Abstract
Vitamin A metabolite, all-trans-retinoic acid (RA), induces cell growth, differentiation, and apoptosis and has an emerging role in gene regulation and alternative splicing events. Protein kinase Cδ (PKCδ), a serine/threonine kinase, has a role in cell proliferation, differentiation, and apoptosis. We reported an alternatively spliced variant of human PKCδ, PKCδVIII that functions as a pro-survival protein (1). RA regulates the splicing and expression of PKCδVIII via utilization of a downstream 5′ splice site of exon 10 on PKCδ pre-mRNA. Here, we further elucidate the molecular mechanisms involved in RA regulation of alternative splicing of PKCδVIII mRNA. Overexpression and knockdown of the splicing factor SC35 (i.e. SRp30b) indicated that it is involved in PKCδVIII alternative splicing. To identify the cis-elements involved in 5′ splice site selection we cloned a minigene, which included PKCδ exon 10 and its flanking introns in the pSPL3 splicing vector. Alternative 5′ splice site utilization in the minigene was promoted by RA. Further, co-transfection of SC35 with PKCδ minigene promoted selection of 5′ splice site II. Mutation of the SC35 binding site in the PKCδ minigene abolished RA-mediated utilization of 5′ splice splice II. RNA binding assays demonstrated that the enhancer element downstream of PKCδ exon 10 is a SC35 cis-element. We conclude that SC35 is pivotal in RA-mediated PKCδ pre-mRNA alternative splicing. This study demonstrates how a nutrient, vitamin A, via its metabolite RA, regulates alternative splicing and thereby gene expression of the pro-survival protein PKCδVIII.
Keywords: Gene Expression, Gene Regulation, Neuron, RNA Splicing, Serine Threonine Protein Kinase, Alternative Splicing, Retinoic Acid, SC35
Introduction
Vitamin A, an important micronutrient and its active metabolite all-trans-retinoic acid (RA)2 influence a broad range of physiological and pathological processes in the embryonic central nervous system and in the mature brain. Protein kinase C (PKC), a serine/threonine kinase family, consists of 11 isoforms and their splice variants and is involved in the regulation of cellular differentiation, growth, and apoptosis (2). Protein kinase Cδ, a member of the novel PKC subfamily, is implicated in both apoptosis (3–10) and cell survival pathways (11–14) Thus, PKCδ has dual effects and represents a switch that determines cell survival and fate. This can be explained by the expression of alternatively spliced variants of PKCδ with distinct functions in the apoptotic cascade. The occurrence of PKCδ isoforms is species-specific. PKCδI is ubiquitously present in all species while PKCδII, -δIV, -δV, -δVI, and -δVII isoforms are present in mouse tissues (15, 16); PKCδIII is present in rats (17) and PKCδVIII is present in humans (1).
An important mechanism of regulating gene expression occurs by alternative splicing which expands the coding capacity of a single gene to produce different proteins with distinct functions (18). It is now established that close to 90% of human genes undergo alternative splicing and encode for at least two isoforms. Divergence observed in gene expression because of alternative splicing may be tissue-specific (19, 20), developmentally regulated (21, 22) or hormonally regulated (23, 24). Of utmost scientific interest is the study of physiological systems in which the splicing pattern changes in response to a stimulus such as a hormone or a nutrient.
Recently, we identified a new splice variant of human PKCδ, PKCδVIII (GenBankTM Accession No. DQ516383). Sequencing and computational analysis of the PKCδVIII sequence indicated that this human splice variant is generated by utilization of an alternative downstream 5′ splice site of PKCδ pre-mRNA exon 10 (1). Further, we demonstrated that RA dramatically increased the expression of PKCδVIII via alternative splicing in NT2 cells. RA promotes hippocampal neurogenesis and spatial memory (25). RA is an early signaling component of the central nervous system (CNS) and acts as a master switch of gene expression. It is well established that the vitamin A metabolite, RA, directly affects transcription of genes. Hence, we sought to elucidate the molecular mechanisms governing this novel observation of RA-mediated alternative splicing of PKCδ pre-mRNA resulting in the expression of the pro-survival protein PKCδVIII.
EXPERIMENTAL PROCEDURES
Cell Culture
The Ntera2 human teratocarcinoma cell line (NT2/D1 cells) is maintained in DMEM, 10% fetal bovine serum (FBS) with fresh medium every 3 days. The cells are supplemented with 10 μm RA as indicated.
Primary Human Neuronal Cells
cDNA from these cells were obtained from Dr. Sanchez-Ramos (James A. Haley Veterans Hospital, Tampa, FL), and the cells were cultured in his laboratory. Patients undergoing anterior temporal lobectomy provided written informed consent allowing the tissue to be used for research. The study was approved by the Institutional Review Board (IRB 102342), University of South Florida. Hippocampal tissue was dissected from the temporal lobe resection, dissociated, and plated for generation of a stem/progenitor cells line using standard methods. Hippocampus biopsies were sterilely removed from a 31-year-old male and transferred to a 35-mm plate containing PBS plus 0.5% BSA. A sterile scalpel was used to finely chop the tissue into small pieces. 0.05% Trypsin/EDTA was added to cells and was incubated at 37 °C for 8–10 min. The pellet was suspended in DMEM/F12 plus 10% FBS, followed by DNase treatment. The final pellet was re-suspended in DMEM/F12, and a cell count for viability was performed. The cells were seeded into a T-75 flask in DMEM/F12 plus 2% FBS, EGF, and bFGF 20 ng/ml. Cells were replated on poly-l-ornithine-coated chamber slides. Digital images of the hippocampal neurons stained with nestin, TuJ1, BrdU, and NeuN were captured using Zeiss confocal microscope and characterized. The cells were maintained at 37 °C in 5% CO2, 95% humidity. As the numbers of proliferating cells reached confluency, aliquots of stem/progenitor cells were frozen for later use. Cells used in experiments described here were plated into 6-well plates.
Western Blot Analysis
Cell lysates (40 μg) were separated on 10% SDS-PAGE. Proteins were electrophoretically transferred to nitrocellulose membranes, blocked with Tris-buffered saline, 0.1% Tween 20 containing 5% nonfat dried milk, washed, and incubated with a polyclonal antibody against either anti-SC35 (26), anti-SF2/ASF, anti-PKCδ (BioSource), or PKCδVIII-specific polyclonal antibody (Patel Laboratory, described in Ref. 1). Following incubation with anti-rabbit IgG-HRP, enhanced chemiluminescence (PierceTM) was used for detection.
Quantitative Real-time RT-PCR
cDNA (2 μl) was amplified by real-time quantitative PCR using Syber (SYBR) Green with an ABI PRISM 7900 sequence detection system (PE Applied Biosystems, Foster City, CA) as described previously to quantify absolute levels of PKCδI and PKCδVIII mRNA in the samples (1). GAPDH was amplified as the endogenous control. Briefly, primers used were as follows: PKCδI sense primer: 5′-GCCAACCTCTGCGGCATCA-3′; antisense primer: 5′-CGTAGGTCCCACTGTTGTC↓TTGCATG-3′; PKCδVIII sense primer: 5′-GCCAACCTCTGCGGCATCA-3′; antisense primer: 5′-CGTAGGTCCCACTGTTGTC↓CTGTCTC-3′. These primers overlap the exon-exon boundary (indicated by the ↓) specific for each transcript. The primers for GAPDH were: sense primer 5′-CTTCATTGACCTCAACTACAT-3′ and antisense primer 5′-TGTCATGGATGACCTTGGCCA-3′. Real time PCR was then performed on samples and standards in triplicates. Absolute quantification of mRNA expression levels for PKCδI and PKCδVIII was calculated by normalizing the values to GAPDH. The results were analyzed with two-tailed Student's t test using PRISM4 statistical analysis software (GraphPad, San Diego, CA). A level of p < 0.05 was considered statistically significant. Significance is determined after three or more experiments.
Transient Transfection of Plasmid DNA
SC35 and SF2/ASF plasmids were obtained from Origene (TrueCloneTM cDNA plasmids). Plasmid DNA (1 to 2 μg) was transfected into cells using Trans-IT® or Lipofectamine® (Invitrogen) per the manufacturer's instructions.
siRNA Transfection
Two siRNAs that target separate areas were used to knockdown expression of SC35. SC35 siRNAs along with its scrambled control were purchased from Ambion® (IDs: 12628 and 12444) and transfected using Ambion's siRNA transfection kit. These were validated for specificity to eliminate off-target gene effects. Ambion's PARIS kit (catalogue 1921) was used to simultaneously isolate proteins and RNA to verify knockdown by siRNA transfection.
RT-PCR
Total RNA was isolated from cells using using RNA-BeeTM (Tel Test, Inc) as per manufacturer's instructions. 2 μg of RNA was used to synthesize first strand cDNA using an Oligo(dT) primer and OmniscriptTM kit (Qiagen). PCR was performed using 2 μl of RT reaction and Takara Taq polymerase. The primers are listed: Human PKCδ sense primer 5′-CACTATATTCCAGAAAGAACGC-3′ and antisense primer 5′-CCCTCCCAGATCTTGCC-3′; PKCδVIII-specific antisense primer 5′-CCCTCCCAGATCTTGCC-3′; SD-SA on pSPL3 sense primer 5′-TCTCAGTCACCTGGACAACC-3′ and antisense primer 5′-CCACACCAGCCACCACCTTCT-3′; SC35 sense primer 5′-TCCAAGTCCAAGTCCTCCTC-3′ and antisense primer 5′-ACTGCTCCCTCTTCTTCTGG-3′; GAPDH sense primer 5′-CTTCATTGACCTCAACTCATG-3′ and antisense primer 5′-TGTCATGGATGACCTTGGCCAG-3′. Using PKCδ primers, PKCδI and PKCδVIII are detected simultaneously: PKCδI is 368 bp and PKCδVIII is 461 bp. Using PKCδVIII-specific primers, PKCδVIII is 424 bp; SC35 is 210 bp; GAPDH is 391 bp; SD-SA: 263 bp; utilization of 5′ splice site I: 419 bp; utilization of 5′ splice site II: 512 bp. 5% of products were resolved on 6% PAGE gels and detected by silver staining. The PCR reaction was optimized for linear range amplification to allow for quantification of products. Densitometric analyses of bands were done using Un-Scan ITTM Analysis Software (Silk Scientific).
Construction of pSPL3-PKCδ Minigenes
The pSPL3 vector (27) contains an HIV genomic fragment with truncated tat exons 2 and 3 inserted into rabbit β-globin coding sequences. The resulting hybrid exons in pSPL3 are globin E1E2-tat exon 2 and tat exon 3-globin E3 separated by more than 2.5 kilobase pairs of tat intron sequence. pSPL3 contains a multiple cloning sequence (MCS) around 300 nucleotides downstream of the tat exon 2 5′ splice site. The SV40 promoter and polyadenylation signal allow for enhanced expression in NT2 cells. There are several cryptic 5′ splice sites, which interfere with minigene splicing and hence sections of the original pSPL3 vector were deleted. First, 874 bp of the tat intronic section lying upstream of SA was deleted. It was designed such that the deletion began 158 bp upstream of SA thereby maintaining the branch point and pyrimidine tract. Primers to amplify genomic PKCδ from NT2 cells were designed using the Gene Tool Software (Bio Tools Inc.) and include the BclI site in the forward primer (in bold type) and BcuI site in the reverse primers (in bold type). The forward primer was designed such that the product will contain the branch point and 3′ splice site. Following amplification of the product, it was ligated into the digested pSPL3 vector. The pSPL3 vector was digested with BamHI (in the MCS) and NheI within the tat intronic sequence which removes an additional 930 bases. The overhangs of the selected restriction enzymes can hybridize and this enabled cloning of the PCR product in the proper orientation. To increase the efficiency and number of positive clones, the ligation reaction was digested with the above restriction enzymes, which cleave any dimers produced by the ligation reaction. The product was verified by restriction digestion and sequencing. The primers used to generate pSPL3-PKCδ minigene were: forward primer 5′ CCTTGATCATGGGAGTTCTGATAATGGTC 3′; reverse primer 5′ CCTACTAGTATCGGGTCTCAGTCTACAC 3′ such that 200 bp of the 5′ intronic sequence was included. The products were ligated into the digested pSPL3 vector and transformed into bacteria using TOP10F cells (Invitrogen). Truncated minigenes were verified by restriction digestion and sequencing.
Site-directed Mutagenesis
The SC35 cis-element (sequence: ggccaaag) identified on the 5′ intronic sequence flanking exon 10 of PKCδ pre-mRNA was mutated in the pSPL3_PKCδ minigene to tagcccata using QuikChange® site-directed mutagenesis kit (Stratagene), which allows for blue-white screening per the manufacturer's instructions. The mutated minigene, pSPL3_PKCδ**, was verified by sequencing.
RNA Binding Assays
The templates used were F1 (which contains PKCδ exon 10 and 120 bp of its 5′ intronic sequence including the putative SC35 binding site); mutated F1 (F1m, same region as F1 but putative SC35 binding site was mutated as described above) and F2 (which is PKCδ exon 10 alone). Single-stranded RNAs were synthesized in vitro using the T7 RNA polymerase and purified on denaturing polyacrylamide gels prior to RNA binding assays. The transcripts were 5′ biotinylated with 0.1 mm biotin-21 as described before (28). RNA gel shift mobility assay was performed with 10 fmol of labeled RNA and 5 ng of recombinant SC35 (ProteinOne) in a 20-μl binding reaction (100 mm Tris, 500 mm KCl, 10 mm dithiothreitol, 2.5% glycerol, 2 units/μl RNAsin) and incubated at 30 °C for 20 min. The complex was run on 8% polyacrylamide gel and transferred to a nylon membrane. Western blot analysis was performed using avidin-HRP conjugate (Pierce).
Statistical Analysis
Gels were densitometrically analyzed using UN-SCAN-ITTM software (Silk Scientific, Inc.) PRISMTM software was used for statistical analysis. The results were expressed as mean ± S.E. of densitometric units or as percent exon inclusion.
RESULTS
Expression of PKCδVIII
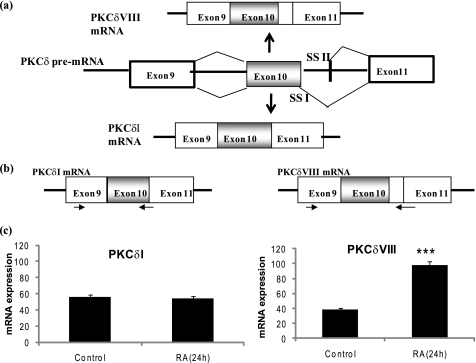
In humans, the PKCδ gene has at least two alternatively spliced variants: PKCδI and PKCδVIII (Fig. 1a). PKCδVIII is generated via alternative splicing of the PKCδ pre-mRNA such that 93 nucleotides are included in the mature PKCδVIII mRNA. This translates to 31 amino acids whose inclusion disrupts the caspase-3 recognition sequence in the hinge region of PKCδVIII protein. Our previous study demonstrated that PKCδVIII functions as a pro-survival protein whereas PKCδI promotes apoptosis. Further, we demonstrated that RA (24 h) significantly increased the expression of PKCδVIII in NT2 cells (1). Here, we sought to extend our findings in primary neuronal cells derived from human adult hippocampus (see “Experimental Procedures”) to demonstrate the physiological significance of the expression pattern of PKCδVIII in human hippocampus and its response to RA. We performed quantitative, two-step real-time RT-PCR using Syber (SYBR) Green technology (29–31). The primers were specific to the exon junctions of PKCδI mRNA and PKCδVIII mRNA as shown in Fig. 1b. Each transcript was normalized to the endogenous control, GAPDH, to obtain absolute quantification. We demonstrate that PKCδVIII increased with RA treatment whereas PKCδI levels remain constant in human primary neuronal cells (Fig. 1c).
FIGURE 1.
a, schematic of alternative 5′ splice site selection in human PKCδ pre-mRNA exon 10 that results in the generation of PKCδI mRNA and PKCδVIII mRNA, which differ by 93 bp in the V3 hinge region. RA promotes expression of PKCδVIII mRNA. SSI: 5′ splice site I; SSII: 5′ splice site II. b, schematic of the primers specific for PKCδI and PKCδVIII used in real time RT-PCR such that they span the exon-exon boundaries. c, primary human neuronal cells from hippocampus were treated with or without RA (10 μm) for 24 h. Total RNA was extracted, and real time RT-PCR analysis using SYBR green was performed in triplicate and repeated three times in separate experiments. The absolute mRNA expression of PKCδI and PKCδVIII transcripts normalized to GAPDH are shown. PKCδVIII expression increases significantly following 24 h of RA treatment; ***, p < 0.0001 (by two-tailed Student's t test).
Concurrent Increases in SC35 and PKCδVIII Levels in RA-mediated PKCδ Alternative Splicing
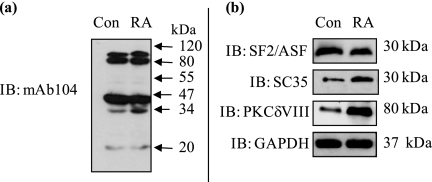
Alternative splicing is regulated by recruiting trans-factors such as serine-arginine-rich (SR) proteins that bind to exonic or intronic splicing enhancers (ESE, ISE) on the pre-mRNA. Hence, the elucidation of trans-factors involved in RA-mediated PKCδ alternative splicing is of critical importance. NT2 cells were treated with or without RA (24 h), and whole cell lysates were analyzed by Western blot analysis using mAb104 antibody that simultaneously detects the phosphoepitopes on all SR proteins. Our results indicated that upon RA treatment, SR protein at ∼30 kDa increased in expression (Fig. 2a). SF2/ASF or SC35 (i.e. SRp30a or SRp30b, respectively) are two SR proteins with molecular masses of ∼30 kDa. Hence, antibodies specific to these individual SR proteins were used next. We observed an increase in SC35 (SRp30b) concurrent with increased PKCδVIII levels in response to RA while SF2/ASF (SRp30a) expression remained relatively constant (Fig. 2b). The observed increase of SC35 with RA reflects total expression levels of SC35. The increases seen with mAb104 antibody, which detects the phosphoepitope, is a reflection of its increased expression rather than increased phosphorylation. SC35, also known as SFRS2 or SRp30b, is a member of the SR splicing protein family and functions as a splicing enhancer (32).
FIGURE 2.
Detection of SR proteins involved in RA-mediated PKCδVIII expression. NT2 cells were treated with RA (10 μm) for 24 h or without RA (control), and Western blot analysis was performed on whole cell lysates using (a) mAb104 antibody that detects all SR proteins and (b) specific antibodies as indicated in the figure. Molecular masses are indicated (kDa). Gels are representative of three separate experiments, and results indicate that SC35 may be involved in increased expression of PKCδVIII by RA. Results demonstrate an increase in SC35 levels concurrent with an increase in PKCδVIII expression upon RA treatment.
SC35 Mimics RA-mediated PKCδVIII Alternative Splicing
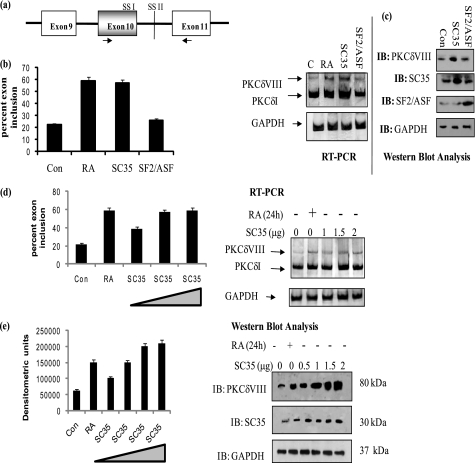
SC35 was transiently transfected into NT2 cells to determine whether it could mimic the effect of RA in increasing the expression of PKCδVIII. SF2/ASF was used as a control and transfected into a separate well. RT-PCR performed using human PKCδ primers which amplified both PKCδI and PKCδVIII products. Simultaneously, Western blot analysis was performed with PKCδVIII-specific antibody. An increase in endogenous PKCδVIII levels in cells overexpressing SC35 was observed (Fig. 3, a–c) while in SF2/ASF transfected cells PKCδVIII expression remained constant. GAPDH was used as internal control for all samples.
FIGURE 3.
SC35 but not SF2/ASF promotes PKCδVIII expression. a, schematic of primer positions used in PCR amplification. These primers detect PKCδI and PKCδVIII simultaneously. b, NT2 cells were transfected with 2 μg of SC35 or SF2/ASF or treated with RA (10 μm) for 24 h. Total RNA was extracted, and RT-PCR was performed using human PKCδ primers as shown above. 5% of the products were separated by PAGE and silver stained for visualization. The graph represents percent exon inclusion calculated as PKCδVIII/(δVIII + δI) × 100 in these samples and is representative of mean ± S.E. in three experiments. c, whole cell lysates were extracted from NT2 cells transfected with 2 μg of SC35 or SF2/ASF. Western blot analysis was performed using specific antibodies as indicated in the figure. The experiments were repeated three times with similar results. d, increasing amounts of SC35 (0 to 2 μg) were transfected into NT2 cells and treated with or without RA (10 μm, 24 h). Total RNA was extracted and RT-PCR was performed using human PKCδ primers as shown above. 5% of the products were separated by PAGE and silver stained for visualization. Graph represents percent exon inclusion calculated as PKCδVIII/(δVIII + δI) × 100 in these samples and is representative of mean ± S.E. in three experiments. e, simultaneously, Western blot analysis was performed on whole cell lysates extracted from NT2 cells transfected with 0–2 μg of SC35, using antibodies as indicated within the figure. The graph represents four experiments performed separately and represents PKCδVIII densitometric units normalized to GAPDH as mean ± S.E. The triangle in the graphs indicates increasing amounts of SC35. Results indicate that SC35 promotes PKCδVIII expression in a dose-dependent manner thereby mimicking the RA response.
To determine whether PKCδVIII expression levels increased in direct proportion with SC35, increasing amounts of SC35 (0–2 μg) were transfected into NT2 cells. Total RNA or whole cell lysates were collected. RT-PCR was performed using PKCδ primers that detect PKCδI and PKCδVIII mRNA, and Western blot analysis was carried out using antibodies for PKCδVIII, SC35, and GAPDH (internal control). As seen in Fig. 3d, PKCδVIII mRNA levels increased with increasing levels of SC35 while PKCδI mRNA levels appeared unaffected. Further, PKCδVIII protein levels (Fig. 3e) increased with increasing doses of SC35 comparable to the increase in PKCδVIII protein seen with RA treatment.
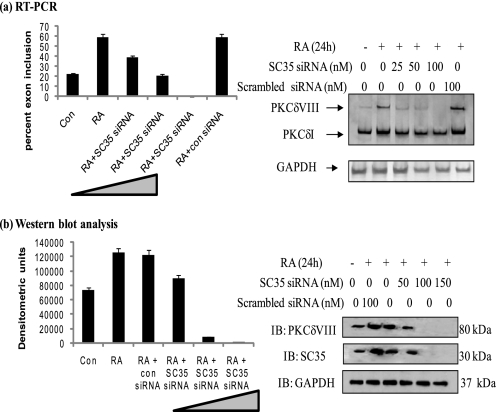
RA Is Unable to Increase Expression of PKCδVIII in the Absence of SC35
To determine the effect of SC35 knockdown on the RA-mediated expression of PKCδVIII, siRNA specific for SC35 were transfected in increasing amounts (0–150 nm) into NT2 cells and treated with RA. Two sets of SC35 siRNA along with its scrambled control were used to validate specificity and eliminate off-target knockdowns. Results indicated similar data with either SC35 siRNAs. Total RNA or whole cell lysates were collected. RT-PCR was performed using PKCδ primers while Western blot analysis was carried out using antibodies for PKCδVIII, SC35, and GAPDH. As seen in Fig. 4a, PKCδVIII mRNA levels decreased with increasing levels of SC35 siRNA while PKCδI mRNA levels appeared unaffected. Further, PKCδVIII protein levels decreased with increasing doses of SC35 siRNA (Fig. 4b). The graph is representative of four individual experiments performed with either SC35 siRNA. The above data confirms that RA cannot promote PKCδVIII expression in the absence of SC35. This demonstrates the involvement of SC35 in RA-mediated alternative splicing of PKCδ pre-mRNA.
FIGURE 4.
Knockdown of SC35 inhibits RA-mediated increased expression of PKCδVIII. Increasing amounts of SC35 siRNA (0–150 nm) were transfected into NT2 cells. Scrambled siRNA was used as a control (con siRNA). Post-transfection, cells were treated with or without RA (10 μm, 24 h). a, total RNA was extracted, and RT-PCR was performed using human PKCδ primers as shown above. 5% of the products were separated by PAGE and silver stained for visualization. Graph represents percent exon inclusion calculated as PKCδVIII/(δVIII + δI) × 100 in these samples and is representative of mean ± S.E. in three experiments. b, simultaneously, whole cell lysates were collected, and Western blot analysis was performed using antibodies as indicated. Graph represents four experiments performed separately and expressed as mean ± S.E. of densitometric units. The triangle in the graphs indicates increasing amounts of SC35 siRNA. Results indicate that knockdown of SC35 inhibits RA-mediated increased expression of PKCδVIII.
Antisense Oligonucleotides Indicate a Role of SC35 cis-Element in PKCδ Alternative Splicing
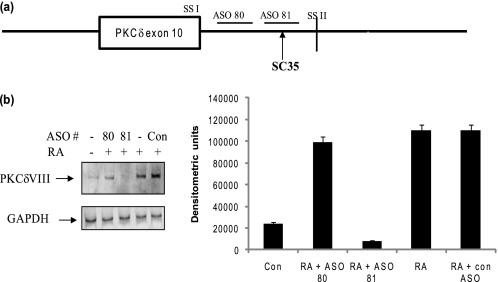
Previous studies identified consensus sequences (33) for several cis-elements present either in the exonic or intronic sequences of pre-mRNA. These consensus sequences serve as a guideline to dissect and analyze putative cis-elements in alternative splicing of pre-mRNA. We combined a web-based resource “ESE finder” (34) and also manually checked for published consensus sequences of cis-elements on PKCδ pre-mRNA to predict putative enhancer and silencer elements that could recruit trans-factors in RA-regulated alternative splicing of PKCδ. To focus on identifying the cis-elements involved in RA-mediated increase in PKCδVIII mRNA levels, antisense oligonucleotides (ASO) (synthesized by Isis Pharmaceuticals, Carlsbad, CA), which are 2′-methoxyethyl-modified, RNase-H resistant were used. These ASOs inhibit binding of trans-factors to their cis-elements without disrupting the splicing event or degrading the mRNA (35, 36).
We transfected a series of 20mer ASOs, which were designed according to predicted enhancer and silencer sites such that they sequentially spanned the unspliced PKCδ pre-mRNA. All wells were also treated with RA and RT-PCR was performed. Transfection of ASO 81 (which spans the putative SC35 binding site) showed a significant decrease in RA-induced PKCδVIII splicing while the other ASOs did not affect the expression of PKCδVIII induced by RA (data not shown). Results (Fig. 5, a and b) shown here represent three experiments performed individually using the scrambled ASO as control, ASO 81 and ASO 80 (which was in close proximity to ASO 81 but did not inhibit RA-mediated PKCδVIII alternative splicing). ASO 81 corresponded to the SC35 binding site as identified by ESE finder and further determined by its consensus sequence, ggccaaag. These results demonstrated that ASO 81 inhibited RA-induced PKCδVIII alternative splicing. This also suggested the position of SC35 cis-element on PKCδ pre-mRNA to be in the intronic region downstream of PKCδ exon 10 and before 5′ splice site II (schematic in Fig. 5a).
FIGURE 5.
Analysis of putative cis-elements and ASO. a, schematic of position of ASOs on PKCδ pre-mRNA. The putative SC35 cis-element lies between 5′ splice site I and II of PKCδ exon 10. SSI: 5′ splice site I; SSII: 5′ splice site II. b, ASOs were transfected into NT2 cells and after overnight incubation cells were treated with or without RA (10 μm, 24 h). The gel represents experiments conducted with scrambled ASO (control), ASO 81 (corresponding to putative SC35 binding site) and ASO 80, which is in close proximity to ASO81. Total RNA was extracted and RT-PCR performed using PKCδVIII-specific primers. 5% products were separated on PAGE and detected by silver nitrate staining. The graph indicates PKCδVIII densitometric units normalized to GAPDH and is representative of mean ± S.E. in three separate experiments. Results indicate that ASO81, which corresponds to the putative SC35 cis-element, inhibits RA-mediated increased expression of PKCδVIII.
Construction of a Heterologous pSPL3_PKCδ-splicing Minigene That Is Responsive to RA
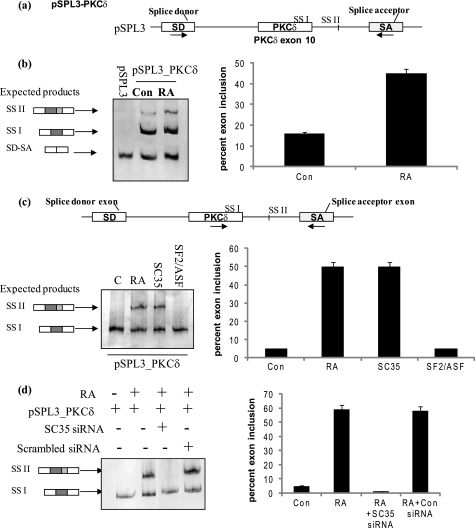
Splicing minigenes are advantageous to identify cis-elements on the pre-mRNA involved in regulated alternative splicing. Further, minigenes aid to correlate the binding of specific SR proteins to individual splicing events. Hence, to dissect the mechanism of RA-mediated regulation of endogenous PKCδ alternative splicing and analyze factors influencing 5′ splice site selection, a PKCδ heterologous minigene was developed. Since the human PKCδ splice variants used alternative 5′ splice sites as determined previously (1), exon 10 of PKCδ pre-mRNA along with its flanking 3′ and 5′ intronic sequences was cloned (as described under “Experimental Procedures”) in the multiple cloning site (MCS) between the splice donor (SD) and splice acceptor (SA) exons of pSPL3, a vector developed to study splicing events (schematic shown in Fig. 6a). 5′ splice site II (which encodes for PKCδVIII mRNA) is 93 bp downstream of PKCδ exon 10, and hence we cloned 200 bp of the 5′ intronic sequence. The minigene also contains a retinoic acid response element (RARE) in its promoter region. The resulting minigene, pSPL3_PKCδ, was confirmed using restriction digestion and sequencing.
FIGURE 6.
Minigene analysis demonstrates that RA promotes utilization of 5′ splice site II on PKCδ exon 10 pre-mRNA. a, schematic represents PKCδ pre-mRNA exon 10 and flanking introns cloned into pSPL3 splicing vector between the SD and SA exons. The resulting minigene is referred to as pSPL3_PKCδ minigene. Arrows indicate position of primers used in RT-PCR analysis. b, pSPL3_PKCδ minigene and pSPL3 empty vector were transfected overnight, and then the cells were treated with or without 10 μm RA for 24 h. Total RNA was extracted and RT-PCR performed using primers as described above. Expected products are SD-SA: constitutive splicing; SSI: usage of 5′ splice site I; SSII: usage of 5′ splice site II. c, 2 μg of SC35 or SF2/ASF was co-transfected along with the pSPL3_PKCδ splicing minigene. In separate wells, 10 μm RA was added for 24 h. Total RNA was extracted and RT-PCR performed using PKCδ exon 10 and SA primers as shown in the schematic. SSI: usage of 5′ splice site I; SSII: usage of 5′ splice site II. d, SC35 siRNA (100 nm) or scrambled siRNA was co-transfected with pSPL3_PKCδ minigene. 10 μm RA was added to wells as indicated. Total RNA was extracted and RT-PCR performed using PKCδ exon 10 and SA primers as shown above in c. 5% of the products were separated by PAGE and silver stained for visualization. Graphs represent percent exon inclusion calculated as SS II/(SS II + SSI) × 100 in the samples and are representative of four experiments performed separately. These results demonstrate that co-transfection of SC35 with the pSPL3_PKCδ minigene promotes utilization of 5′ splice site II. Further, RA is unable to promote utilization of 5′ splice site II on PKCδVIII pre-mRNA in the absence of SC35.
Minigene pSPL3_PKCδ was transfected into NT2 cells; cells were treated with RA (24 h) and RT-PCR performed on total RNA using SD-SA primers. The empty vector pSPL3 with the same modifications used for cloning the minigene, was transfected simultaneously in a separate well. Deletion of intronic sequences between 5′ splice site II and SA exon did not affect RA-mediated utilization of the 5′ splice site II (data not shown) thereby indicating that additional downstream cis-elements were not influencing splice site selection. The predicted products using SD-SA primers are shown (Fig. 6, a and b). RA increased utilization of 5′ splice site II of PKCδ exon 10 in pSPL3_PKCδ minigene thereby mimicking RA-mediated increase in endogenous PKCδVIII expression.
Next, we sought to determine if SC35 could increase the utilization of 5′ splice site II on pSPL3_PKCδ minigene such that it mimics the increase of RA-mediated endogenous expression of PKCδVIII. SC35 or SF2/ASF expression vector (2 μg) was co-transfected along with the pSPL3_PKCδ minigene into NT2 cells. RA was added to a separate well transfected with pSPL3_PKCδ minigene. RT-PCR was performed on total RNA using PKCδ_exon 10 (sense) and SA (antisense) primers as shown (Fig. 6c). SC35 promoted the selection of 5′ splice site II on PKCδ exon 10 in pSPL3_PKCδ splicing minigene thereby mimicking endogenous RA-mediated increased expression of PKCδVIII.
To show that SC35 is crucial for RA-mediated PKCδVIII 5′ splice site selection, SC35 siRNA was co-transfected with pSPL3_PKCδ minigene in NT2 cells. RA was added to the cells as indicated in the figure. RT-PCR was performed on total RNA using PKCδ_exon 10 (sense) and SA (antisense) primers (Fig. 6d). RA treatment could not promote utilization of PKCδVIII 5′ splice site II when SC35 was knocked down. This verified that SC35 was a crucial trans-factor involved in RA-mediated PKCδVIII expression.
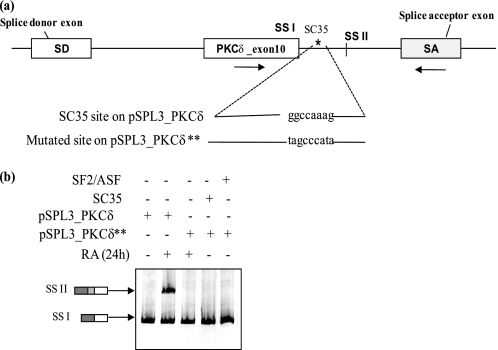
Mutation of SC35 Binding Site on the Heterologous pSPL3-PKCδ Minigene Disrupted Utilization of 5′ Splice Site II
The putative SC35 site identified by its consensus sequence and ASO binding assay (Fig. 5, a and b, above) is in the intronic region between 5′ splice site I and 5′ splice site II of PKCδ exon 10. To establish that the putative sequence was an SC35 cis-element and that it is essential for RA-mediated PKCδVIII alternative splicing, the intronic SC35 cis-element “ggccaaag” was mutated (Fig. 7a). This site was mutated to tagcccata within the pSPL3_PKCδ minigene (described under “Experimental Procedures”) and the mutated pSPL3_PKCδ** minigene was transfected into NT2 cells. The original pSPL3_PKCδ minigene was transfected into a separate well as the control. RA was added for 24 h as indicated in the figure. In separate wells, SC35 or SF2/ASF was transfected along with the mutated pSPL3_PKCδ** minigene, treated with or without RA. RT-PCR was performed on total RNA using PKCδ_exon 10 (sense) and SA (antisense) primers. RA treatment or overexpression of SC35 did not promote the selection of 5′ splice site II on PKCδ exon 10 in the pSPL3_PKCδ**-mutated minigene (Fig. 7b). This experiment demonstrates that the mutated minigene was insensitive to RA treatment and SC35 levels. Further, this indicated that the sequence ggccaaag on PKCδ pre-mRNA was required for RA-mediated PKCδVIII alternative splicing and was a putative binding site for SC35 which is essential for an RA response in PKCδ pre-mRNA 5′ splice site II selection.
FIGURE 7.
Mutation of putative SC35 binding site inhibits RA-mediated utilization of 5′ splice site II utilization on the minigene. a, schematic of the position and sequence of the putative SC35 cis-element on the pSPL3_PKCδ splicing minigene. Arrows indicate the position of primers used in PCR analysis. Putative SC35 binding site ggccaaag was mutated to tagcccaga on the minigene. b, resulting mutated minigene pSPL3_PKCδ** was transfected into NT2 cells. In separate wells, the mutated minigene pSPL3_PKCδ** was co-transfected with either 2 μg of SC35 or SF2/ASF. The original pSPL3_PKCδ splicing minigene was also transfected in a separate well. After overnight transfection, NT2 cells were treated with or without 10 μm RA for 24 h. Total RNA was extracted and RT-PCR performed using primers for PKCδ exon 10 sense and SA antisense as shown. 5% of the products were separated by PAGE and silver stained for visualization. SSI: usage of 5′ splice site I; SSII: usage of 5′ splice site II. Graph represents percent exon inclusion calculated as SS II/(SS II + SSI) × 100 and is representative of three experiments performed separately. Results indicate that mutation of the enhancer element ggccaaag abolishes the ability of RA or SC35 to promote utilization of 5′ splice site II on PKCδ splicing minigene.
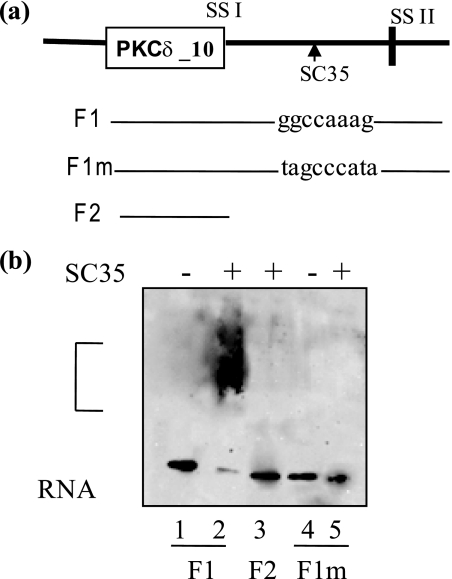
SC35 Binds to the cis-Element on PKCδ Pre-mRNA
The above experiments demonstrated that SC35 is required for RA-mediated increased utilization of 5′ splice site II on PKCδ pre-mRNA, and the enhancer element “ggccaaag” is required for SC35-mediated utilization of 5′ splice site II and hence PKCδVIII alternative splicing. Hence, we sought to determine whether this cis-element is a SC35 binding site by performing RNA gel shift assays. Biotin-labeled RNA fragments were synthesized in vitro and tested for interaction with purified recombinant SC35. The RNA transcript F1 contained PKCδ exon 10 and 120 bp of its flanking 5′ region, which included the putative SC35 cis-element. The RNA transcript F1m has the putative SC35 binding site mutated as described above. RNA transcript F2 contained only the PKCδ exon 10. As shown in Fig. 8, a and b, F2 did not show any gel shift with SC35 indicating that this transcript did not contain a SC35 binding site. There is a gel shift observed with F1 and SC35 indicating that it contains the SC35 binding site and the recombinant SC35 is able to bind to the RNA. There is no binding observed with F1m and SC35 indicating that the SC35 binding site was abolished. These experiments demonstrate that the enhancer element ggccaaag present in the 5′ region of PKCδ exon 10 pre-mRNA is a SC35 cis-element.
FIGURE 8.
Gel mobility assays of F1 and mutated F1 with purified recombinant SC35. a, schematic representation of the position of PKCδ transcripts F1, F1m and F2 used in the gel binding assays. F1 contains exon 10 and 120 bp of flanking 5′ sequence, which includes the enhancer sequence ggccaaag; schematic also indicates its position on the PKCδ pre-mRNA. F1m is the same as F1 with the enhancer sequence mutated to tagcccata. F2 transcript contains PKCδ10 exon only. b, the biotin-labeled in vitro transcribed RNA sequences were incubated with recombinant SC35 at 30 °C for 20 min. The complex was run on an 8% polyacrylamide gel and transferred to a nylon membrane. Western blot analysis was performed using an avidin-HRP conjugate. Lanes are 1: F1; 2: F1 +SC35; 3: F2 +SC35; 4: F1m; 5: F1m +SC35. The bracket indicates RNA-protein complex. The gel represents four experiments performed separately. Results indicate that ggccaaag is an SC35 cis-element on PKCδ pre-mRNA.
DISCUSSION
We have shown here that the splicing factor SC35 plays an important role in RA-mediated alternative splicing of PKCδVIII pre-mRNA. Alternative pre-mRNA splicing generates protein diversity such that humans express more than 100,000 proteins from only about 25,000 protein coding genes. Defective alternative splicing causes a large number of diseases (37–39). Alternative splicing occurs through various mechanisms such as exon skipping, exon inclusion, alternative 3′ splice site usage, alternative 5′ splice site usage, or alternative polyadenylation site usage. The spliceosome catalyzes the pre-mRNA splicing reaction within a large multicomponent ribonucleoprotein complex comprising of small nuclear RNAs (snRNAs) and associated proteins (such as SR proteins).
Exonic or intronic splicing enhancers (ESE, ISE) in the pre-mRNA bind the serine-arginine-rich nuclear factors (SR proteins) to promote the choice of splice sites. Elucidation of the trans-factors involved in regulated alternative splicing is of critical importance because specific cellular stimuli can favor the binding of certain trans-factors over others, thereby changing the splicing pattern. SC35, also known as SFRS2 or SRp30b, is a splicing enhancer and a member of the SR splicing protein family. Here, we have demonstrated that SC35 binds to its cis-element on PKCδ pre-mRNA. SC35 has an N-terminal RNA recognition motif (RRM) domain and a C-terminal arginine/serine-rich (RS) domain. The RRM domain interacts and binds to the target pre-mRNA while the RS domain is highly phosphorylated and is the protein interaction region. SC35 also mediates alternative splicing of CD45 (40), tau exon 10 in Alzheimer disease (41), and neuronal acetylcholinesterase (42).
Our data demonstrate that SC35 enhances the splicing of a pro-survival protein, PKCδVIII in neurons supporting a role on neurogenesis. It is possible that the phosphorylation state of SC35 may further influence splicing of PKCδ pre-mRNA. However, our data indicated that the expression levels of SC35 changed with RA treatment rather than significant changes to its phosphorylation (Fig. 2, a and b). Further, inhibitors of several signaling pathways such as PI3K, JAK/STAT, MAPK did not affect RA-mediated PKCδ splicing (data not shown). Hence, we are currently exploring how RA increases the expression of SC35. RA promotes gene expression via its retinoic acid response element (RARE) in the promoter region of its target genes. Our preliminary studies using computational analysis did not find RAREs on the SC35 promoter region. We are exploring this further in our laboratory.
Our data with primary human neuronal cells demonstrated the physiological significance of the expression pattern of PKCδVIII in human hippocampus and its response to RA. NT2 cells are predominantly used to study neurogenesis, neuronal differentiation, and early development of the nervous system as they represent a culture model for differentiating neurons as well as a potentially important source of cells to treat neurodegenerative diseases (43). Experiments conducted in this study are within the time frame (0–24 h post-RA) in which NT2 differentiation is compared with normal differentiation in the CNS. This mirrors the time frame in which RA regulates the development of CNS and promotes adult neurogenesis. Because these studies required extensive experiment manipulations and repetitions, we conducted our studies in human NT2 cells.
Vitamin A and its metabolite, RA, have multiple therapeutic targets and neuroprotective properties. RA regulates neural development as well as its plasticity and promotes early stages of neurogenesis and increases survival (44). RA also changes the splicing pattern of other genes such as coactivator activator (CoAA) and delta isoform of CaM kinase in P19 embryonal carcinoma stem cells (45, 46). However, the mechanism of RA-induced splicing of genes has not yet been elucidated. We demonstrate here that the splicing factor, SC35 plays a crucial role in mediating RA effects on alternative splicing of PKCδVIII mRNA in neurons. Understanding how RA regulates gene expression thereby increasing the expression of the pro-survival protein PKCδVIII is a step closer to realizing the therapeutic potential of RA in neurodegenerative diseases. To our knowledge, this is the first report linking the trans-factor, SC35 to alternative splicing regulated by RA and the expression of the pro-survival protein PKCδVIII in neurons.
Acknowledgment
We thank the Moffitt DNA Sequencing Core, Tampa, FL, for sequencing of all constructs and minigenes.
This work was supported, in whole or in part, by NIDDK, National Institutes of Health Grant 54393 (to D. R. C.) and a Veteran's Administration MREP Grant (to N. A. P.).
- RA
- all-trans-retinoic acid
- SD
- splice donor
- SA
- splice acceptor
- ASO
- antisense oligonucleotides
- SR
- serine-arginine-rich
- ESE
- exonic splicing enhancers
- ISE
- intronic splicing enhancers.
REFERENCES
- 1.Jiang K., Apostolatos A. H., Ghansah T., Watson J. E., Vickers T., Cooper D. R., Epling-Burnette P. K., Patel N. A. (2008) Biochemistry 47, 787–797 [DOI] [PubMed] [Google Scholar]
- 2.Nishizuka Y. (1986) Science 233, 305–312 [DOI] [PubMed] [Google Scholar]
- 3.Emoto Y., Manome Y., Meinhardt G., Kisaki H., Kharbanda S., Robertson M., Ghayur T., Wong W. W., Kamen R., Weichselbaum R. (1995) EMBO J. 14, 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghayur T., Hugunin M., Talanian R. V., Ratnofsky S., Quinlan C., Emoto Y., Pandey P., Datta R., Huang Y., Kharbanda S., Allen H., Kamen R., Wong W., Kufe D. (1996) J. Exp. Med. 184, 2399–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohtz J. D., Jamison S. F., Will C. L., Zuo P., Lührmann R., Barcia-Blanco M. A., Manley J. L. (1994) Nature 368, 119–124 [DOI] [PubMed] [Google Scholar]
- 6.Anantharam V., Kitazawa M., Wagner J., Kaul S., Kanthasamy A. G. (2002) J. Neurosci. 22, 1738–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyland M. E., Anderson S. M., Matassa A. A., Barzen K. A., Quissell D. O. (1999) J. Biol. Chem. 274, 19115–19123 [DOI] [PubMed] [Google Scholar]
- 8.Denning M. F., Wang Y., Tibudan S., Alkan S., Nickoloff B. J., Qin J. Z. (2002) Cell Death Differ. 9, 40–52 [DOI] [PubMed] [Google Scholar]
- 9.Sitailo L., Tibudan S., Denning M. F. (2004) J. Invest. Dermatol. 123, 1–10 [DOI] [PubMed] [Google Scholar]
- 10.Sitailo L. A., Tibudan S. S., Denning M. F. (2006) J. Biol. Chem. 281, 29703–29710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peluso J. J., Pappalardo A., Fernandez G. (2001) Endocrinology 142, 4203–4211 [DOI] [PubMed] [Google Scholar]
- 12.Kilpatrick L. E., Lee J. Y., Haines K. M., Campbell D. E., Sullivan K. E., Korchak H. M. (2002) Am. J. Physiol. Cell Physiol. 283, C48–C57 [DOI] [PubMed] [Google Scholar]
- 13.Zrachia A., Dobroslav M., Blass M., Kazimirsky G., Kronfeld I., Blumberg P. M., Kobiler D., Lustig S., Brodie C. (2002) J. Biol. Chem. 277, 23693–23701 [DOI] [PubMed] [Google Scholar]
- 14.McCracken M. A., Miraglia L. J., McKay R. A., Strobl J. S. (2003) Mol. Cancer Ther. 2, 273–281 [PubMed] [Google Scholar]
- 15.Sakurai Y., Onishi Y., Tanimoto Y., Kizaki H. (2001) Biol. Pharm. Bull. 24, 973–977 [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi T., Niino Y., Ohtaki H., Kikuyama S., Shioda S. (2006) FEBS Lett. 580, 2458–2464 [DOI] [PubMed] [Google Scholar]
- 17.Ueyama T., Ren Y., Ohmori S., Sakai K., Tamaki N., Saito N. (2000) Biochem. Biophys. Res. Commun. 269, 557–563 [DOI] [PubMed] [Google Scholar]
- 18.Hastings M. L., Krainer A. R. (2001) Curr. Opin Cell Biol. 13, 302–309 [DOI] [PubMed] [Google Scholar]
- 19.Kawahigashi H., Harada Y., Asano A., Nakamura M. (1998) Biochim. Biophys. Acta 1397, 305–315 [DOI] [PubMed] [Google Scholar]
- 20.Libri D., Piseri A., Fiszman M. Y. (1991) Science 252, 1842–1845 [DOI] [PubMed] [Google Scholar]
- 21.Muro A. F., Iaconcig A., Baralle F. E. (1998) FEBS Lett. 437, 137–141 [DOI] [PubMed] [Google Scholar]
- 22.Du K., Peng Y., Greenbaum L. E., Haber B. A., Taub R. (1997) MCB 17, 4096–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chalfant C. E., Mischak H., Watson J. E., Winkler B. C., Goodnight J., Farese R. V., Cooper D. R. (1995) J. Biol. Chem. 270, 13326–13332 [DOI] [PubMed] [Google Scholar]
- 24.Patel N. A., Chalfant C. E., Watson J. E., Wyatt J. R., Dean N. M., Eichler D. C., Cooper D. R. (2001) J. Biol. Chem. 276, 22648–22654 [DOI] [PubMed] [Google Scholar]
- 25.Bonnet E., Touyarot K., Alfos S., Pallet V., Higueret P., Abrous D. N. (2008) PLoS ONE 3, e3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang K., Patel N. A., Watson J. E., Apostolatos H., Kleiman E., Hanson O., Hagiwara M., Cooper D. R. (2009) Endocrinology 150, 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Church D. M., Stotler C. J., Rutter J. L., Murrell J. R., Trofatter J. A., Buckler A. J. (1994) Nat. Genet. 6, 98–105 [DOI] [PubMed] [Google Scholar]
- 28.Gallego M. E., Gattoni R., Stévenin J., Marie J., Expert-Bezançon A. (1997) EMBO J. 16, 1772–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walton H. S., Gebhardt F. M., Innes D. J., Dodd P. R. (2007) J. Neurosci. Methods 160, 294–301 [DOI] [PubMed] [Google Scholar]
- 30.Chand H. S., Ness S. A., Kisiel W. (2006) Int. J. Cancer 118, 1713–1720 [DOI] [PubMed] [Google Scholar]
- 31.Ise R., Han D., Takahashi Y., Terasaka S., Inoue A., Tanji M., Kiyama R. (2005) FEBS Lett. 579, 1732–1740 [DOI] [PubMed] [Google Scholar]
- 32.Liu H. X., Chew S. L., Cartegni L., Zhang M. Q., Krainer A. R. (2000) Mol. Cell. Biol. 20, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladd A. N., Cooper T. A. (2002) Genomic Biol. 3, 1–16 [Google Scholar]
- 34.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R. (2003) Nucleic Acids Res. 31, 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel N. A., Eichler D. C., Chappell D. S., Illingworth P. A., Chalfant C. E., Yamamoto M., Dean N. M., Wyatt J. R., Mebert K., Watson J. E., Cooper D. R. (2003) J. Biol. Chem. 278, 1149–1157 [DOI] [PubMed] [Google Scholar]
- 36.Vickers T. A., Zhang H., Graham M. J., Lemonidis K. M., Zhao C., Dean N. M. (2006) J. Immunol. 176, 3652–3661 [DOI] [PubMed] [Google Scholar]
- 37.D'Souza I., Schellenberg G. D. (2005) Biochim. Biophys. Acta 1739, 104–115 [DOI] [PubMed] [Google Scholar]
- 38.Khoo B., Akker S. A., Chew S. L. (2003) Trends Biotechnol. 21, 328–330 [DOI] [PubMed] [Google Scholar]
- 39.Stamm S. (2002) Hum. Mol. Genet. 11, 2409–2416 [DOI] [PubMed] [Google Scholar]
- 40.Wang H. Y., Xu X., Ding J. H., Bermingham J. R., Jr., Fu X. D. (2001) Mol. Cell 7, 331–342 [DOI] [PubMed] [Google Scholar]
- 41.Hernández F., Pérez M., Lucas J. J., Mata A. M., Bhat R., Avila J. (2004) J. Biol. Chem. 279, 3801–3806 [DOI] [PubMed] [Google Scholar]
- 42.Meshorer E., Bryk B., Toiber D., Cohen J., Podoly E., Dori A., Soreq H. (2005) Mol. Psychiatry 10, 985–997 [DOI] [PubMed] [Google Scholar]
- 43.Misiuta I. E., Anderson L., McGrogan M. P., Sanberg P. R., Willing A. E., Zigova T. (2003) Dev. Brain Res. 145, 107–115 [DOI] [PubMed] [Google Scholar]
- 44.McCaffery P., Zhang J., Crandall J. E. (2006) J. NeuroBiol. 66, 780–791 [DOI] [PubMed] [Google Scholar]
- 45.Yang Z., Sui Y., Xiong S., Liour S. S., Phillips A. C., Ko L. (2007) Nucleic Acids Res. 35, 1919–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donai H., Murakami T., Amano T., Sogawa Y., Yamaguchi T. (2000) Brain Res. Mol. Brain Res. 85, 189–199 [DOI] [PubMed] [Google Scholar]