Abstract
The early B-cell factor (EBF) transcription factors are central regulators of development in several organs and tissues. This protein family shows low sequence similarity to other protein families, which is why structural information for the functional domains of these proteins is crucial to understand their biochemical features. We have used a modular approach to determine the crystal structures of the structured domains in the EBF family. The DNA binding domain reveals a striking resemblance to the DNA binding domains of the Rel homology superfamily of transcription factors but contains a unique zinc binding structure, termed zinc knuckle. Further the EBF proteins contain an IPT/TIG domain and an atypical helix-loop-helix domain with a novel type of dimerization motif. The data presented here provide insights into unique structural features of the EBF proteins and open possibilities for detailed molecular investigations of this important transcription factor family.
Keywords: Cell Differentiation, Crystal Structure, Epigenetics, Helix-loop-helix Transcription Factors, Protein-DNA Interaction, Protein Domains, Transcription Factors, Tumor Suppressor, Early B-cell Factor 1 (EBF1), Early B-cell Factor 3 (EBF3)
Introduction
Transcription factors bind to specific DNA sequences and thereby control the transfer of genetic information from DNA to mRNA. The process of eukaryotic transcription is highly regulated and requires fine-tuned machineries where transcription factors play a pivotal role as DNA binders. In addition, co-activating, chromatin-remodeling, and signaling proteins also play crucial roles in gene regulation without necessarily binding DNA directly but rather through a network of interactions within the transcription module.
The human early B-cell factor (EBF)2 transcription factor family is composed of four members (EBF1–4) characterized by a helix-loop-helix (HLH) motif with resemblance to those found in basic HLH proteins (1, 2). However, rather than a basic HLH, EBF utilizes a unique domain including a zinc coordination motif (3) to bind DNA as dimers to two pseudo-palindromic half-sites separated by two base pairs (4). Although EBF proteins were first identified in B-lymphocytes (1) and olfactory neurons (2), they have now been shown to be involved in the development and function of a large number of specialized cells including adipocytes (5) and osteoclasts (6).
The structured part of the EBF proteins is composed of a conserved N-terminal DNA binding domain (DBD), an IPT/TIG (immunoglobulin, plexins, transcription factors-like/transcription factor immunoglobulin) domain, and an atypical helix-loop-helix motif, more appropriately termed a helix-loop-helix-loop-helix motif (HLHLH) (Fig. 1A) (7). All four members of the EBF family show very high sequence similarity within the structured part, in particular EBF1 and EBF3 with 95% sequence identity over this region (8). An additional, probably unstructured, transactivation region, with lower conservation, is present in the C terminus. We set out to solve structures from all family members, and because of the high degree of sequence identity between the different EBF proteins, the structure of any member of the family members should be representative for the whole family.
FIGURE 1.
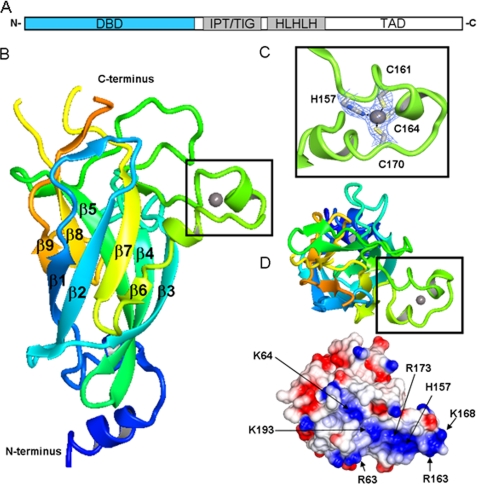
Structural representation of EBF1 DBD. A, schematic illustration of EBF domain composition. DBD depicted in blue. B, schematic representation of the DNA binding domain. The N terminus is colored blue and blends through to the C terminus in orange. The zinc knuckle is represented in the black box. C, close-up of the residues involved in zinc coordination present in the zinc knuckle. D, top-down view of the postulated DNA binding region showing the zinc knuckle region and electrostatic potential surface. Amino acids contributing to potential DNA binding groove are indicated. TAD, transactivation domain.
Here we report the first crystal structures for the EBF family. This is particularly valuable as the EBF family shares very little sequence similarity with other known transcription factors. Because most transcription factors have a well defined modular domain organization with discrete structure and function, we used a multiconstruct approach (9) to produce the different domains of EBF proteins for structural determination by x-ray crystallography. Our structures of the EBF DBD, IPT/TIG, and HLHLH modules represent together a complete coverage of the structured region of these proteins. Surprisingly, the DBD structure reveals striking resemblance to the N-terminal domain of the Rel homology superfamily of transcription factors. The structures of both the IPT/TIG and the HLHLH modules suggest context-dependent functions in dimerization and/or protein-protein interaction.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures can be found in the supplemental material and are available at the Structural Genomics Consortium (SGC) Structures Gallery.
Cloning, Protein Production, and Purification
The DNA sequences corresponding to residues Arg-10–Ser-250 (DBD) and Glu-258–Thr-351 (IPT/TIG) in human EBF1 (gi: 31415878), and Glu-251–His-386 (TIG-HLH) and Ser-250–Val-406 (TIG-HLHLH) in human EBF3 (gi: 53828926) were subcloned into a plasmid also coding for a N-terminal histidine tag and expressed in Escherichia coli cells. Bacterial cells were cultured in Terrific Broth medium. The DNA binding domain was also produced as selenomethionine-labeled using a minimal medium. Purification was performed in two steps using immobilized metal ion affinity chromatography followed by size exclusion chromatography. The N-terminal His tags were proteolytically removed with tobacco etch virus protease. Protein identities were confirmed by mass spectrometry, and samples were stored at −80 °C.
The DBD was complexed with the DNA duplex using primers 5′-GAGAGAGAGACTCAAGGGAATTGTGGCC-3′ and 5′-GGCCACAATTCCCTTGAGTCTCTCTCTC-3′ and subjected to size exclusion chromatography. The peak at a retention volume corresponding to a 2:1 complex was collected and concentrated.
Crystallization, Data Collection, Structure Determination, and Validation
Crystals of the DBD, IPT/TIG, and TIG-HLH were obtained by the sitting drop vapor diffusion method at 4 °C. The crystallization conditions were: DBD, 0.1 m MES, pH 5.8, 2.1 m ammonium sulfate; IPT/TIG, 0.1 m Tris, pH 9, 0.3 m trimethylamine n-oxide, and 23% PEG monomethyl ether 2000; TIG-HLH, 0.1 m Bis-Tris propane, pH 7.5, 0.2 m sodium formate, and 21% PEG 3350. Crystals of TIG-HLHLH were obtained by hanging drop vapor diffusion in 0.1 m Bis-Tris propane, pH 6.9, and 2.9 m NaCl. IPT/TIG crystals were soaked 48 h in ethylmercurithiosalicylate before freezing. All crystals were flash-frozen in liquid nitrogen after quick transfer to well solution containing: DBD, 20% glycerol; IPT/TIG, 1 mm ethylmercurithiosalicylate and 20% glycerol; and TIG-HLH and TIG-HLHLH, 20% ethylene glycol.
Data were collected at DIAMOND (I04), Oxfordshire, England (DBD), and BESSY (BL14-1), Berlin, Germany (IPT/TIG; TIG-HLH; and TIG-HLHLH). Data were indexed and integrated using XDS (10) or iMOSFLM (11) and scaled using XSCALE (10) or SCALA from the CCP4 suite (12). Structures were solved by selenium or mercury single anomalous dispersion using SOLVE (13) or SHELX (14) and by molecular replacement using PHASER (15) or MOLREP (16). Initial models were built using RESOLVE (13) or BUCCANEER (17). Structures were refined using REFMAC (18) or PHENIX (19). Refinement rounds were complemented with manual rebuilding using COOT (20). Data collection and refinement statistics are presented in supplemental Table S1. Structural similarity searches were carried out using the Dali server (21). Z-scores >6 were kept as highly significant. Figures were generated using ccp4mg (22).
Docking
Docking of DBD to DNA was performed using the program HADDOCK (23).
RESULTS
The DNA Binding Domain Reveals Similarities with the Rel Homology Domain Family of Transcription Factors
The structural characterization of the EBF DBD is of particular interest because it shows low or no homology to any other protein. A Dali search (21) with our EBF1 DBD structure returned three highly significant hits: NFAT1 (PDB: 1PZU), the p65 subunit of NF-κB (PDB: 2RAM), and TonEBP (PDB: 1IMH). Interestingly, all three of these proteins belong to the Rel homology domain family (RHD family; Pfam PF00554), which is a member of the p53-like clan of structures (CL0073). The DNA binding domain of EBF shows, at best, only a 14% sequence identity to the p65 subunit of NF-κB, and the structural relationship was not identified based on sequence.
The RHD is a protein domain found in eukaryotic transcription factors composed of two immunoglobulin-like β-barrel subdomains that grip the DNA in the major groove. The first subdomain, referred to as the N-terminal specificity domain, N-RHD, contains recognition loops that interact with DNA bases. The second subdomain, referred to as the C-terminal dimerization domain, C-RHD, contains in addition protein-protein interaction sites and is analogous to the EBF IPT/TIG domain.
The overall structure of EBF DBD reveals an antiparallel β-barrel comprised of nine strands arranged into sheets (Fig. 1B). Although superposition of the EBF DBD β-barrel core with other RHD-containing proteins reveals similar topology, the emanating loops are quite distinct. A characteristic feature in the EBF DBD is the presence of a zinc binding motif termed the “EBF zinc knuckle,” coined by Fields et al. (24). This HCCC zinc binding pattern is one of the more uncommon zinc coordination motifs (25) (Fig. 1C), and searches for similar structural motifs in the PDB with SPASM (26) using either the whole knuckle sequence or only the zinc-coordinating residues did not result in any significant hits.
In addition to the structure maintaining zinc-coordinating residues, mutational studies have identified a set of residues important for DNA interaction; Arg-163 has been shown to be absolutely required for DNA binding, while mutation of Lys-167, Lys-168, Asn-172, and Asn-174, all in the knuckle region, affect DNA binding negatively (24). The EBF zinc knuckle structure shows that the α-α motif positions these residues accessibly for interaction with DNA. Further examination of the electrostatic properties of the surface of the EBF DBD reveals a positively charged groove available for interaction with DNA (Fig. 1D). Residue His-235, also found to be crucial for DNA interaction by mutagenesis, is found in the vicinity of this area.
Structure of the IPT/TIG Domain
Structures were determined for the IPT/TIG domains of both EBF1 and EBF3 (PDB ID: 3MQI and 3MUJ, respectively) (Fig. 2A). Sequence alignment of these two IPT/TIG domains reveals 98% sequence identity, and the resulting crystal structures are very similar, with superposition showing root mean square Cα-Cα distance of 0.72 Å over an 83-residue range. The structures reveal an immunoglobulin-like fold, confirming that they belong to the IPT/TIG domain family (Pfam: TIG: PF01833), which is similar to the C-RHD subdomain (Fig. 2B). This strengthens the structural similarities with the RHD family. TIG domains are typically found in transcription factors but also in cell surface receptors such as Met and Ron (27) and typically exhibit three different topologies (28). The EBF structures presented here reveal a NF-κB-type TIG topology that has several roles; it is involved in DNA binding and/or dimerization but has also been implicated in protein-protein interactions.
FIGURE 2.
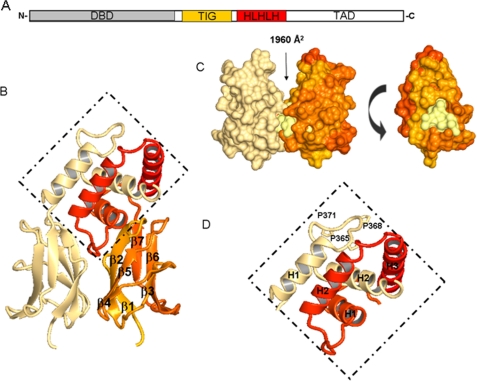
Structural representation of EBF3 IPT/TIG and HLHLH domains. A, schematic illustration of EBF domain composition. IPT/TIG and HLHLH domains are depicted in orange and red, respectively. B, schematic representations of the IPT/TIG and HLHLH domain dimer. Coloring for first chain is the continuation of the rainbow from Fig. 1 where the C terminus is in red, and the other chain is represented in cream. the HLHLH region is presented in the dotted diamond. β-Sheet numbering is shown for the IPT/TIG domain. C, the left-hand side is the representation of the IPT/TIG domain indicating the presence of a buried surface area. The right-hand side is a 90° turn (anticlockwise) of one of the monomers, revealing a hydrophobic core (lemon) situated at the interface of the two molecules. D, close-up of HLHLH region shows packing of α-helix 1 (H1) with α-helix 2 (H2) of adjacent monomers in this region. Proline residues present in the loop between H1 and H2 and available for interaction are also shown. TAD, transactivation domain.
The crystal packing in both the EBF1 and the EBF3 IPT/TIG crystals show formation of a dimer at the interface of strands β1, β2, and β4 (Fig. 2C). The dimer interface has a buried surface area of 1960 Å2 (29) and consists of a hydrophobic core surrounded by electrostatic interactions. This feature supports a role in dimerization and protein-protein interaction.
The closest structural homologue to the EBF IPT/TIG domain is the TIG domain in human calmodulin binding transcription activator 1, CAMTA1 (PDB: 2CXK). CAMTA1 is a member of a transcription factor family composed of a CG-1 DNA binding domain, the TIG domain, calmodulin binding IQ repeats, as well as ankyrin repeats (30). The biology of CAMTA1 is still largely to be explored, and the function of its TIG domain remains unknown.
New Fold for Atypical HLHLH Domain
Unlike the typical HLH motif containing proteins, the HLHLH domain of EBF proteins is not involved in DNA binding but rather in dimerization and transactivation. Evolutionarily, this module has expanded from being a double helix motif to a triple helix motif where the first and either of the following two helices are sufficient for dimer formation (31).
The structure of the two first helices in the HLHLH region of EBF3 reveals a new helical bundle-like fold for which structural homologue searches yield no significant hits (Fig. 2D). In our EBF structure, the first α-helix from one monomer packs with the second helix on the other monomer in an antiparallel fashion. The pair of helices then comes together to form a stable hydrophobic core. In a structure of a protein variant containing all three helices in the HLHLH motif (PDB code 3N50), we do not see the pair of third helices in any of the dimers in the asymmetric unit. Where a third helix is present, it folds over to interact with hydrophobic residues of the adjacent helix from the other monomer. The low occupancy of the third helix in the crystal structure indicates that the interaction of the third helix is weaker than between the two first helices, and its conformation might be affected by crystal contacts. The plasticity of the third helix suggests its involvement in interactions with other proteins.
The HLHLH domain contains a PXXPXXP motif in the loop between H1 and H2 (Fig. 2D). Different polyproline motifs often participate in recognition events, and a similar sequence in the proline repeat domain of p53 was shown to bind the transcriptional co-activator p300 and control p53 acetylation (32). EBF has been shown to have an inhibitory effect on p300/CREB-binding protein through a direct interaction (33), suggesting that the PXXPXXP motif may be the interaction site for p300 or for other partners involved in transactivation.
DISCUSSION
DNA Interaction and the Role of the IPT/TIG Domain
Analogy with members of the Rel homology family suggests that in addition to the DBD, the IPT/TIG domain might be involved in DNA binding. The closest DNA-complexed structural neighbor to EBF is TonEBP (PDB: 1IMH) (21). In TonEBP, the N-RHD and C-RHD domains from two monomers dimerize to almost encircle the DNA (34). Superposition of our DBD and IPT/TIG structures with TonEBP shows that a similar arrangement for the EBFs is plausible (Fig. 3A). However, based on the electrostatic properties of the EBF IPT/TIG surface and the DBD zinc knuckle structure, somewhat different orientations of the domains with respect to the DNA are probable.
FIGURE 3.
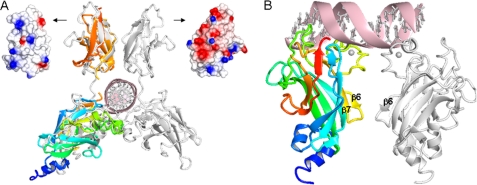
Possible DNA binding modes and IPT/TIG domain DNA interactions in EBF. A, superposition of EBF DBD and IPT/TIG domain (colored as in Figs. 1 and 2) onto the closest structural neighbor, TonEBP (white) in complex with DNA. The dissimilar surface electrostatic potentials of the corresponding TIG domains are shown: left, EBF; right, TonEBP. B, postulated EBF DBD binding to a palindromic consensus DNA binding site based on docking results. DNA contacts are mainly provided by the zinc knuckle area (lime) and the preceding loop (green), whereas dimer forming contacts are found in the zinc knuckle area (lime) and the β6β7 loop (yellow).
Attempts to dock a single or a pair of EBF DBD molecules to a palindromic DNA sequence resulted in several possible binding modes that differ substantially from what is seen in the RHD-DNA complexes. In the top scored model, the EBF zinc knuckle of the first monomer reaches toward the bases in the major groove, and additional DNA contacts are provided by the large preceding loop (Fig. 3B). Salt bridges are formed between the two protein monomers by residues in the zinc knuckle area and loop β6β7. The second DBD molecule is mainly positioned outside the second half-site and is less tightly attached to DNA. This asymmetric binding mode could be valid because the natural EBF binding sites seldom are perfectly palindromic, and EBF is known to tolerate mutations in the consensus site (4).
However, in EBF, the IPT/TIG domain may be dedicated to protein-protein interactions without being involved in DNA interactions. TIG domains have been shown to use the surface of the β-sheets for intermolecular interactions (28, 35), and the complex between CSL and Notch (36) is a well characterized example of such an interaction. A similar type of interaction is possible between the EBFs and Notch, which is known to down-regulate EBF-regulated promoters (37).
Finally, it cannot be excluded that the function of the EBF IPT/TIG domain might be context-dependent with different roles depending on the transcriptional state of the cell. In addition, the complexity is increased by the varied spacing between the DBD and IPT/TIG domains in different isoforms. At least two of the EBF proteins, EBF1 and EBF3, exist in two isoforms where the longer isoform contains two insertions, one shorter between the DBD and the IPT/TIG domains and one longer in the C-terminal transactivation region. The plasticity of the HLHLH domain also allows for interaction-dependent conformations, which can be beneficial in a dynamic transcription module.
Concluding Remarks
We provide the first line of structural information for proteins of the EBF family representing a rather recently identified family of transcription factors. This family was initially thought to be involved in a limited set of defined developmental processes, but it is becoming increasingly evident that these proteins are crucial for normal cell functions in diverse tissues, including lymphocytes, neurons, and adipocytes. In addition, there are increasing amounts of evidence supporting the notion that EBF proteins are targeted in malignant transformation. The structural information covering the conserved domains of the EBF proteins provided in this report will serve as a basis for more detailed molecular investigations of the function of these factors in normal as well as pathological conditions. Future work will show how the EBF proteins interact with both DNA and other proteins in the cell.
Supplementary Material
Acknowledgments
We gratefully acknowledge the beamline personnel at the BESSY (Berlin, Germany) and Diamond (Oxford, United Kingdom) synchrotron radiation facilities. We thank Pär Nordlund and Johan Weigelt for useful discussions. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canada Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and the Wellcome Trust.
This work was supported by a grant from the Academy of Finland (Grant 128322 to L. L.).
The atomic coordinates and structure factors (codes 3LYR, 3MQI, 3MUJ, and 3N50) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Table S1, and references.
- EBF
- early B-cell factor
- DBD
- DNA binding domain
- HLH
- helix-loop-helix
- HLHLH
- helix-loop-helix-loop-helix
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1.Hagman J., Belanger C., Travis A., Turck C. W., Grosschedl R. (1993) Genes Dev. 7, 760–773 [DOI] [PubMed] [Google Scholar]
- 2.Wang M. M., Reed R. R. (1993) Nature 364, 121–126 [DOI] [PubMed] [Google Scholar]
- 3.Hagman J., Gutch M. J., Lin H., Grosschedl R. (1995) EMBO J. 14, 2907–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travis A., Hagman J., Hwang L., Grosschedl R. (1993) Mol. Cell. Biol. 13, 3392–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akerblad P., Lind U., Liberg D., Bamberg K., Sigvardsson M. (2002) Mol. Cell. Biol. 22, 8015–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieslinger M., Folberth S., Dobreva G., Dorn T., Croci L., Erben R., Consalez G. G., Grosschedl R. (2005) Dev. Cell 9, 757–767 [DOI] [PubMed] [Google Scholar]
- 7.Dubois L., Vincent A. (2001) Mech. Dev. 108, 3–12 [DOI] [PubMed] [Google Scholar]
- 8.Wang S. S., Tsai R. Y., Reed R. R. (1997) J. Neurosci. 17, 4149–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gräslund S., Sagemark J., Berglund H., Dahlgren L. G., Flores A., Hammarström M., Johansson I., Kotenyova T., Nilsson M., Nordlund P., Weigelt J. (2008) Protein Expr. Purif. 58, 210–221 [DOI] [PubMed] [Google Scholar]
- 10.Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, Volume 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 12.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 13.Terwilliger T. (2004) J. Synchrotron Radiat. 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 14.Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 15.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vagin A., Teplyakov A. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 17.Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 18.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 19.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21.Holm L., Kääriäinen S., Rosenström P., Schenkel A. (2008) Bioinformatics 24, 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 23.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 24.Fields S., Ternyak K., Gao H., Ostraat R., Akerlund J., Hagman J. (2008) Mol. Immunol. 45, 3786–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel K., Kumar A., Durani S. (2007) Biochim. Biophys. Acta 1774, 1247–1253 [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt G. J. (1999) J. Mol. Biol. 285, 1887–1897 [DOI] [PubMed] [Google Scholar]
- 27.Bork P., Doerks T., Springer T. A., Snel B. (1999) Trends Biochem. Sci. 24, 261–263 [DOI] [PubMed] [Google Scholar]
- 28.Mott H. R., Nietlispach D., Hopkins L. J., Mirey G., Camonis J. H., Owen D. (2003) J. Biol. Chem. 278, 17053–17059 [DOI] [PubMed] [Google Scholar]
- 29.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 30.Bouché N., Scharlat A., Snedden W., Bouchez D., Fromm H. (2002) J. Biol. Chem. 277, 21851–21861 [DOI] [PubMed] [Google Scholar]
- 31.Daburon V., Mella S., Plouhinec J. L., Mazan S., Crozatier M., Vincent A. (2008) BMC Evol. Biol. 8, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dornan D., Shimizu H., Burch L., Smith A. J., Hupp T. R. (2003) Mol. Cell. Biol. 23, 8846–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao F., McCarrick-Walmsley R., Akerblad P., Sigvardsson M., Kadesch T. (2003) Mol. Cell. Biol. 23, 3837–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stroud J. C., Lopez-Rodriguez C., Rao A., Chen L. (2002) Nat. Struct. Biol. 9, 90–94 [DOI] [PubMed] [Google Scholar]
- 35.LaPorte S. L., Juo Z. S., Vaclavikova J., Colf L. A., Qi X., Heller N. M., Keegan A. D., Garcia K. C. (2008) Cell 132, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nam Y., Sliz P., Song L., Aster J. C., Blacklow S. C. (2006) Cell 124, 973–983 [DOI] [PubMed] [Google Scholar]
- 37.Smith E. M., Akerblad P., Kadesch T., Axelson H., Sigvardsson M. (2005) Blood 106, 1995–2001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.