Abstract
Here we identify a previously undescribed protein, HemQ, that is required for heme synthesis in Gram-positive bacteria. We have characterized HemQ from Bacillus subtilis and a number of Actinobacteria. HemQ is a multimeric heme-binding protein. Spectroscopic studies indicate that this heme is high spin ferric iron and is ligated by a conserved histidine with the sixth coordination site available for binding a small molecule. The presence of HemQ along with the terminal two pathway enzymes, protoporphyrinogen oxidase (HemY) and ferrochelatase, is required to synthesize heme in vivo and in vitro. Although the exact role played by HemQ remains to be characterized, to be fully functional in vitro it requires the presence of a bound heme. HemQ possesses minimal peroxidase activity, but as a catalase it has a turnover of over 104 min−1. We propose that this activity may be required to eliminate hydrogen peroxide that is generated by each turnover of HemY. Given the essential nature of heme synthesis and the restricted distribution of HemQ, this protein is a potential antimicrobial target for pathogens such as Mycobacterium tuberculosis.
Keywords: Enzymes, Heme, Metabolism, Porphyrin, Spectroscopy, Genome Analysis, Hemoprotein
Introduction
Heme is an essential compound for the vast majority of living organisms (1). In microorganisms, it is required for energy generation via respiratory chain cytochromes, xenobiotic metabolism by cytochrome P450s, fatty acid desaturation, oxidases, peroxidases, redox (2), and gas sensing (3, 4). More recently, a clear role for heme as a regulatory ligand for a variety of transcription factors such as Bach1 and SREBP has been identified (5–7). In addition, exogenously supplied heme serves as a significant source of iron for many organisms, including pathogenic bacteria (8). Biological heme synthesis is a process that occurs in most organisms that possess heme-containing proteins, and with the exception of only a few organisms, it is an essential biosynthetic pathway. Notable exceptions among eucaryotes are the helminths (9), the cattle tick Boophilus microplus (10), and parasitic Trypanosomatids (11), which appear to lack the pathway completely and acquire heme from their environment. Interestingly, some eucaryotes that lack functional heme biosynthetic pathways maintain bacterial endosymbionts that supply their heme requirement (12). A few procaryotic pathogens and strict anaerobes also lack the ability to synthesize heme (8).
The heme biosynthetic pathway is evolutionarily ancient, and the pathway intermediates are relatively well conserved. However, some differences in pathway enzymes are known to exist between organisms (13, 14). In general three distinctions can be made as follows: (i) plants and most bacteria synthesize the first intermediate, 5-aminolevulinate, from glutamyl-tRNA, although animals and a few bacteria utilize succinyl-CoA and glycine as starting compounds; (ii) anaerobic and facultative bacteria possess oxygen-independent enzymes for two oxidative steps, coproporphyrinogen oxidase and protoporphyrinogen oxidase (PPO),2 that utilize molecular oxygen in aerobes; and (iii) the terminal two enzymes, PPO and ferrochelatase, are membrane-associated in all organisms except for Gram-positive bacteria, where they are soluble.
In the metazoans, all of the pathway enzymes are nuclearly encoded, but the first, 5-aminolevulinate synthase, and last three enzymes, coproporphyrinogen oxidase, PPO, and ferrochelatase, are located in the mitochondria with the remaining four being located in the cytoplasm (1, 13). In eucaryotes, it is now generally accepted that the terminal two enzymes are associated on the opposite sides of the inner mitochondrial membrane so that a transient transmembrane complex exists between PPO and ferrochelatase (15–17). In Gram-negative bacteria, these proteins are also membrane-associated, but their spatial orientations have yet to be determined. It is not known if they are both located on the cytoplasmic side of the cytoplasmic membrane or if they are organized in a fashion similar to what is found in eucaryotes. In Gram-positive organisms, these terminal enzymes are soluble and not membrane-associated (18, 19).
In this study, we undertook the characterization of the terminal enzymes of heme biosynthesis from Gram-positive bacteria to see what ramifications, if any, result from these proteins being soluble rather than membrane-associated. We examined the enzymes from the Actinobacteria, a group of high G + C content Gram-positive bacteria, in particular because of the biomedical importance of this group and because these organisms all have a ferrochelatase (HemH) that possesses a [2Fe-2S] cluster similar to animal ferrochelatases (19). We have also examined the enzymes from Bacillus because the proteins of interest have been structurally characterized to varying degrees (20, 21). By employing bioinformatic approaches, we discovered a previously uncharacterized open reading frame that was found to encode a protein we named HemQ. This protein was found to be essential for heme synthesis in vivo and in vitro. Here, we have expressed HemQ and biochemically characterized the protein.
EXPERIMENTAL PROCEDURES
Cloning
Mycobacterium tuberculosis HemY and HemQ were polymerase chain reacted from genomic DNA (gift of F. Quinn, University of Georgia) and cloned into the NheI/HindIII sites of pTrcHisA (Invitrogen). M. tuberculosis HemH was cloned as described previously (19). Streptomyces coelicolor HemY, HemH, and HemQ were polymerase chain reacted from genomic DNA (gift of J. Westpheling, University of Georgia) and cloned into the NheI/HindIII sites of pTrcHisA. The Propionibacterium acnes HemH/Q fusion was polymerase chain reacted from genomic DNA (gift of A. Strittmatter, Georg-August-Universitat, Gottingen, Germany) and cloned into the NheI/HindIII sites of pTrcHisA. P. acnes HemY was polymerase chain reacted from genomic DNA and was engineered to contain a carboxyl-terminal His6 tag and cloned into the NcoI/HindIII sites of pTrcHisA. Bacillus subtilis HemH and HemY were polymerase chain reacted from genomic DNA (gift of the Dept. of Microbiology, University of Georgia) and cloned into the NheI/HindIII sites of pTrcHisA. B. subtilis HemQ was polymerase chain reacted from genomic DNA and cloned into the NheI/XhoI sites of pTrcHisA. All constructs were sequence-verified by the Georgia Genomics Facility. Site-directed mutants were generated using the QuikChange protocol (Stratagene).
Protein Expression and Purification
Protein production in Escherichia coli and subsequent purification of the recombinant proteins were as described previously (19). For production of heme-loaded HemQ, 1 mg of hemin (Sigma) per ml in DMSO was added to the ultracentrifuge supernatant before loading onto the HisPur (Pierce) metal chelate column. Fast protein liquid chromatography was carried out using an Aktaprime equipped with a Hi-Prep Sephacryl S-300 column (GE Healthcare) under low ionic strength buffer of 10 mm Na-MOPS, pH 7.2.
Complementation Studies
The HemG-deficient E. coli strain SASX38 (22) was made competent for electroporation. Purified plasmid DNA (Roche Applied Science) was electroporated into these cells in the combinations required. The HemH-deficient E. coli strain Δvis (23, 24) was made chemically competent (25) and transformed with plasmid DNAs. In all cases, the transformations were plated on LB-ampicillin plates and incubated at 37 °C overnight. Resulting colonies were inoculated into LB-ampicillin broth and incubated overnight at 37 °C, and plasmid DNA was reisolated to confirm the presence of the expected plasmids.
Enzymatic Assays
Protoporphyrinogen oxidase assays were done as described previously (26) with the addition of 5 mm EDTA to the reaction mixtures. Briefly, 100 nm P. acnes HemY alone or with either 100 nm HemH/Q or 100 nm HemH/Q preloaded with heme was assayed, and activity was monitored at 37 °C using a Synergy HTI plate reader (Biotek, Winooski, VT). The coupled assays of HemQ, HemH, and HemY were done by measuring the amount of heme that was produced in the presence of protoporphyrinogen IX and iron. Reaction mixtures consisted of 1 μm enzymes, 66 mm Tris-HCl, pH 8.0, 5 mm glutathione, 3.3% Tween 20, 100 μm 2-mercaptoethanol, 100 μm ferrous ammonium sulfate, and ∼25 μm protoporphyrinogen IX. In some assays coproporphyrinogen III replaced protoporphyrinogen IX, and in one control, 25 μm protoporphyrin IX was added instead. Heme produced was quantitated as its pyridine hemochromogen (27). Ferrochelatase was assayed by the continuous spectroscopic assay with either protoporphyrin or mesoporphyrin as substrate (28). Catalase activity was monitored by disappearance of hydrogen peroxide spectroscopically at 240 nm (29), and peroxidase activity was monitored with pyrogallol as electron acceptor (30). MALDI-TOF analysis was performed by the proteomics facility at the University of Georgia.
Spectroscopic Methods
Spectroscopic studies were carried out on HemQ samples in 50 mm Tris-MOPS buffer, pH 8.0, with 0.1 mm KCl, 1% sodium cholate, and 20% (v/v) glycerol, unless otherwise indicated. UV-visible spectroscopy was performed using a Cary 1G spectrophotometer (Varian). X-band EPR spectra were recorded using a Bruker ESP-300E spectrometer equipped with an Oxford Instruments ESR-9 flow cryostat. Near-IR MCD measurements were carried out using an Oxford Instruments Spectromag 4000 split-coil superconducting magnet (0–7 T) mated to a Jasco J730 spectropolarimeter (700–2000 nm). Samples for low temperature near-IR MCD studies were exchanged into the equivalent D2O buffer and contained 50% (v/v) d3-glycerol to enable an optical quality glass to form on freezing.
RESULTS
Identification of HemQ as a Required Protein for Heme Synthesis
The terminal two heme biosynthetic synthetic enzymes are PPO and ferrochelatase. In the facultative bacterium E. coli, the PPO step occurs in an oxygen-independent fashion that utilizes HemG, a menadione-dependent flavodoxin (31). The E. coli ΔhemG mutant (originally named SASX38 (22)) grows poorly in culture because it is unable to synthesize heme and must therefore ferment growth substrates. This mutant, however, is functionally complemented to normal wild-type growth when supplied with a Gram-negative bacterial HemY-type PPO or eucaryotic PPO, both of which are FAD-containing, membrane-associated, and oxygen-requiring enzymes that are structurally and functionally similar (32). We discovered that the E. coli ΔhemG mutant is not complemented by plasmid-introduced hemY from Gram-positive organisms, including B. subtilis, M. tuberculosis, S. coelicolor, or P. acnes. In addition, E. coli ΔhemH, which is complemented by ferrochelatases from eucaryotes and Gram-negative bacteria, is not complemented by hemH from these Gram-positive organisms.
These results were of particular interest because HemY and HemH from Gram-positive bacteria are homologous to HemY and HemH from Gram-negative bacteria and eucaryotic PPOs and ferrochelatases, although they differ in that the enzymes from Gram-positive bacteria are soluble rather than membrane-associated. The recombinant proteins are expressed in stable forms by the E. coli expression system and are active when measured in vitro, yet clearly they do not function in vivo in E. coli. These data demonstrate that for functional complementation of the mutant strains of E. coli, a membrane-bound form of HemY or HemH is required and/or another unidentified protein that is not present in E. coli is essential for the Gram-positive forms of these enzymes to be catalytically competent.
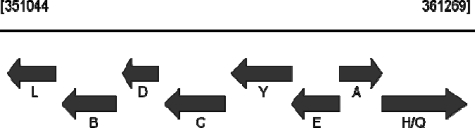
To approach the second possibility, we initially carried out an examination of available genomic data from select members of the Actinobacteria. This approach revealed that an open reading frame, which was annotated as a chlorite dismutase-like hypothetical protein, is routinely found adjacent to either the hemY or hemH (also named hemZ in the Actinobacteria) gene (Fig. 1 and Table 1). In P. acnes, this open reading frame is fused in-frame with the reading frame for hemH. Given the genomic context of the gene for this hypothetical protein as well as the presence of the hemH fusion in P. acnes, it was reasonable to expect that this protein has some connection to heme synthesis in these organisms.
FIGURE 1.
Genomic organization of the heme biosynthetic pathway genes in P. acnes (adapted from NCBI Entrez Gene). The entire heme biosynthetic pathway in this organism is organized into two uber operons (58 and 59). The numbers above the line denote the sequence position of this operon. The letters signify the hem gene names and correspond to the following: hemA, glutamyl tRNA reductase; hemB, porphobilinogen synthase (5-aminolevulinate dehydratase); hemC, hydroxybilane synthase (porphobilinogen deaminase); hemD, uroporphyrinogen synthase; hemE, uroporphyrinogen decarboxylase; hemL, glutamate 1-semialdehyde aminotransferase; hemY, protoporphyrinogen oxidase; and hemH/Q, ferrochelatase-hemQ fusion.
TABLE 1.
Actinobacteria possessing hemQ
Representative list of Actinobacteria containing the gene for hemQ that is adjacent to hemH or hemY.
| Organism | Adjacent gene |
|
|---|---|---|
| hemH | hemY | |
| Corynebacterium jeikeium KY11 | + | |
| Frankia alni | + | |
| M. tuberculosis/avium/leprae | + | |
| Nocardia farcinica | + | |
| P. acnes | + | |
| Rhodococcus sp. RHA1 | + | |
| S. coelicolor/avermitilis | + | |
| Thermobifida fusca YX | + | |
| Trophyeryma whipplei | + | |
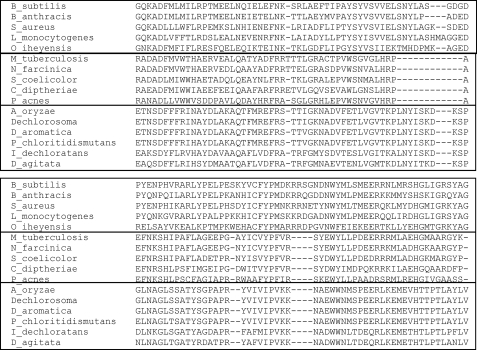
A Blast search of the genome data base using the predicted amino acid sequence for this unknown open reading frame yielded genes for “chlorite dismutase-like” hypothetical proteins that are common among Gram-positive bacteria but not Gram-negative bacteria or eucaryotes. These proteins form the cluster of the orthologous group COG3253. When representative members of this cluster are subjected to ClustalW analysis (33), it becomes clear that three groups exist within this cluster (Fig. 2). These are as follows: 1) proteins for which several group members have been shown to have chlorite dismutase activity; 2) proteins that have no determined function but are not chlorite dismutases, and 3) proteins lacking any known function which are present only in the Actinobacteria.
FIGURE 2.
ClustalW analysis of select COG3253 proteins. The segment shown spans from residues number 71 to 186 of the B. subtilis protein. The sequences are boxed to show HemQ sequences from Gram-positive bacteria on top, HemQ sequences of Actinobacteria in the middle, and chlorite dismutases on the bottom.
As mentioned above, actinobacterial hemY and hemH do not complement E. coli ΔhemG and ΔhemH, respectively. Expression of both HemY and HemH together also do not complement either mutant (Table 2). A role for the Actinobacteria COG3253 protein in heme synthesis in vivo was demonstrated when HemY, HemH, and the COG3253 protein were all expressed simultaneously in either E. coli ΔhemG or ΔhemH. Under these conditions, both E. coli ΔhemG and ΔhemH were complemented to wild-type growth. Expression of the Actinobacteria COG3253, which we now have named HemQ, alone or in combination with either hemY or hemH individually in either mutant strain did not result in complementation. P. acnes possesses a fused hemQ/hemH gene, and expression of this fusion with hemY complemented both mutants. The hemQ/hemH fusion alone did not complement either E. coli ΔhemG or ΔhemH.
TABLE 2.
Complementation of E. coli ΔhemG with various expression plasmids
Plasmids for recombinant protein expression in E. coli were transformed into the hemG-deficient strain SASX38 (22). Positive complementation resulted in the growth of wild-type sized E. coli colonies after overnight incubation at 37 °C.
| Protein expressed from plasmid | Complementation |
|---|---|
| pTrcHisA (empty vector) | − |
| Human protoporphyrinogen oxidase | + |
| S. coelicolor HemY | − |
| S. coelicolor HemY + HemH | − |
| S. coelicolor HemY + HemQ | − |
| S. coelicolor HemY + HemH + HemQ | + |
| M. tuberculosis HemY | − |
| M. tuberculosis HemY + HemH | − |
| M. tuberculosis HemY + HemQ | − |
| M. tuberculosis HemY + HemH + HemQ | + |
| P. acnes HemY | − |
| P. acnes HemY + HemH/Q | + |
| B. subtilis HemY | − |
| B. subtilis HemY + HemH | − |
| B. subtilis HemY + HemQ | − |
| B. subtilis HemY + HemH + HemQ | + |
| P. acnes HemY + M. tuberculosis HemH | − |
| P. acnes HemY + M. tuberculosis HemQ | − |
| P. acnes HemY + M. tuberculosis HemQ +HemH | + |
| P. acnes HemH/Q and M. tuberculosis HemY | + |
| M. tuberculosis HemY + HemH + B. subtilis HemQ | + |
| M. tuberculosis HemH + HemQ + B. subtilis HemY | + |
| M. tuberculosis HemY +HemQ + B. subtilis HemH | + |
| B. subtilis HemY + HemH + M. tuberculosis HemQ | + |
Given that the non-chlorite dismutase COG3253 proteins in other Gram-positive bacteria are distinct enough from actinobacterial HemQ to cluster apart from HemQ in ClustalW analysis and that the genes for these proteins were not found in association with heme biosynthesis proteins in Bacillus and Staphylococcus, it was not immediately obvious that HemQ existed in all heme-synthesizing Gram-positive organisms and not just in the Actinobacteria. However, examination of all currently available Gram-positive genome sequences with the Signature Tool in the SEED data base (34) along with co-localization of genes and data from Staphylococcus aureus microarray gene expression experiments (35) identified the COG3253 as a putative heme biosynthesis-related protein in all Gram-positive organisms. To confirm this relationship, B. subtilis COG3253 protein was expressed along with the B. subtilis hemY and hemH in E. coli ΔhemG and ΔhemH in the same fashion as described for the Actinobacteria hemY/Q/H experiments. The data obtained (Table 2) were identical to those described above. In addition, we tested mixed combinations of the B. subtilis and M. tuberculosis hemY/Q/H and found that HemQ was interchangeable despite the significant sequence difference.
Expression and Isolation of HemQ
To explore how HemQ may participate in heme synthesis, it was necessary to clone, express, and purify HemY, HemH, and HemQ from representative organisms. Previously, we had expressed and characterized HemH from M. tuberculosis (19) and S. coelicolor.3 For this study, HemY from these organisms was cloned with an amino-terminal His6 tag, expressed, purified, and characterized. It was found that HemY from these organisms is a soluble monomeric protein with a molecular weight of ∼47,000 and a bound FAD. This is similar to HemY of B. subtilis (36) but distinct from HemY of Gram-negative bacteria and eucaryotic PPOs, which are membrane-associated homodimers (37).
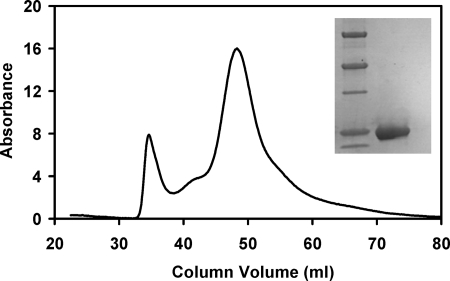
Various cloned HemQs were engineered to have an amino-terminal His6 tag to allow for purification by metal affinity chromatography as is done with HemY and HemH (19, 36). The molecular mass of recombinant HemQ by SDS-PAGE (Fig. 3) and the predicted amino acid sequence are ∼26,000 daltons. Nondenaturing gel filtration of purified M. tuberculosis HemQ is consistent with a homo-multimeric protein (Fig. 3), which is similar to other COG3253 proteins (38, 39). Although the predicted molecular size of the multimer on gel exclusion chromatography corresponds to a globular, soluble protein of ∼200,000 daltons, the fact that all COG3253 proteins for which a crystal structure exists clearly show that the homo-oligomer is donut-shaped with a large central hole is consistent with HemQ being a hexameric or pentameric molecule.
FIGURE 3.
Sephacryl S-300 size exclusion chromatography and SDS-PAGE of purified HemQ from M. tuberculosis. Details are under “Experimental Procedures.” The 1st eluted peak corresponds to the column void volume. The major protein peak elutes at a position consistent with a soluble globular protein of ∼200,000 daltons. The inset shows purified HemQ adjacent to molecular weight markers (Bio-Rad Precision Plus, where the darker bands correspond to 75,000, 50,000, and 25,000, respectively) in an SDS-polyacrylamide gel. HemQ migrates with an apparent molecular weight of 26,000.
HemY and the HemQ/HemH fusion protein from P. acnes were also cloned, expressed, and characterized. By SDS-PAGE, the fusion protein was of the expected size, although presumed proteolytic products the size of HemH and HemQ were also noted. The P. acnes protein was confirmed by MALDI-TOF as a fusion of HemH and HemQ (data not shown). HemY (36) and HemH (40) of B. subtilis have previously been expressed, purified, and characterized. HemQ from this organism was expressed and purified as described for all other HemQs.
The possibility that HemY, HemQ, and HemH exist as a stable multimeric complex was examined in two ways. First, all three proteins were co-expressed in the E. coli ΔhemG strain, isolated by metal chelate chromatography and then subjected to gel filtration. Second, all three proteins were individually expressed, purified, and then mixed together before being subjected to gel filtration. In both instances, the individual components eluted in separate fractions and not as a complex. Using apo-HemQ versus heme-loaded HemQ gave identical results. Although these data do not provide experimental support for the presence of a stable complex of the recombinant proteins, it does not rule out the existence of a transient complex, or the possibility that an in vivo stable complex is not stable to isolation with the purification protocol.
Characterization of Heme Binding by HemQ
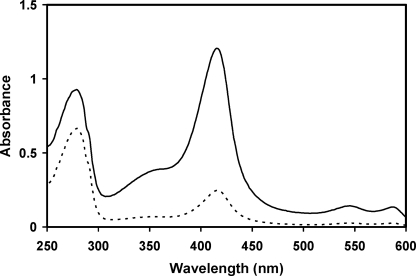
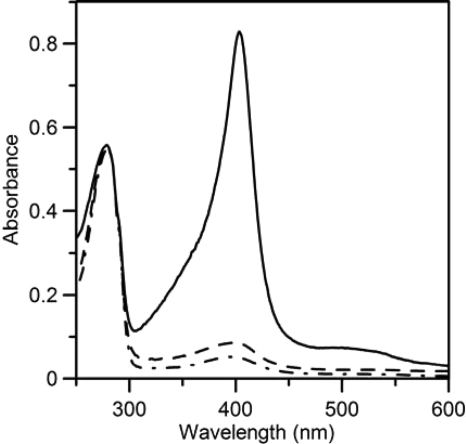
As isolated, the recombinant HemQ has minor absorbance at 410 nm that is attributable to the presence of a small and variable amount of heme (Fig. 4). Given that HemQ has significant sequence similarity to chlorite dismutase and contains all of the residues previously identified as being required for chlorite dismutase to possess a bound heme (38), the possibility that HemQ also has a heme-binding site was examined. When a solution of hemin was added to crude cell extracts of HemQ-expressing E. coli prior to protein purification via metal chelate chromatography, the HemQ protein that was isolated was a bright red hemoprotein. The visible absorbance was typical of a noncovalently bound heme b-containing protein (Fig. 4).
FIGURE 4.
UV-visible absorption spectra of M. tuberculosis HemQ. The figure shows the spectra of HemQ as purified in the presence of 250 mm imidazole with (solid line) and without (dashed line) added hemin. The base line of the two scans is shifted for clarity.
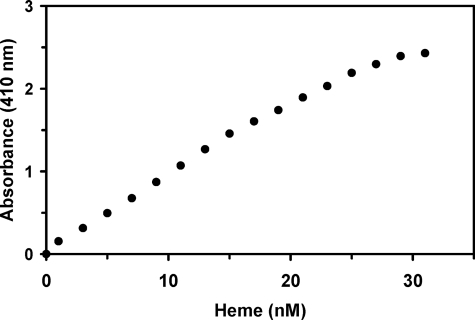
It was possible to titrate the apo-HemQ with heme (either ferro- or ferri-heme) and follow this spectroscopically (Fig. 5). The data obtained do not yield a sharp transition point suggesting that HemQ binds heme poorly, is capable of binding more than one heme, or that the individual subunits in the multimeric structure do not have a uniform binding affinity. Direct titration followed by an increase in the Soret band proved problematic because the peak absorbance began shifting from that characteristic of the hemoprotein to that of free heme prior to the addition of stoichiometric amounts of heme. Furthermore, dialysis of the heme-loaded HemQ results in release of bound heme so that the isolation of a fully heme-loaded HemQ in the absence of free heme is not possible. Without an accurate extinction coefficient for the hemoprotein, an accurate binding constant could not be determined. A best estimate based upon available data would suggest a kd of ∼30–40 μm.
FIGURE 5.
Titration of apo-HemQ with hemin. Purified HemQ (19.5 μm) was repetitively scanned following successive additions of freshly prepared hemin (100 μm stock solution in 1% (w/v) Triton X-100). Absorbance at 410 nm was plotted against the final concentration of hemin added.
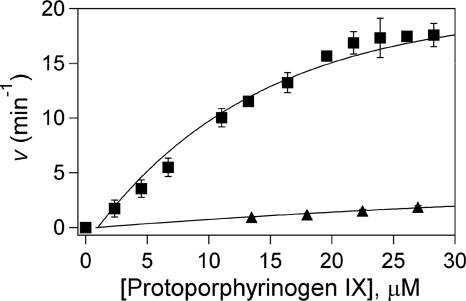
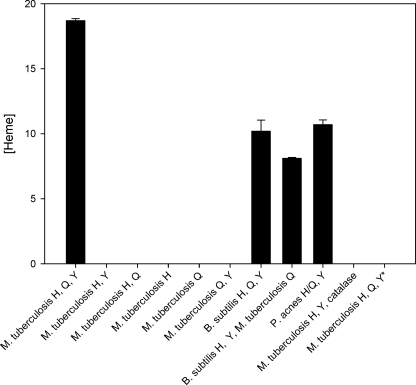
HemY of P. acnes assayed in vitro has a Vmax of 3.0 min−1, which is above that reported for B. subtilis (0.05 min−1) (36) but lower than that of other reported PPOs (41, 42). The Km for protoporphyrinogen is 19 μm. Addition of holo-HemQ (with heme bound) increased the measured Vmax to 19.8 min−1 and decreased the Km to 10.1 μm. HemQ without added heme had a minimal impact on HemY activity (Fig. 6). HemH from M. tuberculosis assayed in vitro with iron and mesoporphyrin had a specific activity of 1.1 min−1. Addition of holo- or apo-HemQ had no impact on HemH activity. In vitro HemY plus HemH together had no detectable activity when protoporphyrinogen and ferrous iron were added as substrates. However, in the presence of HemY+, HemQ+, and HemH, there was significant enzyme activity with protoporphyrinogen and ferrous iron as substrates (Fig. 7). If the HemQ that was added to this last experiment was apo-HemQ, the overall activity was lower than what was found when heme-loaded HemQ was used. However, given that preparations of “apo-HemQ” always have some small amount of bound heme and that any heme formed during the reaction could bind to any apo-HemQ and convert it to the holo-hemoprotein, it is not possible to determine whether apo-HemQ is functional in the overall reaction. No heme was formed if protoporphyrin, rather than protoporphyrinogen, was present with ferrous iron. Additionally, if coproporphyrinogen, rather than protoporphyrinogen, and iron were supplied, no heme was formed in vitro suggesting that HemQ either does not have coproporphyrinogen oxidase activity or that this activity requires the presence of some compound not present in the reaction mixture.
FIGURE 6.
Stimulation of HemY activity by HemQ. HemY from P. acnes was assayed as described in the text with HemQ that did not have added heme (lower curve) and with heme-loaded HemQ (upper curve).
FIGURE 7.
Heme synthesis from protoporphyrinogen IX and ferrous iron. The bar graph shows heme formation in vitro by 1 nmol of HemY and HemH. When heme-loaded HemQ was employed in the assay, the amount of heme (present in holo-HemQ) added was quantitated and accounted for less than 5% of total heme in assays with heme forming activity. In one experiment (far right) HemQ was replaced with catalase, and in one experiment protoporphyrin, rather than the porphyrinogen, was supplied. In neither of these instances was heme formation observed.
HemQ variants in which the conserved histidine (His-156 in M. tuberculosis HemQ), a potential axial ligand for the heme iron based on the crystal structure of chlorite dismutase from Azospira oryzae (38), is mutated to alanine possessed minimal activity (less than 10% of wild-type HemQ, data not shown) in the three protein system but did complement E. coli ΔhemG. However, it should be recognized that E. coli requires only minimal amounts of heme to functionally complement the mutant and that other studies have shown that human ferrochelatase variants that possess only a few percent of wild-type activity will readily complement E. coli ΔhemH. Mutation of this histidine to cysteine resulted in a protein that lacked detectable activity in vitro and did not complement E. coli ΔhemG.
Because other COG3253 proteins have been found to have low peroxidase activity, we also assayed holo-HemQ from both M. tuberculosis and B. subtilis for peroxidase activity with both pyrogallal and guaiacol as substrates with hydrogen peroxide. Holo-HemQ has low peroxidase activity with specific activities of 380 and 350 min−1 with pyrogallal (B. subtilis and M. tuberculosis, respectively) and 10 min−1 with guaiacol for M. tuberculosis. These values are at least 3 orders of magnitude below those reported for typical peroxidases. These HemQs were also assayed for catalase activity and found to have turnovers of 1.4 × 104 and 1.7 × 104 min−1 for B. subtilis and M. tuberculosis, respectively.
Spectroscopic Characterization of the Heme Center in HemQ
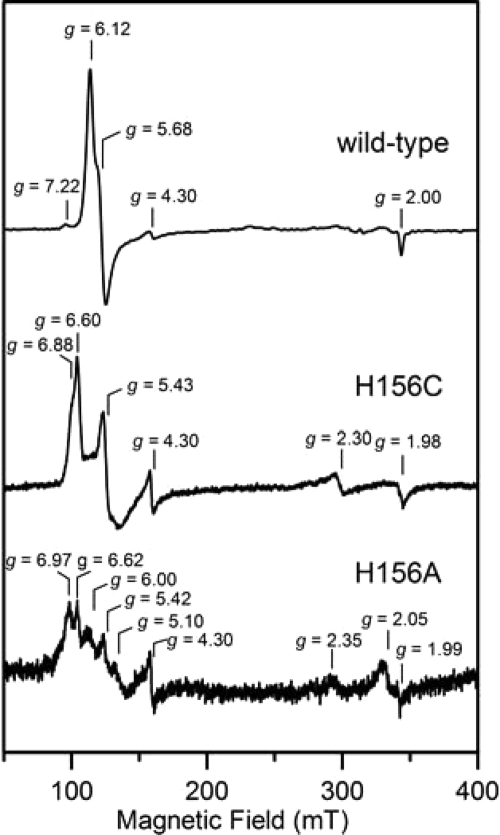
Recombinant forms of heme-reconstituted wild type, H156C, and H156A M. tuberculosis HemQ were initially purified in the presence of 250 mm imidazole, which was used to elute the protein from the metal affinity column. The UV-visible absorption and EPR spectra after extensive dialysis to remove imidazole are shown in Figs. 8 and 9, respectively. Removal of the imidazole from wild-type HemQ shifts the heme Soret band from 414 to 403 nm and the αβ bands from 588 and 548 nm to a broad band centered near 510 nm (Figs. 4 and 8). These spectral shifts are indicative of conversion from a six-coordinate low spin ferric heme to a five-coordinate high spin ferric heme, a conclusion that is confirmed by EPR studies (see below). The H156C and H156A variants both showed similar Soret band maxima to that of wild-type HemQ. However, the variant samples contain <10% of the bound heme found in wild type, indicating that these mutations dramatically reduce heme binding and suggest that His-156 is a heme axial ligand in HemQ. After removal of imidazole, EPR studies of wild type, H156C, and H156A HemQ demonstrate that all three contain high spin ferric heme centers as evidenced by resonances from the lowest Ms = ±1/2 doublet of the S = 5/2 ground state (Fig. 9). Wild-type HemQ has a near-axial major high spin ferric heme component with effective g values of 6.12, 5.68, and 2.00 and a variable intensity more rhombic minor component with effective g values of 7.22, ∼5.8, and ∼1.9. In accord with the lower heme content in H156C and H156A HemQ and the conclusion that His-156 is a heme ligand in wild-type HemQ, both variants exhibit distinct and substantially weaker high spin ferric EPR signals. H156C HemQ exhibits two similar high spin ferric heme resonances with effective g values of 6.60, 5.42, 1.98, 6.88, ∼5.20, and ∼1.97. H156A HemQ is more heterogeneous with at least three resolved high spin ferric heme resonances evident in the low field region.
FIGURE 8.
UV-visible absorption spectra of heme-reconstituted wild-type (solid line), H156C (dot-dashed line), and H156A (dashed line) M. tuberculosis HemQ after dialysis to remove imidazole. The spectra have been normalized to give the same absorbance at 280 nm.
FIGURE 9.
EPR spectra of heme-reconstituted wild-type, H156C, and H156A M. tuberculosis HemQ after dialysis to remove imidazole. The EPR spectra were recorded at 10 K with 1 milliwatt microwave power using a microwave frequency of 9.60 GHz and a modulation amplitude of 0.65 mT.
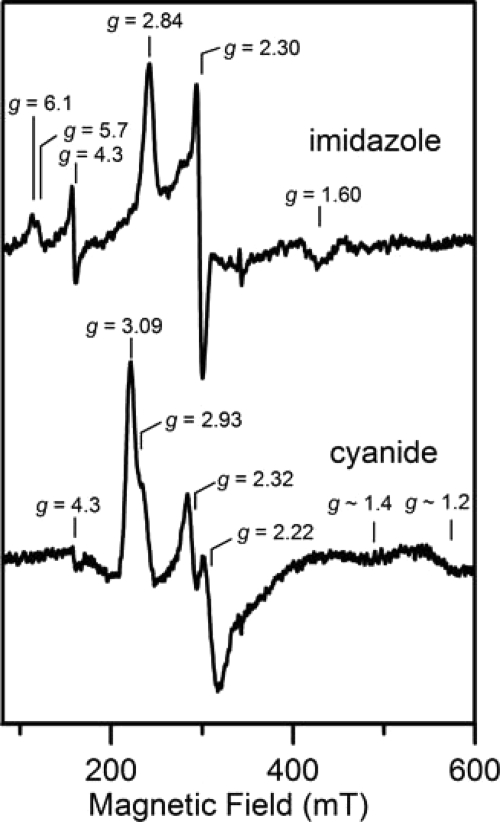
The high spin ferric heme species in HemQ is indicative of a pentacoordinate iron center or a hexacoordinate iron center with a weak sixth ligand such as H2O. Hence, the heme iron is likely to be accessible to small molecule substrates and other small molecule ligands such as imidazole and cyanide. This was confirmed by UV-visible absorption, EPR, and low temperature MCD studies of oxidized HemQ in the presence of excess imidazole or cyanide, which clearly demonstrate ligand binding at the vacant iron coordination site and conversion to a low spin (S = 1/2) ferric heme. The absorption spectrum in the presence of excess imidazole is shown in Fig. 4, and excess cyanide effects similar changes in the heme absorption bands with the Soret band shifting to 420 nm and the αβ bands to 585 and 545 nm, respectively (data not shown). In both cases, EPR studies demonstrate the loss of the high spin ferric resonances and the appearance of S = 1/2 low spin ferric resonances (Fig. 10). The imidazole-bound form of ferric HemQ has g values of 2.84, 2.30, and 1.60 and is similar to that reported for the imidazole-bound ferric form of chlorite dismutases (g ∼2.96, 2.25, and 1.51) (38, 43). The cyanide-bound form is a mixture of two low spin ferric species, a major component with g = 3.09, 2.22, and ∼1.2 and a minor component with g = 2.93, 2.32, and ∼1.4. Cyanide binds with much greater affinity than imidazole as the addition of a 25-fold excess of cyanide to samples containing a 250-fold excess of imidazole results in complete conversion to the cyanide-bound form.
FIGURE 10.
EPR spectra of heme-reconstituted M. tuberculosis HemQ in the presence of 250-fold excess of imidazole and a 25-fold excess of sodium cyanide. The EPR spectra were recorded at 10 K with 5 milliwatt microwave power using a microwave frequency of 9.60 GHz and a modulation amplitude of 0.65 mT.
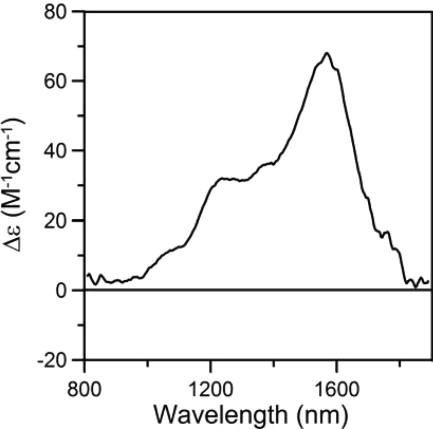
The combination of EPR and near-IR MCD spectroscopy has proven a powerful methodology for the determining the axial ligands of low spin ferric hemes (44). The energy of porphyrin(π)-to-ferric charge transfer bands that are best observed in the near-IR MCD spectrum at liquid helium temperatures and high magnetic fields serve a sensitive indicator of the nature of axial ligands, particularly when used in conjunction with the observed g value anisotropy in the S = 1/2 EPR signal. Fig. 11 shows the near-IR MCD spectrum of imidazole-bound HemQ at 1.8 K with an applied field of 6 T. The spectrum is characteristic of a low spin ferric heme and includes two positive temperature-dependent MCD bands that arise from porphyrin (π)-to-ferric charge transfer transitions. The lower energy band is invariably narrower and more intense and provides a convenient marker of axial heme ligation. For imidazole-bound HemQ, this band is observed at 1564 nm, the range identified for a low spin ferric heme with bis-imidazole ligation (1500–1660 nm) (44, 45). Taken together with the EPR-determined g value anisotropy (g = 2.84, 2.30, and 1.60), which is also indicative of bis-imidazole ligation (44, 45), the results indicate that the heme center in imidazole-bound HemQ is axially coordinated by an indigenous histidine ligand and an exogenous imidazole ligand. As discussed above, mutagenesis results identify the indigenous histidine ligand as the rigorously conserved histidine, His-156, in M. tuberculosis HemQ.
FIGURE 11.
Near-IR low temperature MCD spectrum of heme-reconstituted M. tuberculosis HemQ in the presence of 250-fold excess of imidazole. The spectrum was recorded with an applied magnetic field of 6 T at 1.8 K.
DISCUSSION
Since the pioneering work of Shemin and Rittenberg (46), the enzymes of heme synthesis have been studied to varying extents, and until recently, it has long been assumed that all of the pathway enzymes have been identified. With the advent of genomic data for an increasing number of organisms, it is now clear that some of the enzymes of the pathway are “missing” in some bacteria (47). In particular, the proteins catalyzing the antepenultimate and penultimate steps have yet to be identified for a variety of bacteria. Even so, for bacteria that possess all known heme biosynthetic enzymes, there was no reason to anticipate that additional essential proteins might exist. This work demonstrates that for at least the Gram-positive bacteria an additional protein, HemQ, is an essential component of heme synthesis in these organisms.
Gram-positive bacteria differ from other organisms in that they possess monomeric, soluble protoporphyrinogen oxidases (HemY) and ferrochelatases (HemH) rather than homodimeric, membrane-associated enzymes. There has been considerable research on HemH of B. subtilis, including numerous structural studies (20, 48). HemY from this organism has been studied with respect to its resistance to photosensitizing herbicides (36), and there is a recent crystal structure of the protein (21). However, there are little data on these enzymes in other Gram-positive bacteria. In this study, we examined the terminal enzymes of heme biosynthesis in the Actinobacteria and B. subtilis. The Actinobacteria are a group of bacteria of particular interest because many of them are animal or plant pathogens. Early in our studies, we noted that these proteins, when expressed in mutants of E. coli that lacked the ability to make either a functional protoporphyrinogen oxidase or ferrochelatase, did not complement the heme auxotrophy. This was unexpected because these mutant strains are effectively complemented by all other protoporphyrinogen oxidases or ferrochelatases, whether from animals or Gram-negative bacteria.
During bioinformatic investigations of hemY and hemH in Mycobacterium, Propionibacterium, and Streptomyces species, it was noted that the gene adjacent to either hemH or hemY was annotated as a chlorite dismutase-like hypothetical protein. Uber-operon analysis (49) for Mycobacterium and Propionibacterium reveals that this gene, which we name hemQ, exists along with all other identified heme synthesis genes in one of two uber-operons. In Mycobacterium, it is in uber 55 with hemA–C, CysG(D), -E, and -Y, with hemH (Z) located in uber 68. In Propionibacterium, hemQ is a fusion with hemH in uber 59 along with hemA. The genes hemB–E, -L, and -Y are in uber 58. This linkage strongly suggests that hemQ is involved in heme synthesis in the Actinobacteria. Although hemQ is always found in the operons for heme synthetic pathway enzymes of the Actinobacteria, it is infrequently found in heme synthesis-related operons in other Gram-positive organisms. Interestingly, and consistent with what has been found with other Gram-positive organisms, there is no identifiable hemF (coproporphyrinogen oxidase) gene in these organisms.
Although the structure of HemQ from an Actinobacteria is not yet available, the structure of two putative HemQs are presently available annotated as members of the COG3253 family as follows: Protein Data Bank code 1T0T of Geobacillus stearothermophilus and Protein Data Bank code 1VDH of Thermus thermophilus (39). Both are crystallographic homopentamers but as isolated they do not contain heme, and the available structures are of the apoproteins. No biochemical data exist to allow for functional assignment of either protein. One bona fide chlorite dismutase from Azospira oryzae has been crystallized with heme bound and is well studied (38). In the crystal form, this enzyme is present as a hexamer although in solution it exists as a pentamer. The Protein Data Bank codes 1T0T and 1VDH proteins are pentameric and form a donut-like structure with a central hole and putative heme-binding pockets on the outer surface at one end of the structure. In chlorite dismutase, the heme binding pocket is located in the same spatial region, but one helical segment that forms a lip to the pocket in the HemQ proteins is positioned over the heme in chlorite dismutase and encloses most of the outside of the pocket. Unfortunately, the chlorite dismutase structure lacks density on the inside of the pocket so it is not possible to determine whether the heme is exposed on the inside of the donut-like structure. In all structures, the conserved histidine that is the heme iron ligand in chlorite dismutase is located in the same position.
HemQ as isolated has minimal bound heme but readily binds exogenously supplied heme and iron-free protoporphyrin. Given that the heme-containing holo-HemQ stimulates HemY and is required along with HemY + HemH to catalyze the conversion of protoporphyrinogen and iron into heme, it is clear that HemQ functions as a hemoprotein and not simply as a carrier of protoporphyrin from HemY to HemH or as a heme chaperone to remove the heme from HemH. In addition, the level of catalase activity of HemQ, although lower than other characterized catalases, is clearly sufficient to eliminate the three molecules of hydrogen peroxide produced by HemY for each molecule of protoporphyrinogen oxidized. The spectroscopic and mutagenesis studies are also consistent with a specific enzymatic function as they indicate that the bound heme is high spin ferric iron and axially ligated by the rigorously conserved histidine ligand, His-156 in M. tuberculosis HemQ, with the sixth coordination site available for binding small molecule substrates or ligands. However, HemQ would need to correspond to a new class of catalase/peroxidase enzymes as it does not have axial tyrosinate heme ligation, which is the hallmark of classical catalases, and it shows no significant sequence homology with classical peroxidases or the bacterial catalase/peroxidase enzymes found in pathogens such as M. tuberculosis, which also have axial histidine heme ligation. Moreover, because catalase added to the in vitro assay did not replace the requirement for HemQ (Fig. 7), and the fact that the E. coli strains used in this study possess both hydroperoxidases/catalases I and II (50), it is clear that functions for HemQ other than the observed catalase activity cannot be ruled out at present.
Given that all three proteins must be present to obtain measurable HemH activity and to synthesize heme, the possibility that these proteins exist in vivo as a complex must be considered. The finding of a fusion protein of HemQ/HemH in P. acnes adds support to this notion. In addition, in silico binding exercises (51, 52) of B. subtilis HemH with the predicted HemQ of G. stearothermophilus result in models of HemH bound to the heme-binding pocket of HemQ. Because of the lack of crystallographic structure surrounding the face of the active site for B. subtilis HemY (21), such exercises are not possible with HemQ plus HemY. Our present inability to isolate a complex may result from improper stoichiometries with the recombinant protein as produced in E. coli, weakness of protein-protein interactions, or the possibility that the complex is dynamic and transient during catalysis as has been suggested to occur with mammalian ferrochelatase and protoporphyrinogen oxidase in situ. Clearly the determination of the putative complex binding constants and/or the structure of the HemY-HemQ-HemH complex will be necessary to answer this question and better understand the role HemQ plays in catalysis.
The identification of HemQ as an essential protein for heme synthesis in the firmicutes and Actinobacteria has significant implications. This protein is a member of the COG3253 family of proteins, and no members of this family are found in eucaryotes, including man. Given that heme synthesis is essential for the viability of aerobic organisms and that HemQ is essential for heme synthesis in Gram-positive bacteria, this protein/gene becomes a viable antibiotic target. With the international socioeconomic and public health impact of tuberculosis in particular and the limited arsenal of tuberculosis drugs, the availability of a target such as HemQ could have significant positive consequences.
This work was supported, in whole or in part, by National Institutes of Health Grants DK032303 (to H. A. D.) and GM62524 (to M. K. J.).
H. A. Dailey, unpublished data.
- PPO
- protoporphyrinogen oxidase
- MCD
- magnetic circular dichroism
- T
- tesla.
REFERENCES
- 1.Ponka P. (1999) Am. J. Med. Sci. 318, 241–256 [DOI] [PubMed] [Google Scholar]
- 2.Ortiz de Orué Lucana D., Groves M. R. (2009) Amino Acids 37, 479–486 [DOI] [PubMed] [Google Scholar]
- 3.Roberts G. P., Kerby R. L., Youn H., Conrad M. (2005) J. Inorg. Biochem. 99, 280–292 [DOI] [PubMed] [Google Scholar]
- 4.Spiro T. (2008) ACS Chem. Biol. 3, 673–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schläfli P., Borter E., Spielmann P., Wenger R. H. (2009) Cell. Mol. Life Sci. 66, 876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mense S. M., Zhang L. (2006) Cell Res. 16, 681–692 [DOI] [PubMed] [Google Scholar]
- 7.Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. (2007) Science 318, 1786–1789 [DOI] [PubMed] [Google Scholar]
- 8.Wilks A., Burkhard K. A. (2007) Nat. Prod. Rep. 24, 511–522 [DOI] [PubMed] [Google Scholar]
- 9.Rao A. U., Carta L. K., Lesuisse E., Hamza I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braz G. R., Coelho H. S., Masuda H., Oliveira P. L. (1999) Curr. Biol. 9, 703–706 [DOI] [PubMed] [Google Scholar]
- 11.Lara F. A., Sant'anna C., Lemos D., Laranja G. A., Coelho M. G., Reis Salles I., Michel A., Oliveira P. L., Cunha-E-Silva N., Salmon D., Paes M. C. (2007) Biochem. Biophys. Res. Commun. 355, 16–22 [DOI] [PubMed] [Google Scholar]
- 12.Wu B., Novelli J., Foster J., Vaisvila R., Conway L., Ingram J., Ganatra M., Rao A. U., Hamza I., Slatko B. (2009) PLoS Negl. Trop. Dis. 3, e475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dailey H. A. (1997) J. Biol. Inorg. Chem. 2, 411–417 [Google Scholar]
- 14.O'Brian M. R., Thöny-Meyer L. (2002) Adv. Microb. Physiol. 46, 257–318 [DOI] [PubMed] [Google Scholar]
- 15.Ferreira G. C., Andrew T. L., Karr S. W., Dailey H. A. (1988) J. Biol. Chem. 263, 3835–3839 [PubMed] [Google Scholar]
- 16.Koch M., Breithaupt C., Kiefersauer R., Freigang J., Huber R., Messerschmidt A. (2004) EMBO J. 23, 1720–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proulx K. L., Woodard S. I., Dailey H. A. (1993) Protein Sci. 2, 1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dailey H. A., Dailey T. A. (2003) in The Porphyrin Handbook (Kadish K. M., Smith K. M., Guilard R. eds) pp. 93–122, Academic Press, San Diego [Google Scholar]
- 19.Dailey T. A., Dailey H. A. (2002) J. Bacteriol. 184, 2460–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Karadaghi S., Hansson M., Nikonov S., Jönsson B., Hederstedt L. (1997) Structure 5, 1501–1510 [DOI] [PubMed] [Google Scholar]
- 21.Qin X., Sun L., Wen X., Yang X., Tan Y., Jin H., Cao Q., Zhou W., Xi Z., Shen Y. (2010) J. Struct. Biol. 170, 76–82 [DOI] [PubMed] [Google Scholar]
- 22.Sãsãrman A., Chartrand P., Lavoie M., Tardif D., Proschek R., Lapointe C. (1979) J. Gen. Microbiol. 113, 297–303 [DOI] [PubMed] [Google Scholar]
- 23.Miyamoto K., Nakahigashi K., Nishimura K., Inokuchi H. (1991) J. Mol. Biol. 219, 393–398 [DOI] [PubMed] [Google Scholar]
- 24.Nakahigashi K., Nishimura K., Miyamoto K., Inokuchi H. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 10520–10524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung C. T., Niemela S. L., Miller R. H. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 2172–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepherd M., Dailey H. A. (2005) Anal. Biochem. 344, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dailey H. A., Fleming J. E. (1983) J. Biol. Chem. 258, 11453–11459 [PubMed] [Google Scholar]
- 28.Najahi-Missaoui W., Dailey H. A. (2005) Blood 106, 1098–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beers R. F., Jr., Sizer I. W. (1952) J. Biol. Chem. 195, 133–140 [PubMed] [Google Scholar]
- 30.Chance B., Maehly A. C. (1955) Methods Enzymol. 2, 773–775 [Google Scholar]
- 31.Boynton T. O., Daugherty L. E., Dailey T. A., Dailey H. A. (2009) Biochemistry 48, 6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dailey T. A., Dailey H. A., Meissner P., Prasad A. R. (1995) Arch. Biochem. Biophys. 324, 379–384 [DOI] [PubMed] [Google Scholar]
- 33.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 34.McNeil L. K., Reich C., Aziz R. K., Bartels D., Cohoon M., Disz T., Edwards R. A., Gerdes S., Hwang K., Kubal M., Margaryan G. R., Meyer F., Mihalo W., Olsen G. J., Olson R., Osterman A., Paarmann D., Paczian T., Parrello B., Pusch G. D., Rodionov D. A., Shi X., Vassieva O., Vonstein V., Zagnitko O., Xia F., Zinner J., Overbeek R., Stevens R. (2007) Nucleic Acids Res. 35, D347–D353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson K. L., Roberts C., Disz T., Vonstein V., Hwang K., Overbeek R., Olson P. D., Projan S. J., Dunman P. M. (2006) J. Bacteriol. 188, 6739–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrigall A. V., Siziba K. B., Maneli M. H., Shephard E. G., Ziman M., Dailey T. A., Dailey H. A., Kirsch R. E., Meissner P. N. (1998) Arch. Biochem. Biophys. 358, 251–256 [DOI] [PubMed] [Google Scholar]
- 37.Dailey T. A., Dailey H. A. (1998) J. Biol. Chem. 273, 13658–13662 [DOI] [PubMed] [Google Scholar]
- 38.de Geus D. C., Thomassen E. A., Hagedoorn P. L., Pannu N. S., van Duijn E., Abrahams J. P. (2009) J. Mol. Biol. 387, 192–206 [DOI] [PubMed] [Google Scholar]
- 39.Ebihara A., Okamoto A., Kousumi Y., Yamamoto H., Masui R., Ueyama N., Yokoyama S., Kuramitsu S. (2005) J. Struct. Funct. Genomics 6, 21–32 [DOI] [PubMed] [Google Scholar]
- 40.Hansson M., Hederstedt L. (1994) Eur. J. Biochem. 220, 201–208 [DOI] [PubMed] [Google Scholar]
- 41.Dailey H. A., Dailey T. A. (1996) J. Biol. Chem. 271, 8714–8718 [DOI] [PubMed] [Google Scholar]
- 42.Dailey T. A., Dailey H. A. (1996) Protein Sci. 5, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagedoorn P. L., De Geus D. C., Hagen W. R. (2002) Eur. J. Biochem. 269, 4905–4911 [DOI] [PubMed] [Google Scholar]
- 44.Cheesman M. R., Greenwood C., Thomson A. J. (1991) in Advances in Inorganic Chemistry (Sykes A. G. ed) pp. 201–255, Academic Press, New York [Google Scholar]
- 45.Finnegan M. G., Knaff D. B., Qin H., Gray K. A., Daldal F., Yu L., Yu C. A., Kleis-San Francisco S., Johnson M. K. (1996) Biochim. Biophys. Acta 1274, 9–20 [DOI] [PubMed] [Google Scholar]
- 46.Shemin D., Rittenberg D. (1946) J. Biol. Chem. 166, 621–625 [PubMed] [Google Scholar]
- 47.Panek H., O'Brian M. R. (2002) Microbiology 148, 2273–2282 [DOI] [PubMed] [Google Scholar]
- 48.Lecerof D., Fodje M., Hansson A., Hansson M., Al-Karadaghi S. (2000) J. Mol. Biol. 297, 221–232 [DOI] [PubMed] [Google Scholar]
- 49.Che D., Li G., Mao F., Wu H., Xu Y. (2006) Nucleic Acids Res. 34, 2418–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loewen P. (1996) Gene 179, 39–44 [DOI] [PubMed] [Google Scholar]
- 51.Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) Nucleic Acids Res. 32, W96–W99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]