Abstract
The role of the cancer/testis antigen CAGE in drug resistance was investigated. The drug-resistant human melanoma Malme3M (Malme3MR) and the human hepatic cancer cell line SNU387 (SNU387R) showed in vivo drug resistance and CAGE induction. Induction of CAGE resulted from decreased expression and thereby displacement of DNA methyltransferase 1(DNMT1) from CAGE promoter sequences. Various drugs induce expression of CAGE by decreasing expression of DNMT1, and hypomethylation of CAGE was correlated with the increased expression of CAGE. Down-regulation of CAGE in these cell lines decreased invasion and enhanced drug sensitivity resulting from increased apoptosis. Down-regulation of CAGE also led to decreased anchorage-independent growth. Down-regulation of CAGE led to increased expression of p53, suggesting that CAGE may act as a negative regulator of p53. Down-regulation of p53 enhanced resistance to drugs and prevented drugs from exerting apoptotic effects. In SNU387R cells, CAGE induced the interaction between histone deacetylase 2 (HDAC2) and Snail, which exerted a negative effect on p53 expression. Chromatin immunoprecipitation assay showed that CAGE, through interaction with HDAC2, exerted a negative effect on p53 expression in Malme3MR cells. These results suggest that CAGE confers drug resistance by regulating expression of p53 through HDAC2. Taken together, these results show the potential value of CAGE as a target for the development of cancer therapeutics.
Keywords: DNA Methylation, DNA Methyltransferase, Drug Resistance, Histone Deacetylase, Oncogene, p53, CAGE, Snail
Introduction
CAGE is a cancer/testis gene that was isolated by the screening of recombinant cDNA expression libraries using the sera of patients with gastric cancers (1). CAGE is a typical cancer/testis antigen in that its expression in normal tissues is restricted to testis while showing wide expression in various tumor tissues and cancer cell lines (1). CAGE is localized to the X chromosome and exists as single copy (1). CAGE contains helicase and DEAD box domain (1). DEAD box family proteins are known to play important roles in RNA metabolism, cellular growth, and spermatogenesis (2, 3).
A positive rate of anti-CAGE antibody in 7 of 13 (53.8%) patients with microsatellite instability-positive endometrial cancer and in 1 of 3 patients with atypical endometrial hyperplasia (4) suggests that CAGE can be useful for prognosis or early diagnosis of patients with microsatellite instability-positive endometrial cancers.
The expression of CAGE is cell cycle-regulated (1) and under epigenetic regulation (5). 5′-Aza-2′-deoxycytidine, an inhibitor of DNA methyltransferase I, restores expression of CAGE in cancer cell lines that do not express CAGE (5). 5′-Aza-2′-deoxycytidine has been shown to act as an inducer of proteasomal degradation of nonchromatin-bound DNMT1 (6). The methylation of CAGE promoter sequences in premalignant lesions suggests that the expression status of CAGE can be a useful diagnostic marker for early detection of cancer (5). DNA hypomethylation of CAGE was frequently found in cervical carcinoma cells (7). The fact that CAGE expression does not completely correlate with the methylation status (5) suggests that some other factors regulate expression of CAGE as well. Previously, we reported that CAGE promotes cell motility by activating extracellular signal-regulated kinase (ERK) and p38 MAPK (8). As is the case for many other cancer/testis antigens, CAGE-derived peptides were shown to enhance cytolytic T lymphocyte activity (9).
Currently, there have not been reports concerning role of CAGE in drug resistance. In this study, we examined the role of CAGE in drug resistance as well as the mechanism of CAGE-promoted drug resistance.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Cancer cell lines used in this study were cultured in Dulbecco's modified minimal essential medium (Invitrogen) supplemented with heat-inactivated 10% fetal bovine serum (Invitrogen) and antibiotics at 37 °C in a humidified incubator with a mixture of 95% air and 5% CO2.
Human umbilical vein endothelial cells were isolated from human umbilical cord veins by collagenase treatment and used in passages 3–6. The cells were grown in M199 medium supplemented with 20% fetal bovine serum, 100 units/ml penicillin G, 100 μg/ml streptomycin, 3 ng/ml basic fibroblast growth factor (Upstate), and 5 units/ml heparin at 37 °C under 5% CO2, 95% air.
Drug-resistant cancer cell lines were established by stepwise addition of celastrol to SNU387 human hepatic cancer cells and Malme3M human melanoma cells. Cells surviving the cytotoxic effects of drugs (attached fraction) were collected and subjected to further selection.
Materials
Anti-p53, anti-Bcl-2, and anti-HDAC2 antibodies were purchased from Cell Signaling Technology. Polyclonal anti-CAGE (anti-DDX53) antibody was purchased from Sigma. We used monoclonal anti-CAGE antibody throughout this study. Both polyclonal and monoclonal anti-CAGE antibody showed the same results. All other antibodies used in this study were purchased from Santa Cruz Biotechnology. All chemicals used in this study were purchased from Sigma. Anti-mouse and anti-rabbit IgG-horseradish peroxidase conjugate antibody was purchased from Pierce. An enhanced chemiluminescence (ECL) kit was purchased from Amersham Biosciences. Lipofectamine and PlusTM reagent were purchased from Invitrogen. The transwell chamber system was purchased from Costar (Acton, MA). Bioneer (Daejeon, Korea) synthesized all primers used in this study. The dominant negative PKCα3 and PKCδ constructs were kindly provided by Prof. Soh Jaewon (Inha University, Korea).
Tissue Array Analyses
Tissue arrays were purchased from US Biomax, Inc. Tumor slides were deparaffinized and rehydrated using xylene and alcohol; for immunoperoxidase labeling, endogenous peroxidase was blocked with 0.3% H2O2 in absolute methanol for 15 min at room temperature. Primary anti-CAGE antibody was reacted with the tissue for 2 h in a humid chamber at room temperature and washed with phosphate-buffered saline for 10 min, and the sections were incubated for 20 min at room temperature with secondary antibody. After additional incubation with streptavidin-horseradish peroxidase for 10 min, immunoreactive sites were visualized using 3,3′-diaminobenzidine for 5 min. The sections were counterstained with Harris' hematoxylin, dehydrated, and mounted with coverslips.
Tumorigenic Potential of CAGE
Stable transfectant of HeLa cells (1 × 106) expressing CAGE under the control of doxycycline were suspended in growth factor-reduced Matrigel matrix (BD Bioscience) and injected into both flanks of athymic nude mice. Doxycycline (1 μg/ml) was injected via tail vein three times a week. Tumor volumes were measured at the indicated days after injection of HeLa cells. Procedures involving animals were carried out with guidelines set out by the animal ethics committee at Kangwon National University.
In Vivo Drug Resistance
Athymic nude mice (BALB/c nu/nu, 5–6-week-old females) were obtained from Orient Bio Inc. (Seoul, Korea) and were maintained in a laminar air-flow cabinet under aseptic conditions. Drug-sensitive or drug-resistant cancer cells (1 × 106) were injected subcutaneously into the dorsal flank area of the mice. Following the establishment of tumor, celastrol was administered via tail vein at a dose of 1 mg/kg twice per week. Tumor volume was determined by direct measurement with calipers and calculated by the following formula: (large diameter) × (small diameter) × 0.52.
Preparation of siRNA Duplexes and Transfection
The siRNA duplexes were constructed with the following target sequences: CAGE, sense (5′-AACTCTGTCAACCTAAGAAGCCCTGTCTC-3′), and antisense (5′-AAGCTTCTTAGGTTGACAGAGCCTGTCTC-3′); CAGE-1, sense (5′-AACTCTGTCAACCTAAGAAGCCCTGTCTC-3′), and antisense (5′-AAGCTTCTTAGGTTGACAGAGCCTGTCTC-3′); CAGE-2, sense (5′-AAGACTGAATTGGCGTTGGCTCCTGTCTC-3′), and antisense (5′-AAAGCCAACGCCAATTCAGTCCCTGTCTC-3′); scrambled CAGE-1, sense (5′-AATTAATGATCGCCCAGAACCCCTGTCTC-3′), and antisense (5′-AAGGTTCTGGGCGATCATTAACCTGTCTC-3′); scrambled CAGE-2, sense (5′-AAAGAGTCGTATGGTGTCCTGCCTGTCTC-3′), and antisense (5′-AACAGGACACCATACGACTCTCCTGTCTC-3′); DNMT-1, sense (5′-AATTTTCCCTTGCCCTTCCCTCCTGTCTC-3), and antisense (5′-AAAGGGAAGGGCAAGGGAAAACCTGTCTC-3′); SNAIL, sense (5′-AAACAGAGTCCCAGATGAGCACCTGTCTC-3′), and antisense (5′-AATGCTCATCTGGGACTCTGTCCTGTCTC-3′); p53, sense (5′-AACTTTTGAGAAGCTCAAAACCCTGTCTC-3′), and antisense (5′-AAGTTTTGAGCTTCTCAAAAGCCTGTCTC-3′); scrambled p53, sense (5′-ACGGGTTTAAGCATATCACACTATTCCCA-3′), and antisense (5′-TTAAGATTCACGCTTATGCCTAGTATGCC-3′); and control, sense (5′-AATTCTCCGAACGTGTCACGTCCTGTCTC-3′), and antisense (5′-AAACGTGACACGTTCGGAGAACCTGTCTC-3′). Control siRNA sequences were derived from green fluorescent protein sequences. The construction of siRNAs was performed according to the instruction manual provided by the manufacturer (Ambion, Austin, TX). The transfection of the siRNA construct was performed by using Lipofectamine 2000 (Invitrogen). Transfection of plasmids was carried out by Lipofectamine Plus reagent and Lipofectamine reagent (Invitrogen).
Methylation-specific PCR
Genomic DNAs from various samples were subjected to sodium bisulfite modification. For detection of the methylated alleles, the sense and antisense primers 5′-TTTTATACGATTCGGAATTCGAC-3′ and 5′-CAAATCTACGACCTATTTCCCG-3′, respectively, were used for the amplification of the methylated allele. The sense and antisense primers 5′-GTTTTTTATATGATTTGGAATTTGAT-3′ and 5′-AATTCAAATCTACAACCTATTTCCCA-3′, respectively, were used for the amplification of the unmethylated allele. PCR was performed for 30 cycles at 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 1 min.
Western Blot Analysis
For PAGE and Western blot, cell lysates were prepared using lysis buffer (62.5 mm Tris-HCl, pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 50 mm dithiothreitol, 0.01% (w/v) bromphenol blue, 10 mm NaF, 1% (v/v) protease inhibitor mixture, 1 mm sodium orthovanadate). The samples were boiled for 5 min, and equal amounts of protein (20 μg/well) were analyzed on a 10% SDS-PAGE. After electrophoresis, proteins were transferred onto a nitrocellulose membrane and subjected to immunoblotting. The dilution of each primary antibody was empirically determined. After extensive washing, blots were further incubated with an anti-mouse or anti-rabbit IgG-horseradish peroxidase-conjugated antibody at a 1:3,000 dilution for 1 h at room temperature and were developed using an enhanced chemiluminescence kit (Amersham Biosciences).
Gelatin Zymography
For gelatin zymography, conditioned medium from each cell line cultured in serum-free medium was mixed 3:1 with substrate gel sample buffer (40% (v/v) glycerol, 0.25 m Tris-HCl, pH 6.8, and 0.1% (w/v) bromphenol blue) and loaded onto a 7.5% SDS-PAGE containing type I gelatin (2 mg/ml).
Cell Viability Determination
The cells were assayed for their growth activity using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma) test.
Anchorage-independent Growth Assay
Anchorage-independent growth assays were performed according to the manufacturer's instructions (Millipore). The assays were done in 96-well plates, and the plates were incubated at 37 °C for 21–28 days. Anchorage-independent growth was evaluated by using the cell stain solution. Stained colonies were counted using a microscope, and intensity of staining was quantified by measuring absorbance at 490 nm.
ChIP Assays
Assays were performed according to manufacturer's instructions (Upstate). Briefly, one-fourth of the chromatin solution was reserved for total input. The remaining solution was precleared with protein A-agarose, subsequently incubated with respective antibody (2 μg/ml each) for 12 h at 4 °C with shaking, and then further incubated with protein A-Sepharose for 2 h. The immunoprecipitates were reverse cross-linked. PCR was done on the phenol/chloroform-extracted DNA. For detection of DNMT1 binding to CAGE promoter sequences, specific primers of CAGE promoter (5′-GTGAAGATGCTTATGGCACA-3′ (sense) and 5′-ATGCTGATGGTGTCAACTGG-3′ (antisense)) were used. To check specificity of DNMT1 binding, mutant primers of CAGE promoter (5′-GCCATTATAGCGGGCTTTAG-3′ (sense) and 5′-GGGGTAGGTATGTGAAGGCT-3′ (antisense)) were used. For detection of CAGE, HDAC2, or Snail binding to p53, promoter sequences 5′-GTTGATGGGATTGGGGTTTT-3′ (sense) and 5′-GTGTCACCGTCGTGGAAAG-3′ (antisense) were used. For detection of c-fos binding to p53 promoter sequences, 5′-CAGAATTTTCCACCCCAAAA-3′ (sense) and 5′-TGGCACAAAGCTGGACAGT-3′ (antisense) were used.
Immunoprecipitation
Cells (1 × 107) were lysed in immunoprecipitation buffer (50 mmol/liter HEPES, pH 7.6, 150 mmol/liter NaCl, 5 mmol/liter EDTA, 0.1% Nonidet P-40). After centrifugation (10 min at 15,000 × g) to remove particulate material, the supernatant was incubated with each antibody (2 μg/ml) with constant agitation at 4 °C. The immunocomplexes were precipitated with protein A/G-Sepharose (Sigma) and analyzed by Western blot.
Cellular Fractionation
Nuclear and cytosolic extract was prepared with a nuclear/cytosol fractionation kit (Biovision, Mountain View, CA). Cells were collected by centrifugation at 600 × g for 5 min at 4 °C. Cell pellets were washed twice with ice-cold phosphate-buffered saline, followed by the addition of 0.2 ml of Cytosol Extraction Buffer A and vigorous mixing for 5 s. Ice-cold Cytosol Extraction Buffer B (11 μl) was then added to the solution. After mixing, nuclei and cytosolic fractions were separated by centrifugation at 16,000 × g for 5 min (supernatants were cytosolic fraction). Nuclear extraction buffer was added to the nuclei. After vortexing for a total of 40 min, nuclei were centrifuged at 16,000 × g for 10 min. Supernatants thus obtained were the nuclear fraction. Protein concentration of each fraction was determined using the DC protein assay kit (Bio-Rad). Equal amounts of nuclear/cytosolic extracts were loaded for SDS-PAGE, and Western blot analysis was performed. Purity of the cytosolic and nuclear fraction was confirmed by glyceraldehyde-3-phosphate dehydrogenase and histone H1, respectively.
For fractionation of cytosol and membrane, cells were resuspended in a buffer containing 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml leupeptin and were lysed by sonication. The lysates were then centrifuged at 100,000 × g for 1 h at 4 °C. The supernatants constitute the cytosolic fraction. The pellet was resuspended in the above buffer, which also contained 0.1% Triton X-100, and the mixture was lysed by sonication and centrifuged at 100,000 × g for 1 h at 4 °C to obtain the membrane fraction (supernatant). Purity of the cytosol and membrane fraction was confirmed by glyceraldehyde-3-phosphate dehydrogenase and focal adhesion kinase, respectively.
Histone Deacetylase Activity Assays
Histone deacetylase activity was measured according to the manufacturer's instructions (Cayman Chemical, Ann Arbor, MI). The activity was measured according to the manufacturer's instructions. For immunoprecipitation, cells were lysed with ice-cold buffer (10 mm Tris-HCl, pH 7.4, 10 mm NaCl, 15 mm MgCl2, 250 mm sucrose, 0.12 mm EDTA, 0.5% Nonidet P-40, and a mixture of protease inhibitors). The lysates were suspended with nuclear extraction buffer (50 mm HEPES, pH 7.5, 420 mm NaCl, 0.5 mm EDTA, 0.1 mm EGTA, and 10% glycerol), sonicated for 30 s, and centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant containing the nuclear extract was immunoprecipitated with anti-CAGE (2 μg/ml), anti-HDAC2 (2 μg/ml), or anti-IgG antibody (2 μg/ml). The immunoprecipitants were incubated with 200 μm acetylated fluorometric substrate for 30 min at 37 °C, and 40 μl of developer was added. After 15 min, the fluorescence was measured using an excitation wavelength of 340–360 nm and an emission wavelength of 440–460 nm.
Chemoinvasion Assay
The invasive potential was determined by using a transwell chamber system with 8-μm pore polycarbonate filter inserts (CoSTAR, Acton, MA). The lower and upper sides of the filter were coated with gelatin and Matrigel, respectively. Trypsinized cells (2 × 104) in the serum-free RPMI 1640 medium containing 0.1% bovine serum albumin were then added to each upper chamber of the transwell. RPMI 1640 medium supplemented with 10% fetal bovine serum was placed in the lower chamber, and cells were incubated at 37 °C for 16 h. The cells were fixed with methanol, and the invaded cells were stained and counted. Results were analyzed for statistical significance using the Student's t test. Differences were considered significant when p < 0.05.
Tube Formation Assays
Tube formation assays were performed to examine the effect of CAGE on the angiogenic potential of cancer cell lines. For this, growth factor-reduced Matrigel was added to 24-well plates (200 μl Matrigel per well) and polymerized for 30 min at 37 °C. Human umbilical vein endothelial cells were first incubated in M199 containing 1% fetal bovine serum for 1 h, followed by the addition of conditioned medium of each cell line. After 6–8 h of incubation at 37 °C in a 95:5% (v/v) mixture of air/CO2, the endothelial cells were photographed using an inverted microscope (magnification, ×100; Olympus). Three independent experiments were performed.
Caspase-3 Activity Assays
Caspase-3 activity was measured according to the manufacturer's instructions (BioVision, Palo Alto, CA). Cells were lysed in 0.1 m HEPES buffer, pH 7.4, containing 2 mm dithiothreitol, 0.1% CHAPS, and 1% sucrose. Cell lysates were incubated with a colorimetric substrate, 200 μm Ac-DEVD-p-nitroanilide, for 30 min at 30 °C. The fluorescence was measured at 405 nm using a microtiter plate reader.
Annexin V-FITC Staining
Apoptosis determination was carried out by using annexin V-FITC according to the manufacturer's instructions (Biovision). To determine the effect of CAGE on apoptosis, drug-resistant cancer cells were transiently transfected with control siRNA (10 nm) or CAGE siRNA (10 nm). At 48 h after transfection, cells were treated with or without celastrol (1 μm) or taxol (1 μm) for 16 h. Ten thousand cells were counted for three independent experiments.
CD8+ T Cell Activity Assays
CD8+ T cells were positively isolated from whole blood by using the CD8 positive isolation kit (Dynal Biotech) according to the manufacturer's instructions. Isolated CD8+ T cells were incubation with plate-bound 10 μg/ml anti-CD3 monoclonal antibody (eBioscience, San Diego) for 48 h. The target cells (1 × 104 per well) were placed in each well of 96-well plates and were co-cultured with effecter cells for 48 h. Cytotoxic effect of CD8+ T cells was determined by MTT assay.
RESULTS
Tissue Array Analyses of CAGE Expression
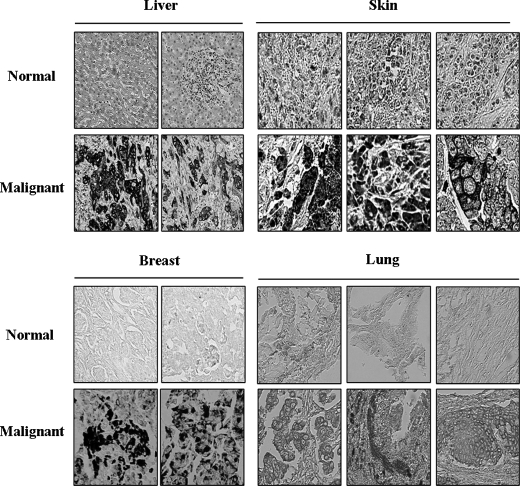
Previously, we examined the expression levels of CAGE in various tumor tissues and cancer cell lines (5). However, in this study, CAGE expression was determined by reverse transcription-PCR. Therefore, we examined the expression of CAGE protein by immunohistochemistry using various tissue arrays each consisting of 100 core tissues. Immunohistochemistry analysis shows expression of CAGE in various tumor tissues, such as melanoma, hepatoma, and breast tumors (Fig. 1). CAGE is expressed in 15 of 76 malignant melanomas (19.7%), in 26 of 70 malignant hepatic cancers (37.1%), in 40 of 84 lung cancers, and in 46 of 88 (52.3%) breast cancers. Expression of CAGE among these tumor tissues ranges from 19 to 52%, and its expression in normal tissues is negligible. This establishes the expression pattern of CAGE and suggests a potential role for CAGE in tumorigenesis.
FIGURE 1.
Expression analyses of CAGE in various tumor tissues. Immunohistochemical analysis for CAGE was performed by using tissue arrays. CAGE was detected using the diaminobenzidine method.
Establishment and Characterization of Drug-resistant Cancer Cell Lines
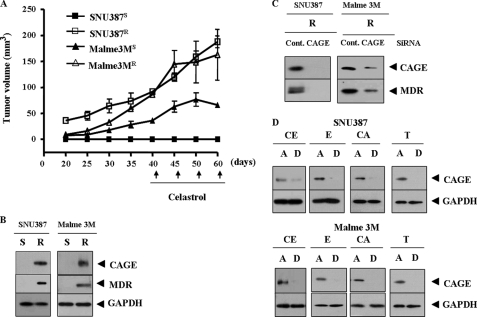
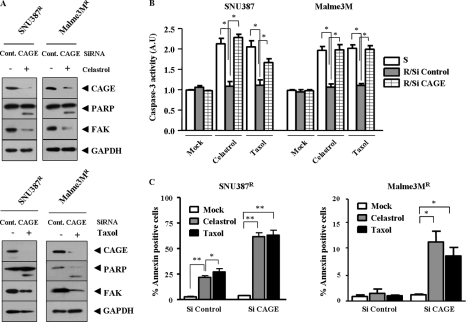
Because CAGE is present in various tumor tissues, we hypothesized that CAGE plays a role in tumorigenesis and drug resistance. To examine the role of CAGE in drug resistance, we established drug-resistant cancer cell lines by stepwise addition of celastrol, an inhibitor of NF-κB, to human melanoma Malme3M and human hepatic SNU387 cancer cells. Celastrol is known to suppress vascular endothelial growth factor-induced angiogenesis via inhibition of Akt/mammalian target of rapamycin pathway (10). SNU387R (Fig. 2A) and Malme3MR cells (Fig. 2B) show relatively higher resistance to drugs such as celastrol and taxol. Taxol is known to induce microtubule polymerization to exert anti-cancer activity. This enhanced drug resistance is inversely correlated with caspase-3 activity (Fig. 2, C and D). We next examined whether these cell lines would display in vivo drug resistance. For this, Malme3MR and SNU387R cells were injected via the tail vein into athymic nude mice. SNU387R and Malme3MR cell lines show enhanced tumorigenic potential and resistance to celastrol (Fig. 3A). CAGE is overexpressed in SNU387R and Malme3MR cells (Fig. 3B). These cell lines also show increased expression of multidrug resistance protein (MDR), and down-regulation of CAGE exerts a negative effect on the induction of MDR (Fig. 3C). Various drugs induce expression of CAGE in the attached fraction of SNU387 and Malme3M cells (Fig. 3D). These results further suggest a role for CAGE in drug resistance. Next, we investigated the mechanism of CAGE induction in these drug-resistant cancer cell lines.
FIGURE 2.
Establishment of drug-resistant cancer cell lines. A, comparison of drug sensitivity between SNU387 and SNU387R cells. Each cell line was treated with various concentrations of each drug for 24 h. Cell numbers were determined by MTT assays. Each value represents an average of three independent experiments, and each experiment was performed in triplicate. B, comparison of drug sensitivity between Malme3M and Malme3MR. C, comparison of caspase-3 activity in response to drugs between SNU387 and SNU387R cells. D, comparison of caspase-3 activity was made between Malme3M and Malme3MR. S denotes drug-sensitive, and R denotes drug-resistant. A.U., arbitrary units.
FIGURE 3.
Drug-resistant cancer cell lines show in vivo drug resistance and induction of CAGE. A, cancer cells were injected into athymic nude mice via tail vein. Celastrol (1 mg/kg) was injected into each nude mouse after the tumor reached a certain size. Tumor volume was measured as described under “Experimental Procedures.” Five mice were used for the injection of each cell line. Each value represents an average obtained from each mouse. B, Western blot analysis shows induction of CAGE and MDR in SNU387R and Malme3MR cells. S denotes drug-sensitive and R denotes drug-resistant. C, drug-resistant cancer cells were transiently transfected with control siRNA (10 nm) or CAGE siRNA (10 nm). At 48 h after transfection, cell lysates were prepared and subjected to Western blot analysis. D, SNU387 or Malme3M cells were treated with various anticancer drugs for 16 h (each at 1 μm). Cells were divided into attached (A) and detached (D) fraction. Cell lysates from each fraction were subjected to Western blot analysis. CE, celastrol; E, etoposide (DNA-damaging agent); CA, camptothecin (an inhibitor of topoisomerase); T, taxol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
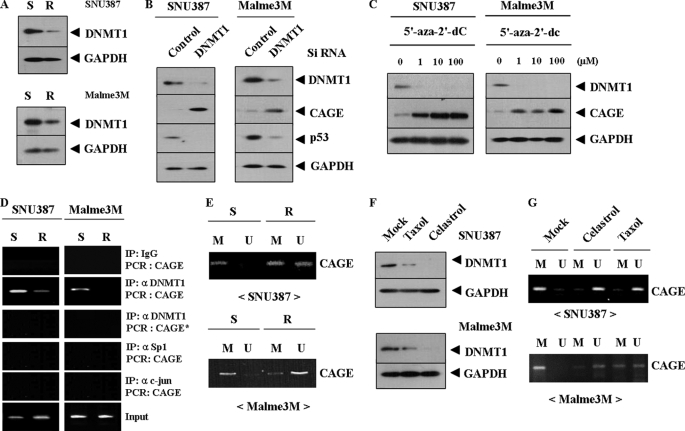
Down-regulation of DNA Methyltransferase 1 (DNMT1) Is Responsible for Induction of CAGE
The expression level of CAGE is under epigenetic regulation in various cancer cell lines (5). We hypothesized that DNMT1 would play a role in the induction of CAGE. In SNU387R and Malme3MR cells, the expression level of DNMT1 is decreased (Fig. 4A), which leads to increased expression of CAGE (Fig. 4B). This suggests that DNMT1 may directly regulate expression of CAGE. 5′-Aza-2′-deoxycytidine, an inhibitor of DNMT1, induces expression of CAGE in both SNU387R and Malme3MR cells (Fig. 4C), further suggesting that CAGE expression may be under epigenetic regulation. Because down-regulation of DNMT1 leads to increased expression of CAGE, the decreased expression of p53 (Fig. 4B), the effect of DNMT1 on cellular proliferation was examined. Down-regulation of DNMT1 decreases sensitivity to celastrol and taxol (supplemental Fig. 1, A and C), which is accompanied by decreased caspase-3 activity (supplemental Fig. 1, B and D).
FIGURE 4.
Down-regulation of DNMT1 is responsible for induction of CAGE drug-resistant cancer cells. A, Western blot analysis shows decreased expression of DNMT1 in SNU387R and Malme3MR cells. S denotes drug-sensitive and R denotes drug-resistant. B, SNU387 or Malme3M cells were transiently transfected with control siRNA (10 nm) or DNMT1 siRNA (10 nm). At 48 h after transfection, cell lysates were prepared and subjected to Western blot analysis. C, SNU387 or Malme3M cells were treated with various concentrations of 5′-aza-2′-deoxycytidine for 72 h, followed by Western blot analysis. D, ChIP assays show a lack of DNMT1 binding to CAGE promoter sequences in SNU387R and Malme3MR cells. * denotes ChIP assays employing mutant CAGE promoter primer. IP, immunoprecipitation. E, methylation-specific PCR of genomic DNA shows that the lack of CAGE expression is correlated with methylation of CAGE promoter sequences. M denotes methylation and U denotes unmethylation. S denotes drug-sensitive and R denotes drug-resistant. F, SNU387 (upper panel) or Malme3M (lower panel) was treated with celastrol (1 μm) or taxol (1 μm) for 16 h, followed by Western blot analysis. G is the same as F except that methylation-specific PCR was performed. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
ChIP assay was performed to examine direct binding of DNMT1 to CAGE promoter sequences. The lack of DNMT1 binding to CAGE promoter sequences is evident in both SNU387R and Malme3MR (Fig. 4D). CAGE promoter sequences do not contain binding sites for Sp-1 or AP-1. ChIP assay shows lack of binding of Sp-1 or c-Jun to CAGE promoter sequences (Fig. 4D). ChIP assays using mutant CAGE promoter primers show a lack of DNMT1 binding to CAGE promoter sequences (Fig. 4D). Methylation-specific PCR shows hypomethylation of CAGE in SUN387R (Fig. 4E, upper panel) and Malme3MR cells (Fig. 4E, lower panel). Because drugs increase the expression level of CAGE (Fig. 3D), the effect of drugs on DNMT1 expression was examined. Celastrol and taxol decrease the expression of DNMT1 in both SNU387 (Fig. 4F, upper panel) and Malme3M cells (Fig. 4F, lower panel). This decreased expression of DNMT1 is accompanied by hypomethylation of CAGE promoter sequences (Fig. 4G). These results suggest that methylation of CAGE promoter sequences is at least partially responsible for the lack of its expression in drug-sensitive SNU387 and Malme3M cells. These results suggest that epigenetic regulation exerted by DNMT1 confers resistance to drugs.
CAGE Regulates Drug Sensitivity
To further examine the role of CAGE in drug resistance, HeLa cells stably expressing CAGE under control of doxycycline were established. Induction of CAGE leads to increased expression of MDR, BCL-2, and VEGFR1, as well as decreased expression of p53 and activation of EGFR (supplemental Fig. 2A). Induction of CAGE confers resistance to taxol (supplemental Fig. 2B). HeLa cells expressing CAGE show enhanced tumorigenic potential when injected into athymic nude mice (supplemental Fig. 2C). Supernatants from HeLa cells that were induced to express CAGE by doxycyline (1 μg/ml) enhance tube formation in human umbilical vein endothelial cells (supplemental Fig. 2D). These results suggest that drug resistance conferred by CAGE is closely related with its tumorigenic and angiogenic potential.
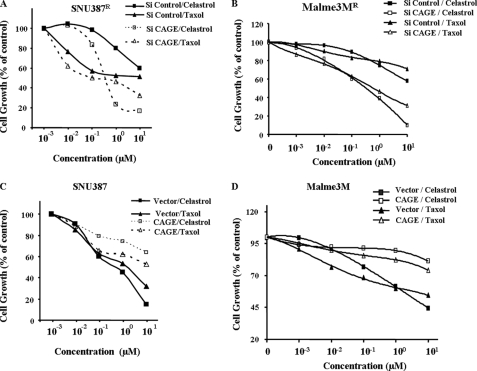
The effect of CAGE on drug sensitivity was examined. Down-regulation of CAGE enhances sensitivity to various anti-cancer drugs, such as celastrol and taxol in both SNU387R (Fig. 5A) and Malme3MR cells (Fig. 5A). Overexpression of CAGE confers resistance to drugs such as celastrol and taxol in both SNU387 (Fig. 5C) and Malme3M cells (Fig. 5D). These results suggest that expression level of CAGE may regulate drug sensitivity.
FIGURE 5.
CAGE regulates sensitivity to drugs. SNU387R cells (A) or Malme3MR cells (B) were transiently transfected with control siRNA (10 nm) or CAGE siRNA (10 nm). The next day, cells were treated with or without drugs at various concentrations for 24 h as indicated. Cell viability was determined by MTT assay. SNU387 cells (C) or Malme3M cells (D) were transiently transfected with control vector (1 μg) or CAGE cDNA (1 μg). The next day, these cells were treated with or without drugs at various concentrations for 24 h as indicated. Cell viability was measured by MTT assays.
Down-regulation of CAGE Enhances Sensitivity to Drugs by Inducing Apoptosis
Down-regulation of CAGE leads to enhanced sensitivity to drugs (Fig. 5, A and B). We therefore examined whether this enhanced sensitivity was related to apoptosis. Down-regulation of CAGE induces cleavage of poly(ADP-ribose) polymerase and focal adhesion kinase in response to celastrol (Fig. 6A, upper panel) and taxol (Fig. 6A, lower panel) in both SNU387R and Malme3MR cells. Celastrol and taxol increase caspase-3 activity in SNU387 and Malme3M cells, as expected (Fig. 6B). Down-regulation of CAGE leads to an increased caspase-3 activity in response to drugs in SNU387R and Malme3MR cells (Fig. 6B). These results suggest that down-regulation of CAGE induces apoptosis to sensitize drug-resistant cancer cells to drugs. SNU387R (supplemental Fig. 3A) and Malme3MR cells (supplemental Fig. 3B) show low levels of caspase-3 activity in response to drugs. Overexpression of CAGE exerts a negative effect on caspase-3 activity increased by celastrol and taxol in both SNU387 and Malme3M cells (supplemental Fig. 3, C and D). Down-regulation of CAGE by various CAGE siRNAs, but not by scrambled CAGE siRNAs, enhances sensitivity to drugs (supplemental Fig. 4B) as well as caspase-3 activity in response to drugs (supplemental Fig. 4C). Down-regulation of CAGE leads to enhanced apoptosis as evidenced by annexin V-FITC staining in SNU387R and Malme3MR cells (Fig. 6C). These results suggest that down-regulation of CAGE exerts apoptotic effects to enhance drug sensitivity.
FIGURE 6.
Down-regulation of CAGE exerts apoptotic effects. A, drug-resistant SNU387R or Malme3MR cells transiently transfected with control siRNA (10 nm) or CAGE siRNA (10 nm) treated with or without celastrol (1 μm) (upper panel) or with or without taxol (1 μm) for 16 h (lower panel), followed by Western blot analysis; FAK, focal adhesion kinase; PARP, poly(ADP-ribose) polymerase; Cont, control. B, down-regulation of CAGE leads to enhanced caspase-3 activity in drug-resistant cancer cell lines. Each value represents an average of three independent experiments. A.U., arbitrary units. *, p < 0.05. C, apoptosis was measured by annexinV-FITC staining method. Drug-resistant cancer cells were transiently transfected with control siRNA (10 nm) or CAGE siRNA (10 nm), followed by treatment with drugs (each at 1 μm) for 24 h. *, p < 0.05; **, p < 0.005.
CAGE Enhances the Invasion Potential of Cancer Cell Lines and Anchorage-independent Cell Growth
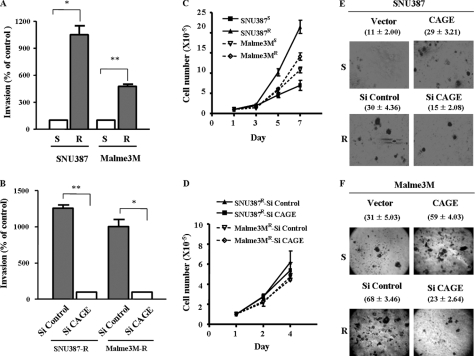
Multidrug resistance is closely related with invasion potential (11, 12). Therefore, we examined whether drug resistance conferred by CAGE would be related with enhanced invasion potential. SNU387R and Malme3MR cell lines show increased expression and secretion of MMP-2, but not MMP-9 (data not shown). Down-regulation of CAGE exerts a negative effect on the expression and secretion of MMP-2 in these drug-resistant cancer cell lines (data no shown). These drug-resistant cell lines show enhanced invasion potential (Fig. 7A), which is decreased by down-regulation of CAGE (Fig. 7B). Moreover, down-regulation of CAGE by various CAGE siRNAs, but not by scrambled CAGE siRNAs, exerts a negative effect on the invasion potential of SNU387R and Malme3MR cells (supplemental Fig. 5B). Drug-resistant cancer cell lines show higher cellular proliferation rates than their counterparts (Fig. 7C). However, down-regulation of CAGE does not affect cellular proliferation of these cancer cell lines (Fig. 7D). Down-regulation of CAGE exerts a negative effect on anchorage-independent growth of SNU387R cells (Fig. 7E) and Malme3MR cells (Fig. 7F), suggesting that CAGE may confer cellular growth advantage under anchorage-independent conditions. These results suggest that CAGE-promoted drug resistance is correlated with enhanced invasion potential and anchorage-independent growth.
FIGURE 7.
Down-regulation of CAGE leads to decreased invasion potential and decreased anchorage-independent growth. A, invasion potentials of drug-sensitive and drug-resistant cancer cell line were compared using a transwell chamber. *, p < 0.05; **, p < 0.005. S denotes drug-sensitive and R denotes drug-resistant. B, down-regulation of CAGE decreases the invasion potential of drug-resistant cancer cells. Cells were transiently transfected with control siRNA (10 nm) or CAGE siRNA (10 nm). At 48 h after transfection, cellular invasion assays were performed. C, drug-resistant cancer cells show increased cellular proliferation. D, down-regulation of CAGE does not affect cellular proliferation of drug-resistant cancer cells. SNU387 (E) or Malme3M cells (F) were transiently transfected with various constructs as indicated. At 48 h after transfection, cells were harvested, counted, resuspended in 0.2% soft agar, and seeded onto 0.4% soft agar supplemented with 10% fetal bovine serum (500 cells/well for SNU387 cells and 1000 cells for Malme3M cells). After 4 weeks, colonies were stained and counted. Numbers in parentheses represent anchorage-independent colonies.
CAGE Exerts a Negative Effect on p53 and Down-regulation of p53 Induces Drug Resistance
p53 enhances apoptosis by regulating Fas (FAS) (13) and plays important role for various cellular activities, including DNA repair and drug resistance (13, 14). We therefore hypothesized that p53 might be involved in CAGE-promoted drug resistance. We first examined whether CAGE would affect the expression level of p53. By using various CAGE siRNAs and nonfunctional CAGE siRNAs (scrambled CAGE siRNAs), we found that CAGE acted as a negative regulator of p53 expression in SNU387R and Malme3MR cells (Fig. 8A). Next, we examined whether p53 affected drug resistance. Down-regulation of p53 induces resistance to celastrol and taxol in SNU387 (Fig. 8B) and Malme3M cells (Fig. 8B). Down-regulation of p53 exerts a negative effect on cleavage of poly(ADP-ribose) polymerase and focal adhesion kinase, apoptotic marker proteins, by celastrol and taxol (Fig. 8C). Down-regulation of p53 exerts a negative effect on apoptotic effect by celastrol and taxol in both SNU387 and Malme3M cells, as assessed by annexin V-FITC staining (Fig. 8D). These results suggest that CAGE may regulate p53 expression in conferring drug resistance.
FIGURE 8.
Down-regulation of p53 leads to enhanced resistance to celastrol and taxol. A, down-regulation of CAGE leads to restoration of p53 expression in SNU387R and Malme3MR cells. Each cell line was transiently transfected with control siRNA (10 nm), CAGE siRNA (each at 10 nm), or scrambled CAGE siRNA (each at 10 nm). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, SNU387 cells or Malme3M cells were transiently transfected with control siRNA (10 nm), p53 siRNA (10 nm), or scrambled p53 siRNA (each at 10 nm). The next day, cells were treated with or without drugs at various concentrations for 24 h as indicated. Cell viability was determined by MTT assay. Each value represents average of three independent experiments. C, Western blot analysis shows that down-regulation of p53 exerts a negative effect on apoptotic cleavage of poly(ADP-ribose) polymerase (PARP) and focal adhesion kinase (FAK) by drugs. D, annexin V-FITC staining shows that down-regulation of p53 decreases apoptotic cell death. Each value represents average of three independent experiments. **, p < 0.005.
HDAC2 Is Necessary for the Repression of p53 Expression by CAGE
Because down-regulation of p53 enhances resistance to celastrol and taxol (Fig. 8B), we investigated the mechanism by which CAGE regulates the expression of p53. MAGEA2, a cancer/testis antigen, interacts with HDAC3 to exert transcriptional repression on p53 and confer resistance to drugs (15). Down-regulation of HDAC1 was shown to induce P-glycoprotein to confer resistance to various drugs (16). We therefore hypothesized that histone deacetylase(s) may be involved in CAGE-promoted drug resistance.
HDAC2 interacts with Snail to exert transcriptional repression on various genes (17–19). Because p53 promoter contains Snail-binding sequences, it is possible that Snail may also regulate expression of p53. Down-regulation of Snail was shown to restore expression of p53 in both SNU387R and Malme3MR cells (data not shown). HDAC2 modulates p53 transcriptional activities (20). We therefore hypothesized that HDAC2 may have a role in the regulation of p53 expression in relation with CAGE. We first examined expression levels of HDAC2 in both SNU387 and Malme3M cells. Expression level of HDAC2 is increased in SNU387R cells but not in Malme3MR cells (data not shown). Down-regulation of CAGE decreases expression level of HDAC2 in SN387R cells but not in Malme3MR cells (Fig. 9A). Down-regulation of CAGE decreases HDAC2 activity in both SNU387R cells and Malme3MR cells (Fig. 9B). CAGE is not present in the nucleus and therefore does not interact with HDAC2 or contain HDAC2 activity in SNU387R cells (data not shown). Cellular fractionation shows that CAGE is present in the nucleus in Malme3MR cells (Fig. 9C), suggesting different mode of regulation of p53 by CAGE in Malme3MR cells from SNU387R cells.
FIGURE 9.
HDAC2 is responsible for repression of p53 by CAGE. A, down-regulation of CAGE decreases expression of HDAC2 in SNU387R cells but not in Malme3MR cells. B, down-regulation of CAGE decreases HDAC2 activity in both SNU387R cells (left panel) and Malme3MR cells (right panel). C, cell lysates from each fraction were immunoprecipitated with anti-CAGE antibody (2 μg/ml) or anti-HDAC2 antibody (2 μg/ml), followed by Western blot analysis. Western blot analysis shows the interaction between CAGE and HDAC2 in Malme3MR cells. S denotes sensitive and R denotes resistant. D, in vitro deacetylation assay using immunoprecipitated (IP) complex shows CAGE contains HDAC2 activity in Malme3MR cells. *, p < 0.05. IB, immunoblot. E, in vitro deacetylation assay of acetylated fluorometric substrate by immunoprecipitated complex (by anti-HDAC2 antibody or anti-CAGE antibody) shows increased HDAC2 activity in Malme3MR cells. Each value represents average of three independent experiments. ***, p < 0.0001; *, p < 0.05. F, ChIP assays show binding of HDAC2 to p53 promoter sequences in SNU387R cells and in Malme3MR cells. G, ChIP assay shows that down-regulation of CAGE inhibits binding of HDA2 to p53 promoter sequences in SNU387R cells and Malme3MR cells.
CAGE interacts with HDAC2 in the nucleus of Malme3MR cells (Fig. 9C). Exogenous transfection of green fluorescent protein-CAGE confirms the interaction between CAGE and HDAC2 (data not shown). The above results suggest that CAGE may exert repression of p53 expression through interaction with HDAC2 in Malme3MR cells. CAGE contains HDAC2 activity in Malme3MR cells (Fig. 9D). This is reasonable in that CAGE shows interaction with HDAC2 in Malme3MR cells (Fig. 9C). Although down-regulation of CAGE does not affect expression level of HDAC2, interaction between HDAC2 and CAGE and interaction between HDAC2 and Snail are inhibited (Fig. 9E), suggesting that CAGE and Snail are necessary for HDAC2 activity in Malme3MR cells. ChIP assay shows that HDAC2 binds to the p53 promoter sequences in SNU387R cells and Malme3MR cells (Fig. 9F). Down-regulation of CAGE inhibits binding of HDAC2 to p53 promoter sequences in SNU387R cells and Malme3MR cells (Fig. 9G). The inhibition of HDAC2 by trichostatin A (TSA), an inhibitor of histone deacetylase(s), inhibits binding of HDAC2 to p53 promoter sequences in both SNU387R cells and Malme3MR cells (data not shown). TSA restores expression and acetylation of p53 in Malme3MR cells (data not shown), implying that acetylation of p53 is necessary for p53 function and for sensitizing cells to drugs. Taken together, these results suggest that CAGE affects, directly or indirectly, HDAC2 to repress expression of p53 and confer resistance to drugs.
DISCUSSION
We examined the role of CAGE in drug resistance and studied the mechanisms associated with it. By employing tissue arrays, we found expression of CAGE in various tumor tissues but not in normal tissues (Fig. 1). The relationship between CAGE expression and clinical parameters such as metastasis is now under investigation. We previously reported the oncogenic potential of CAGE (8). In this study, we further examined tumorigenic potential of CAGE. For this, stable transfectants of HeLa cells expressing CAGE under the control of doxycycline were injected into athymic nude mice. Here, inducible expression of CAGE led to increased phosphorylation of EGFR, increased expression of MDR and BCL-2, and decreased expression of p53 (supplemental Fig. 2A) in HeLa cells as well as increased tumorigenic potential of HeLa cells (supplemental Fig. 2C). Induction of CAGE in HeLa cells also led to phosphorylation of RB as well as increased expression of cell cycle-related proteins, such as cyclin A, B, and D.4 With the oncogenic potential of CAGE established, we hypothesized that CAGE might confer resistance to drugs. With the exception of MAGE-A2, there have not been reports concerning role for cancer/testis antigen in drug resistance. Celastrol is an active ingredient of traditional Chinese herbal medicine. Celastrol acts as inducer of heat shock response (21), a suppressor of human prostate cancer growth (22), and an inhibitor of proteasome and NF-κB to enhance apoptosis (23). Celastrol was shown to potentiate radiotherapy by impairment of DNA damage processing and enhancing apoptosis in prostate cancer PC3 (24), and it was shown to exert a negative effect on the growth of human glioma xenograft by suppressing vascular endothelial growth factor receptor expression (25). Therefore, to examine the role of CAGE in drug resistance, we established drug-resistant cancer cell lines displaying resistance to celastrol and taxol (Fig. 2, A and B). In vivo drug resistance was also confirmed (Fig. 3A). Previously, we reported that the methylation status of CAGE promoter sequences was correlated with CAGE expression (5). This was supported by the fact that celastrol and taxol induced expression of CAGE in SNU387 and Malme3M cells (Fig. 3D). The induction of CAGE by drugs implies a role of CAGE in drug resistance. The CAGE promoter contains binding sites for transcription factors such as ELK-1 and GATA-1, and ChIP assay showed binding of ELK-1 in SNU387R cells and GATA-1 in Malme3MR cells, respectively (data not shown). Overexpression of DNMT1 was related with a multimodality-resistant phenotype in tumor cells (26), and moreover, overexpression of DNMT1 was seen in c-Fos-overexpressing tumor cells (26). In our study, we actually found a decreased expression of DNMT1 in SNU387R and Malme3MR cells (Fig. 4A). Inhibition of DNMT1 led to decreased expression of p53 (data not shown), suggesting that DNMT1 may be involved in drug resistance. Therefore, we examined whether down-regulation of DNMT1 would affect the expression level of CAGE. Down-regulation of DNMT1 led to the induction of CAGE in both SNU387 and Malme3M cells (Fig. 4B). Additionally, DNMT1 was shown to regulate expression of various cancer/testis genes, such as MAGE-A1, NY-ESO-1, and XAGE-1 in colorectal cancer cells (27). Down-regulation of DNMT1 also led to the activation and hypomethylation of MAGE-A1 in melanoma cells (28). 5′-Aza-2′-deoxycytidine was shown to induce DNMT-1 degradation through the proteasomal pathway (29). 5′-Aza-2′-deoxycytidine was shown to induce expression of CAGE in both SNU387 and Malme3M cells (Fig. 4C). This indicates that lack of CAGE expression in SNU387 and Malme3M cells might be due to methylation of CAGE promoter sequences. ChIP assay showed a lack of binding of DNMT1 to CAGE promoter sequences in these drug-resistant cancer cells (Fig. 4D), suggesting that demethylation is at least partially responsible for the induction of CAGE in drug-resistant cancer cell lines. Methylation-specific PCR analysis showed hypomethylation of CAGE promoter sequences in SNU387R and Malme3MR cells (Fig. 4E), which is therefore possibly responsible for drug resistance. Down-regulation of DNMT1 confers drug resistance (supplemental Fig. 1, A and C) and decreases caspase-3 activity in response to drugs (supplemental Fig. 1, B and D). This confirms that induction of CAGE by demethylation of its promoter leads to drug resistance. CAGE-1, another cancer/testis antigen gene (30), was shown to be under epigenetic regulation in primary adenocarcinomas and signet ring cell carcinomas of the urinary bladder (31).
We investigated mechanisms of drug resistance conferred by CAGE. Melanoma antigen-11 was shown to activate the hypoxic response by inactivating hypoxia-inducible factor prolyl hydroxylase 2 (32), suggesting possible involvement of cancer/testis antigens in angiogenesis. Overexpression of CAGE enhanced the angiogenic potential of SNU387 and Malme3M cells (supplemental Fig. 6, A and B). Overexpression of VEGFR1 was seen in SNU387R and Malme3MR cells, and down-regulation of CAGE decreased expression of VEGRR1 in these cells (supplemental Fig. 6C). Vascular endothelial growth factor receptor plays important roles in tumor growth and resistance to drugs (33). It is probable that enhanced angiogenic potential is closely related with the drug resistance conferred by CAGE. B7-H1 is expressed on some tumor cells and interacts with programmed death 1 protein (PD1) to exert a negative effect on cytotoxic function of CD8+ T cells (34).
Oncogenic kinase NPM/ALK induces immunosuppressive proteins such as B7-H1 and PD1 (35). We hypothesized that drug resistance maybe correlated with expression level of B7-H1. We therefore examined whether CAGE overexpression would confer resistance to cell death by CD8+T lymphocytes. SNU387R and Malme3MR cells show increased expression of B7-H1, an anti-apoptotic receptor (supplemental Fig. 7A). Down-regulation of CAGE leads to decreased expression of B7-H1 (supplemental Fig. 7A). SNU387R cells and Malme3MR cells show resistance to cytotoxic effect of CD8+ T cells (supplemental Fig. 7B). Overexpression of CAGE induces resistance of SNU387 and Malme3M cells to CD8+ T cells, whereas down-regulation of CAGE increases sensitivity of SNU387R and Malme3MR cells to CD8+ T cells (supplemental Fig. 7C). This suggests that drug resistance conferred by CAGE is closely related with resistance to cytotoxic effect of CD8+ T cells.
Up-regulation of MMP-2 is closely related with enhanced invasion of drug-resistant leukemia cells (36), and its activity is correlated with overexpression of MDR in breast cancer cells (37). Up-regulation of MMP-2 was seen in SNU387R and Malme3MR cells.4 Down-regulation of CAGE led to decreased expression of MMP-2.4 These results suggest that CAGE-induced drug resistance is closely related with enhanced invasion potential. Down-regulation of p53 increases the EGFR promoter activity (38). EGFR acts as a negative regulator of p53 (39). Notch signaling is known to regulate the expression of EGFR through p53 (40). p53 promoter contains binding sites for various transcription factors such as AP-1 and SNAIL. Because down-regulation of p53 leads to enhanced resistance to drugs (Fig. 8, C and D), it is possible that p53 may act as a negative regulator of EGFR.
It is known that Snail binds to p53 promoter sequences for the repression of p53 function (41). Snail antagonizes p53-mediated apoptosis (42). SLUG, an EMT-related gene, negatively regulates p53-mediated apoptosis by inhibiting p53-up-regulated mediator of apoptosis, a target of p53 (43).
Overexpression of CAGE leads to the induction of Snail SNU387 cells (supplemental Fig. 8A). HDAC2 interacts with Snail in SNU387R cells (supplemental Fig. 8B). We also found overexpression of Snail in SNU387R and Malme3MR cells that was reversed upon down-regulation of CAGE.4 Down-regulation of Slug led to enhanced apoptosis in neuroblastoma (12). HDAC2 interacts with Snail to exert transcriptional repression of E-cadherin and the promotion of tumor metastasis (19, 44). HDAC2 confers resistance to etoposide in pancreatic cancer cells by activating BH3-only NOXA (45). In our data, HDAC2 interacts with Snail in Malme3MR cells (Fig. 9C) and SNU387R cells (supplemental Fig. 8B). This suggests that HDAC2-SNAIL complex may repress p53 expression. Down-regulation of Snail restores expression of p53 in SNU387R cells and Malme3MR cells (supplemental Fig. 8C), suggesting that HDAC2-SNAIL complex may indeed repress p53 expression.
PKCϵ confers resistance to TRAIL through the Akt-dependent down-regulation of p53 (46). PKCα protects against apoptosis by inducing stabilization of Bcl-2 (47). PKCδ confers resistance to cisplatin in thyroid cells (48), and PKCη also confers protection against apoptosis (49). In our data, CAGE interacts with PKCα in SNU387 and CAGE interacts with PKCδ in Malme3M.4 Both PKCα and PKCδ mediate the effect of CAGE on drug resistance by regulating HDAC2 activity and p53.4 It may be necessary to determine the domain of CAGE that interacts with PKCα or PKCδ. Heptapeptide derived from the binding domain of PKCδ to HSP27 was able to restore sensitivity to radiation and cisplatin in NCI-H1299 lung cancer cells (50). This approach could be employed for the development of CAGE-derived peptides to enhance sensitivity to drugs. In this study, we found a novel role of CAGE in drug resistance in relation with HDAC2 and p53.
Taken together, these studies indicate tumorigenic potential of CAGE as well as its role in drug resistance. Therefore, it is possible that CAGE can be employed as a target of cancer immunotherapy.
Supplementary Material
This work was supported by grants from the Korea Research Foundation (C1001478-01-01, C1006272-01-01, and 2009-0063908) and by a grant (FG06-2-23) from the 21C Frontier Functional Human Genome Project from the Ministry of Science & Technology in Korea. This work was also supported by a grant from the National Research Foundation (C00016, to Y. K.) and by the Korea Research Foundation Grant funded by the Korean Government (MEST) (The Regional Research Universities Program/Medical & Bio-Materials Research Center).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8.
Y. Kim, H. Park, D. Park, Y.-S. Lee, J. Choe, J.-H. Hahn, H. Lee, Y.-M. Kim, and D. Jeoung, unpublished data.
- PKC
- protein kinase C
- ChIP
- chromatin immunoprecipitation
- DNMT1
- DNA methyltransferase 1
- EGFR
- epidermal growth factor receptor
- HDAC
- histone deacetylase
- MSP
- methylation-specific PCR.
REFERENCES
- 1.Cho B., Lim Y., Lee D. Y., Park S. Y., Lee H., Kim W. H., Yang H., Bang Y. J., Jeoung D. I. (2002) Biochem. Biophys. Res. Commun. 292, 715–726 [DOI] [PubMed] [Google Scholar]
- 2.Wortham N. C., Ahamed E., Nicol S. M., Thomas R. S., Periyasamy M., Jiang J., Ochocka A. M., Shousha S., Huson L., Bray S. E., Coombes R. C., Ali S., Fuller-Pace F. V. (2009) Oncogene 28, 4053–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai-Morris C. H., Sheng Y., Lee E., Lei K. J., Dufau M. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6373–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwata T., Fujita T., Hirao N., Matsuzaki Y., Okada T., Mochimaru H., Susumu N., Matsumoto E., Sugano K., Yamashita N., Nozawa S., Kawakami Y. (2005) Clin. Cancer Res. 11, 3949–3957 [DOI] [PubMed] [Google Scholar]
- 5.Cho B., Lee H., Jeong S., Bang Y. J., Lee H. J., Hwang K. S., Kim H. Y., Lee Y. S., Kang G. H., Jeoung D. I. (2003) Biochem. Biophys. Res. Commun. 307, 52–63 [DOI] [PubMed] [Google Scholar]
- 6.Datta J., Ghoshal K., Denny W. A., Gamage S. A., Brooke D. G., Phiasivongsa P., Redkar S., Jacob S. T. (2009) Cancer Res. 69, 4277–4285 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Lee T. S., Kim J. W., Kang G. H., Park N. H., Song Y. S., Kang S. B., Lee H. P. (2006) Ann. N.Y. Acad. Sci. 1091, 218–224 [DOI] [PubMed] [Google Scholar]
- 8.Shim H., Shim E., Lee H., Hahn J., Kang D., Lee Y. S., Jeoung D. (2006) Mol. Cells 21, 367–375 [PubMed] [Google Scholar]
- 9.Shim E., Shim H., Bae J., Lee H., Jeoung D. (2006) Biotechnol. Lett. 28, 515–522 [DOI] [PubMed] [Google Scholar]
- 10.Pang X., Yi Z., Zhang J., Lu B., Sung B., Qu W., Aggarwal B. B., Liu M. (2010) Cancer Res. 70, 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q. Q., Xu J. D., Wang W. J., Cao X. X., Chen Q., Tang F., Chen Z. Q., Liu X. P., Xu Z. D. (2009) Clin. Cancer Res. 15, 2657–2665 [DOI] [PubMed] [Google Scholar]
- 12.Vitali R., Mancini C., Cesi V., Tanno B., Mancuso M., Bossi G., Zhang Y., Martinez R. V., Calabretta B., Dominici C., Raschellà G. (2008) Clin. Cancer Res. 14, 4622–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rho J. K., Choi Y. J., Ryoo B. Y., Na I. I., Yang S. H., Kim C. H., Lee J. C. (2007) Cancer Res. 67, 1163–1169 [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., Borchert G. L., Surazynski A., Phang J. M. (2008) Oncogene 27, 6729–6737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monte M., Simonatto M., Peche L. Y., Bublik D. R., Gobessi S., Pierotti M. A., Rodolfo M., Schneider C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11160–11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S. N., Kim N. H., Lee W., Seo D. W., Kim Y. K. (2009) Mol. Cancer Res. 7, 735–744 [DOI] [PubMed] [Google Scholar]
- 17.Rountree M. R., Bachman K. E., Baylin S. B. (2000) Nat. Genet. 25, 269–277 [DOI] [PubMed] [Google Scholar]
- 18.Lai A., Lee J. M., Yang W. M., DeCaprio J. A., Kaelin W. G., Jr., Seto E., Branton P. E. (1999) Mol. Cell. Biol. 19, 6632–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peinado H., Ballestar E., Esteller M., Cano A. (2004) Mol. Cell. Biol. 24, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harms K. L., Chen X. (2007) Cancer Res. 67, 3145–3152 [DOI] [PubMed] [Google Scholar]
- 21.Westerheide S. D., Bosman J. D., Mbadugha B. N., Kawahara T. L., Matsumoto G., Kim S., Gu W., Devlin J. P., Silverman R. B., Morimoto R. I. (2004) J. Biol. Chem. 279, 56053–56060 [DOI] [PubMed] [Google Scholar]
- 22.Yang H., Chen D., Cui Q. C., Yuan X., Dou Q. P. (2006) Cancer Res. 66, 4758–4765 [DOI] [PubMed] [Google Scholar]
- 23.Sethi G., Ahn K. S., Pandey M. K., Aggarwal B. B. (2007) Blood 109, 2727–2735 [DOI] [PubMed] [Google Scholar]
- 24.Dai Y., DeSano J. T., Meng Y., Ji Q., Ljungman M., Lawrence T. S., Xu L. (2009) Int. J. Radiat. Oncol. Biol. Phys. 74, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y., Zhou Y., Fan Y., Zhou D. (2008) Cancer Lett. 264, 101–106 [DOI] [PubMed] [Google Scholar]
- 26.Mishra M. V., Bisht K. S., Sun L., Muldoon-Jacobs K., Awwad R., Kaushal A., Nguyen P., Huang L., Pennington J. D., Markovina S., Bradbury C. M., Gius D. (2008) Mol. Cancer Res. 6, 243–249 [DOI] [PubMed] [Google Scholar]
- 27.James S. R., Link P. A., Karpf A. R. (2006) Oncogene 25, 6975–6985 [DOI] [PubMed] [Google Scholar]
- 28.Loriot A., De Plaen E., Boon T., De Smet C. (2006) J. Biol. Chem. 281, 10118–10126 [DOI] [PubMed] [Google Scholar]
- 29.Ghoshal K., Datta J., Majumder S., Bai S., Kutay H., Motiwala T., Jacob S. T. (2005) Mol. Cell. Biol. 25, 4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Park S., Lim Y., Lee D., Cho B., Bang Y. J., Sung S., Kim H. Y., Kim D. K., Lee Y. S., Song Y., Jeoung D. I. (2003) Biochim. Biophys. Acta 1625, 173–182 [DOI] [PubMed] [Google Scholar]
- 31.Kunze E., Schlott T. (2007) Int. J. Mol. Med. 20, 557–563 [PubMed] [Google Scholar]
- 32.Aprelikova O., Pandolfi S., Tackett S., Ferreira M., Salnikow K., Ward Y., Risinger J. I., Barrett J. C., Niederhuber J. (2009) Cancer Res. 69, 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naumov G. N., Nilsson M. B., Cascone T., Briggs A., Straume O., Akslen L. A., Lifshits E., Byers L. A., Xu L., Wu H. K., Jänne P., Kobayashi S., Halmos B., Tenen D., Tang X. M., Engelman J., Yeap B., Folkman J., Johnson B. E., Heymach J. V. (2009) Clin. Cancer Res. 15, 3484–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karim R., Jordanova E. S., Piersma S. J., Kenter G. G., Chen L., Boer J. M., Melief C. J., van der Burg S. H. (2009) Clin. Cancer Res. 15, 6341–6347 [DOI] [PubMed] [Google Scholar]
- 35.Marzec M., Zhang Q., Goradia A., Raghunath P. N., Liu X., Paessler M., Wang H. Y., Wysocka M., Cheng M., Ruggeri B. A., Wasik M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20852–20857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J. H., Kim S. H., Cho D., Lee I. K., Kim H. J., Kim T. S. (2009) Int. J. Cancer 125, 1074–1081 [DOI] [PubMed] [Google Scholar]
- 37.Yang J. M., Xu Z., Wu H., Zhu H., Wu X., Hait W. N. (2003) Mol. Cancer Res. 1, 420–427 [PubMed] [Google Scholar]
- 38.Bheda A., Creek K. E., Pirisi L. (2008) Oncogene 27, 4315–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolev V., Mandinova A., Guinea-Viniegra J., Hu B., Lefort K., Lambertini C., Neel V., Dummer R., Wagner E. F., Dotto G. P. (2008) Nat. Cell Biol. 10, 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purow B. W., Sundaresan T. K., Burdick M. J., Kefas B. A., Comeau L. D., Hawkinson M. P., Su Q., Kotliarov Y., Lee J., Zhang W., Fine H. A. (2008) Carcinogenesis 29, 918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S. H., Lee S. J., Jung Y. S., Xu Y., Kang H. S., Ha N. C., Park B. J. (2009) Neoplasia 11, 22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurrey N. K., Jalgaonkar S. P., Joglekar A. V., Ghanate A. D., Chaskar P. D., Doiphode R. Y., Bapat S. A. (2009) Stem Cells 27, 2059–2068 [DOI] [PubMed] [Google Scholar]
- 43.Wu W. S., Heinrichs S., Xu D., Garrison S. P., Zambetti G. P., Adams J. M., Look A. T. (2005) Cell 123, 641–653 [DOI] [PubMed] [Google Scholar]
- 44.von Burstin J., Eser S., Paul M. C., Seidler B., Brandl M., Messer M., von Werder A., Schmidt A., Mages J., Pagel P., Schnieke A., Schmid R. M., Schneider G., Saur D. (2009) Gastroenterology 137, 361–371 [DOI] [PubMed] [Google Scholar]
- 45.Fritsche P., Seidler B., Schüler S., Schnieke A., Göttlicher M., Schmid R. M., Saur D., Schneider G. (2009) Gut 58, 1399–1409 [DOI] [PubMed] [Google Scholar]
- 46.Shankar E., Sivaprasad U., Basu A. (2008) Oncogene 27, 3957–3966 [DOI] [PubMed] [Google Scholar]
- 47.Villar J., Quadri H. S., Song I., Tomita Y., Tirado O. M., Notario V. (2009) Cancer Res. 69, 102–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muscella A., Urso L., Calabriso N., Vetrugno C., Rochira A., Storelli C., Marsigliante S. (2009) Br. J. Pharmacol. 156, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rotem-Dai N., Oberkovitz G., Abu-Ghanem S., Livneh E. (2009) Exp. Cell Res. 315, 2616–2623 [DOI] [PubMed] [Google Scholar]
- 50.Kim E. H., Lee H. J., Lee D. H., Bae S., Soh J. W., Jeoung D., Kim J., Cho C. K., Lee Y. J., Lee Y. S. (2007) Cancer Res. 67, 6333–6341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.