flow-through fraction; lane 3, the fraction eluted with imidazole from the column. (D), 800 MHz TROSY-[HN-15N]-HSQC NMR spectrum of purified YaiZ in 25 mM MES buffer, pH 6.0, containing 10% LOPG in 95 % H2O / 5 % 2H2O. (E), 800 MHz TROSY-[HN-15N]-HSQC NMR spectrum of purified YnaJ in 25 mM MES buffer, pH 6.0, containing 10% LOPG in 95 % H2O / 5 % 2H2O. (F) The superimposition of spectra A (black) and D (red). (G) The superimposition of spectra B (black) and E (red).
NMR spectroscopy of YaiZ and YnaJ purified by Ni-NTA affinity chromatography
Since both YaiZ and YnaJ, expressed with the pColdI(SP4) vector, have a hexa-His purificaiton tag at their N-terminal ends, they can each be purified using a Ni-NTA column. YaiZ and YnaJ were extracted from the membrane fraction by incubating in 25 mM MES buffer (pH 6.0) containing 10% LOPG at room temperature for 1 hr, followed by centrifugation at 135,000 × g, 4 °C, for 30 min (lanes 1, Figure 4c). The supernatant was then purified by Ni-NTA affinity purification. Almost all of the YaiZ (or YnaJ) binds to the column while all other proteins were collected in the flow-through fraction (lanes 2, Figure 4c). Purified YaiZ (or YnaJ) was eluted with a buffer containing 10% LOPG (lanes 3, Figure 4c). Far ultraviolet circular dichroism (CD) spectra of Ni-NTA purified YaiZ, YnaJ were recorded to confirm that the proteins expressed by the SPP system are folded into well-ordered structures (see supporting information, Figure 2a and 2b). TROSY [HN -15N]]-HSQC spectra of these samples of purified 15N-enriched YaiZ and YnaJ were collected on 800 MHz spectrometer at 40 °C (Figures 4d and 4e).
It is of interest to note that the spectra for both purified and unpurified proteins look very similar, however, a few differences can be observed (Figure 4f and 4g). The quality of 2D TROSY 15N-HSQC of purified samples is greatly improved as the peaks become sharper with narrower line-widths and better signal-to-noise. One factor contributing towards this improvement is that the sample becomes more homogenous on purification. Purification also removes phospholipids and proteins that may interact with the protein of interest, resulting in line broadening. In addition, some of the peaks that are observed in the unpurified spectra but not in the purified one may be due to the fact that a minor amount of the isotope-labeled impurities. Also, subtle changes in the chemical shifts between the purified and unpurified peaks may be due to the difference in the chemical environments of purified and unpurified proteins and/or subtle structural differences resulting from the purification process.
Discussion
High yields of four E. coli integral membrane proteins were achieved by the cSPP system, as well as KcsA and E1B19K (see supporting information, Figure 1). This is the first demonstration of obtaining isotope-filtered NMR spectra (i.e., 2D TROSY [HN-15N]-HSQC spectra) of isotope-enriched membrane proteins directly extracted from the membrane fraction without any additional purification steps. We have shown previously, using a cold-shock vector for selective expression of a target protein, that a 13C, 15N-enriched soluble cytoplasmic protein could be produced and used to obtain extensive backbone resonance assignments and secondary structure using whole cell extracts without purification (Qing et al., 2004). The present study demonstrates, using the cSPP system, selective isotope-enrichment and NMR analysis of a single targeted membrane protein.
Choice of detergent plays an important role towards obtaining a good sample suitable for NMR studies of membrane proteins. To screen detergents that yield the best NMR spectra for membrane proteins, we used a high-throughput detergent screening method using a microNMR probe. Zhang et al have previously shown the advantages of using microscale detergent screening to identify the best detergent condition for the NMR studies of purified proteins(Zhang et al., 2008). Importantly, the cSPP system allows one to screen detergent without protein purification. Using unpurified proteins for detergent screening ensures a faster method for screening a variety of detergents thereby reducing both the cost and time involved to purify the proteins. 1D and 2D 15N-TROSY HSQC screening for 12 different detergents is typically around 18-20 hr with a sample volume of about 30 μl. Once the best detergent condition is identified NMR samples can be prepared by simply dissolving the membrane fractions from prepared by the cSPP methods in appropriate detergents, a well-resolved TROSY-HSQC spectrum could be obtained.
Though unpurified samples gave reasonably good spectra, a one-step purification significantly improves the quality of the spectra. Unpurified proteins contain phospholipids and other proteins which can contribute towards line-broadening of the peaks. Therefore, one-step purification by the affinity column chromatography helps to eliminate these unwanted interaction maintaining membrane proteins in an environment that is closer to its physiological state. Notably, traditional methods used for purification of membrane proteins are often tedious and time-consuming. Proteins obtained through this one-step purification process yield high quality HSQC spectra which is likely to generate good membrane protein structures.
The SPP-NMR technology for membrane proteins is quite general, and can be applied in a wide-range of NMR-based structure-function studies. For example, backbone resonance assignments obtained in this manner would provide the locations of alpha helices and beta strands in integral membrane protein structures, and key information about biochemical function and mechanisms. The high quality of these TROSY-HSQC spectra (Figures 4a and 4b) further suggests that it may be possible to determine complete three-dimensional structures of membrane proteins without substantial purification, avoiding the need for reconstitution of certain membrane proteins for structural studies. TROSY-HSQC spectra of 15N-enriched membrane proteins, obtained without substantial purification, can also be used to optimize detergent conditions for use in subsequent purification of the target protein for biochemical studies, antibody production, and NMR studies, as well as for crystallization and three-dimensional structure determination by X-ray crystallography.
In summary, the cSPP technology used provides a number of advantages over the conventional methods used for the structural studies of membrane proteins. First, NMR structure determination of membrane proteins can be carried out without purification resulting in nearly 100% protein yield for NMR structural study. Secondly, a substantial reduction of the cost for isotopes (more than 99%) may be achieved, as the cell culture can be condensed up to 150 fold without affecting protein yields. Thirdly, in the cSPP system, 2H2O has no inhibitory effect on protein production, yielding high-quality 2H,13C,15N-enriched samples of soluble proteins(Schneider et al., 2009). For small soluble proteins, such perdeuterated samples allow reliable automated analysis of resonance assignments and 3D structures. Indeed, not only deuterium but also other toxic amino acid analogues such as selenomethionine and fluorophenylalanine do not exhibit any inhibitory effects on the protein production, since cell growth is completely arrested in the SPP cells (Suzuki et al., 2006). Fourthly, Solid-state NMR is one of the very few non-invasive methods that can be used for membrane protein studies(Schneider et al., 2009). The membrane fraction prepared by the cSPP method is ideal for solid-state NMR study of membrane proteins. Fifthly, the cSPP system allows preparation of membrane proteins in a minimally altered form. Sixthly, the cSPP-NMR system, with selective isotope-enrichment of target protein, provides means for screening detergent buffer conditions to find conditions suitable for protein purification prior to actually initiating the purification process. Taking together, the work shown in the present paper and applicable technologies described above demonstrate that the cSPP-NMR system has great potential for structural studies of membrane proteins, and in addressing some of the most challenging problems in modern structural biology.
Supplementary Material
Acknowledgments
We thank Dr. M. Suzuki for his efforts in initial phases of this work, and Dr. A. McDermott for providing a clone of the gene for KcsA. This work was supported by the National Institutes of Health Grants U54 GM074958 (G.T.M. and M. I.), U54 GM75026 (G.T.M. and M. I.), 1R01 GM085449 (M.I.), and R37 CA53370 (E. W.), and partially by a research fund from Takara-Bio., Inc.
Abbreviations used
- LMPC
1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine
- LMPG
1-myristoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)]
- LOPC
1-oleoyl-2-hydroxysn-glycero-3- phosphocholine
- LOPG
1-oleoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)]
- LPPG
1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)]
- βOG
n-octyl-β-D-glucopyranoside
- Zwittergent 3-16
Hexadecyl-N-sulfobetaine
- cSPP
condensed single protein production system
References
- Aramini JM, Rossi P, Anklin C, Xiao R, Montelione GT. Microgram-scale protein structure determination by NMR. Nat Methods. 2007;4:491–493. doi: 10.1038/nmeth1051. [DOI] [PubMed] [Google Scholar]
- Arora A, Abildgaard F, Bushweller JH, Tamm LK. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Adeishvili K, Wuthrich K. Transverse relaxation-optimized NMR spectroscopy with the outer membrane protein OmpX in dihexanoyl phosphatidylcholine micelles. Proc Natl Acad Sci U S A. 2001;98:2358–2363. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G, Ma LC, Khorchid A, Swapna GV, Mal TK, Takayama MM, Xia B, Phadtare S, Ke H, Acton T, et al. Cold-shock induced high-yield protein production in Escherichia coli. Nat Biotechnol. 2004;22:877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- Schneider WM, Tang Y, Vaiphei ST, Mao L, Shen Y, Raman S, Baker D, Inouye M, Roth MJ, Montelione GT. Efficient Production of Perdeuterated Soluble and Membrane Proteins for NMR Studies. Nature Methods. 2009 doi: 10.1007/s10969-010-9083-x. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgen PL, Hu Y, Guan L, Kaback HR, Girvin ME. An approach to membrane protein structure without crystals. Proc Natl Acad Sci U S A. 2002;99:14037–14040. doi: 10.1073/pnas.182552199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Mao L, Inouye M. Single protein production (SPP) system in Escherichia coli. Nat Protoc. 2007;2:1802–1810. doi: 10.1038/nprot.2007.252. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Roy R, Zheng H, Woychik N, Inouye M. Bacterial bioreactors for high yield production of recombinant protein. J Biol Chem. 2006;281:37559–37565. doi: 10.1074/jbc.M608806200. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Zhang J, Liu M, Woychik NA, Inouye M. Single protein production in living cells facilitated by an mRNA interferase. Mol Cell. 2005;18:253–261. doi: 10.1016/j.molcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Horst R, Geralt M, Ma X, Hong WX, Finn MG, Stevens RC, Wuthrich K. Microscale NMR screening of new detergents for membrane protein structural biology. J Am Chem Soc. 2008;130:7357–7363. doi: 10.1021/ja077863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


