Abstract
In the mammalian brain high affinity nicotine-binding sites are composed of at least the α4 and β2 subunits. Additional nicotinic acetylcholine receptor subunits that are often co-expressed with α4+β2 include α5. The introduction of α5 into 293 cells expressing α4+β2 strongly favors assembly of α4+α5+β2 receptors, increases constitutive ligand binding density as measured using [3H]epibatidine, but reduces the magnitude of up-regulation in response to chronic nicotine. In contrast, when β4 is substituted for β2, α5 interferes with the assembly of these receptors, demonstrating an important role for the β subunit in this process. When cells co-express α4+α5+β2+β4, over 50% of the subunit associations include all four subunits, but they fail to be detected using [3H]epibatidine binding. However, complexes of α4+α5+β2 do preferentially emerge from these subunit mixtures, and these mixtures bind ligand. In previous studies of α4+β2+β4 co-expression by 293 cells, the inflammatory cytokines IL-1β and TNFα influenced the outcome of receptor assembly (Gahring, L. C., Days, E. L., Kaasch, T., González de Mendoza, M., Owen, L., Persiyanov, K., and Rogers, S. W. (2005) J. Neuroimmunol. 166, 88–101). When α5 is included in this subunit mixture, and cells are exposed to either inflammatory cytokine, subunit association is no longer altered. These findings suggest that α5 is an influential modulator of α4+β2 nicotinic acetylcholine receptor assembly and stabilizes their expression in response to fluctuations in external conditions.
Keywords: Inflammation, Nicotinic Acetylcholine Receptors, Protein Assembly, Receptor Regulation, Tumor Necrosis Factor (TNF), Up-regulation, α5
Introduction
In the mammalian central nervous system a key nicotinic receptor (nAChR)2 is the high affinity nicotine-binding site that undergoes up-regulation in response to chronic exposure to ligands such as nicotine (1–4). Up-regulation can increase high affinity ligand binding densities 3–6-fold, and this change has been directly related to many unique features of nAChR biology including the addiction phenotype. The high affinity nicotine-binding site that is up-regulated by chronic ligand exposure was revealed through immunological methods to be composed almost entirely of receptors containing α4 and β2 subunits (4). This result was confirmed by mouse genetic approaches where ablation of either of these subunits effectively eliminates almost all high affinity binding sites in the brain (5, 6). However, the mechanisms governing the up-regulation process without significant compromise to their normal physiological roles or in extreme environments as during exposure to chronic nicotine or at sites of inflammation remain to be clarified (for discussions see Refs. 7 and 8).
Heterologous expression systems, which closely parallel results from in vivo studies (8), have been employed to show that nAChRs from subunit combinations such as α4+β2, α4+β4, or α4+β2+β4 (9–13) readily interact to form functional receptors. But in the brain, subunit expression and possible interactions are not this simple. For example, in the adult brain β4 expression is limited to only a few brain regions, whereas other subunits such as α5 are widely expressed. Although described as an α subunit, α5 lacks key residues important for ligand binding and is therefore considered a structural subunit (8). Nevertheless, its inclusion impacts upon many receptor functions. For example, in the absence of α5 there is a shift in receptor efficacy for multiple pharmacological agents, changes in channel ion permeability, and alterations to neurotransmitter release (8, 15, 16). These differences extend to the mouse where the absence of α5 impacts upon sensitivity to nicotine-induced locomotor activity and seizure (17) as well as the susceptibility to inflammatory bowel disease (18). In humans genetic findings suggest that a polymorphism in the cytoplasmic domain of α5 corresponds with susceptibility to early life abuse of nicotine and subsequent addiction (19, 20). Some of these effects may be explained by the finding that the majority of α4+β2 receptors in the rodent brain also contain α5, and these receptors exhibit limited up-regulation in response to nicotine (14). However, differing cell types and brain regions do differ quantitatively in α5 inclusion, indicating that regional heterogeneity in the effect by this subunit is likely to be present (8, 14, 21–24). Thus, understanding the impact of α5 on nAChR assembly and response to the environment is relevant to understanding basic mechanisms contributing to the receptor assembly process.
In this study we examined the role of α5 in modulating α4+β2 receptor assembly in the transiently transfected 293 cell model system as measured by the binding of [3H]epibatidine ([3H]EB). To summarize, α5 readily enters into stable interactions with α4+β2 as measured by [3H]EB binding. Although α5 incorporation strongly favors binding sites composed of α4+β2+α5 subunits, it interferes with expression of ligand-binding sites if β4 is substituted for β2. In terms of up-regulation to nicotine, α5 lessens this response. Finally, the co-expression of α5 with α4+β2, α4+β4, or α4+β2+β4 effectively abolished any impact by either TNFα or IL-1β on nAChR assembly, which is observed when α5 is not present (13). These findings reveal an additional and possibly modulatory role by α5 in stabilizing nAChR assembly to rapid shifts in external conditions such as those produced by nicotine or pro-inflammatory cytokines.
EXPERIMENTAL PROCEDURES
Materials
All of the reagents were identical to those reported previously (19). In all cases nAChR subunit-specific antibodies were prepared for epitopes in the cytoplasmic domain, and their use has been described in detail (4, 9, 12, 14, 24–28). In this study rabbit polyclonal 5009 or 4964 (α4), 4842 or 305 (β2), 4886 (β4), and 4889 (α5) were used. The monoclonal antibody to the R3b epitope (2F5) and construction of epitope-tagged α4 (termed α4R3b) were also described (13). Secondary antibodies were from Jackson ImmunoResearch (West Grove, PA).
Expression Vectors
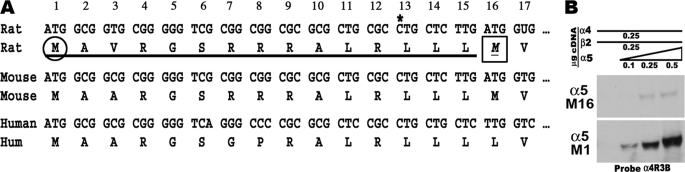
In all cases rat nAChR subunit cDNAs within the pcDNA3.1+ expression vector (Invitrogen) were as used and described previously (13). The epitope-tagged nAChR-β2HA and nAChR-β4HA expression vectors were generously provided by Dr. William Green (University of Chicago, Chicago, IL). During the course of these studies, we identified a problem in the sequence of the rat α5 cDNA expression construct PC989E(4) (provided by James Patrick, Baylor College of Medicine, Houston TX) (29) (Fig. 1). This expression construct deleted the initial 15 amino acids of the native protein, resulting in an incorrect start methionine. To correct this, standard RT-PCR-based methods were used to generate the original rat α5 5′-end from a rat brain cDNA pool, which was then subcloned into the pcDNA3.1 expression vector to generate a rat α5 expression cDNA construct with the sequence shown in Fig. 1 that is also consistent with the cDNA of other α5 subunits from mouse and human. When either of the α5 constructs is co-transfected with α4 and β2 into 293 cells, either methionine is competent as a translational start, and the product interacts with other nAChR subunits. However, translational efficiency of the repaired Met16 construct, as determined using co-immunoprecipitation and measurement of association with α4, was on the order of 10-fold more than constructs with the second methionine (Met16; Fig. 1b). All of the experiments reported in this study used the corrected α5 cDNA, whose translation is initiated from the first methionine.
FIGURE 1.
Definition of the α5 leader sequence used for expression in transfected cells. (A) In the original rat expression clone of α5 (PC989E(4)), there is an omission of the cytosine base identified by an asterisk that lead to a subcloned variant that lacked the first methionine (circled), thus forcing sole translation of the α5 cDNA protein product to initiate from the second methionine (boxed and identified as Met16). This sequence was corrected through RT-PCR methods of native α5 mRNA (see “Experimental Procedures”) to generate a cDNA that includes the longer α5, including an initiating methionine at residue 1 (circled). This also brings the α5 cDNA into agreement with those from the mouse (GenBankTM DQ318788 and C57BL6/J) and human (GenBankTM NM_000745), respectively. (B) As shown using Western blot analysis of 293 cells transfected with α4+β2 and either the corrected α5 or the Met16 start methionine, expression of α5 can proceed from either methionine, although the first initiating methionine is clearly favored.
Construction of the α5R3b epitope-tagged subunit was done as described for α4 (for details see Ref. 13), where the GluR3R3b sequence was introduced into the α5 subunit in place of the cysteine loop. These subunits are expressed, and they function similarly to the wild-type counterparts in these measurements (13). Because α5R3b is significantly smaller than α4R3b, both subunits can be detected simultaneously on Western blots using an anti-R3b epitope-defined monoclonal antibody (2F5).
Cell Culture and Transfection
Human embryonic kidney 293 cells were grown as before (13) under standard tissue culture conditions in DMEM (Cellgro, obtained from Fisher) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 1% penicillin/streptomycin (Cellgro, obtained from Fisher), and 1% sodium pyruvate. The jet-PEI system (QBiogene, Irvine, CA) was used for all transfections of 293 cells. Treatment with nicotine and/or optimized pro-inflammatory cytokines (25 ng/ml IL-1β or 25 ng/ml TNFα; both human recombinant) was initiated 2 h after transfection (13). All of the cells were harvested within 24 h after transfection.
Immunoprecipitation and Western Blots
As before (13), the cells were rinsed with Tris-buffered saline and solubilized in 10 mm Tris, 150 mm NaCl, 0.25% Nonidet P-40, 0.2% Triton X-100, pH 7.4, containing protease inhibitors using 10 passes with a glass Dounce homogenizer. Immunoprecipitation and Western blot analysis were done as described in detail elsewhere (4, 13, 14).
Sucrose Gradients
The methods used for sucrose gradient analysis were adapted from other investigators and have been described previously (13, 30, 31). Briefly, 293 cells transiently transfected with various nAChR subunits were solubilized in Triton X-100 and cleared by centrifugation before being layered onto a 5-ml sucrose gradient (continuous 5–20% (w/w) in 20 mm Tris, pH 7.5) prepared using a Gradient Master 107ip (BioComp Instruments Inc., Fredericton, Canada). The gradients were centrifuged at 54,000 rpm for 2 h (SW55Ti rotor; Beckman Instruments, Fullerton, CA) at 4 °C. The fractions were collected using an FC203B fraction collector with Auto Densiflow (Labconco, Kansas City, KS). The Svedberg markers run in a separate gradient from the samples were bovine serum albumin, 4.2 S (Pierce); β-amylase, 8.9 S (Sigma); catalase, 11.2 S (Sigma); and purified 20 S proteosome subunit, 20 S (a gift from Dr. M. Rechsteiner, University of Utah, Salt Lake City, UT).
Two-dimensional Blue Native Gel Electrophoresis (BNG)
Samples were prepared from transfected 293 cells as described previously (13). Briefly, crude membranes were suspended in buffer (aminocaproic acid, 50 mm Bis-Tris, pH 7.0, 2% dodecylmaltoside, 0.5 mm EDTA, and 5% (w/v) Coomassie Blue G), and these samples were resolved using blue native gradient gel (4–20% acrylamide) electrophoresis. This gel was soaked in a SDS + dithiothreitol solution, rotated 90 °C, and placed onto the second denaturing SDS-PAGE gel for size fractionation. Analyses thereafter used standard Western blot methods. Molecular weight markers were from the Amersham Biosciences high molecular weight calibration kit.
[3H]Epibatidine Binding
Radioligand binding of transiently transfected 293 cells was as before (32). Crude membrane fractions were prepared from transfected cells following brief homogenization and collected by centrifugation for 20 min at 20,000 × g at 40 °C. Binding was for 4 h at room temperature of 5 nm of [3H]epibatidine (PerkinElmer Life Sciences) to 20 μg of protein/sample. All of the samples were measured in triplicate. Nonspecific binding included an additional sample set where the membranes were preincubated for 30 min with nicotine (300 μm) before adding the radiolabeled compound (26). The samples were vacuum-filtered through GF/C glass filters (Whatman International), and standard methods of scintillation counting using a Beckman scintillation counter with internal standards followed. Specifically bound counts were calculated by subtracting counts remaining in the nicotine preincubated control sample from the total bound fraction.
RESULTS
The Association of α5 with α4 Varies According to Which β Subunit Is Co-expressed
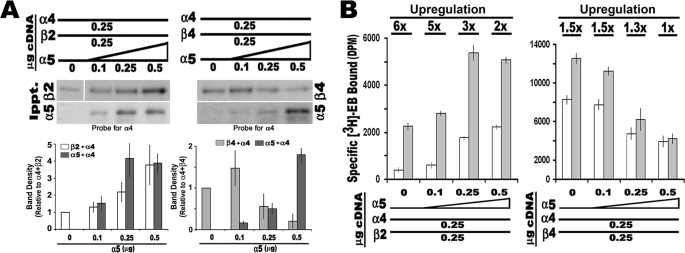
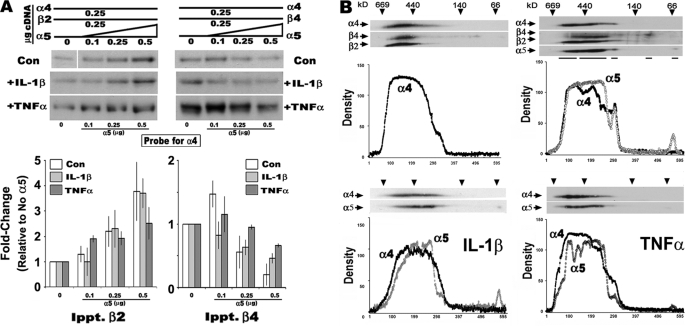
To determine whether α5 modifies nAChR assembly and influences the impact of nicotine or cytokines on this process, we began by examining how α5 interacts to form receptors. To measure this, we used [3H]EB binding to membranes from cells transfected with either α4+β2 or α4+β4. In these experiments (Fig. 2), the cDNA concentrations of the α4 and relevant β subunit were held constant (0.25 μg/transfection, respectively), whereas the concentration of α5 input cDNA was varied from 0, 0.1, 0.25, to 0.5 μg, respectively. After incubation, the cells were solubilized, divided into equal fractions, and subjected to immunoprecipitation with the specified anti-β or anti-α5 subunit antibody (see “Experimental Procedures”). Immunoprecipitates were fractionated by SDS gel electrophoresis, and the association with α4 was revealed using Western blot analysis (Fig. 2A). In starting mixtures of α4+β2, increasing the amount of α5 corresponds with more α5 in association with α4 as well as between α4 and β2. The increased association of α5 with α4 and β2 also corresponds with increased [3H]EB binding, indicating that receptor assembly was promoted by α5 (Fig. 2B). When input cDNA concentrations were equivalent for all three subunits (25 μg each), the amount of total binding detected was ∼4-fold greater than transfections of α4+β2 not containing α5. To determine whether α5 altered agonist-mediated up-regulation, this experiment was also repeated in the presence of chronic nicotine (1 μm) during the duration of the transfection. As shown in Fig. 2B, the relative proportion of increased [3H]EB binding in cells treated with nicotine versus those not exposed to nicotine validates that these complexes do undergo nicotine-mediated up-regulation. However, the relative degree of up-regulation diminished from almost 6-fold in non-α5-transfected cells to only 2-fold in cells co-expressing α5 (0.5 μg α5 cDNA transfected). Thus, α5 increases the number of ligand-binding sites in receptors composed of α4+β2 subunits, but the relative amount of nicotine-mediated up-regulation is diminished. This result is consistent with similar reports of ligand binding but poor up-regulation of α4α5β2 receptors in the rodent brain (14).
FIGURE 2.
The α5 subunit enters into stable complexes with α4 and up-regulation to nicotine is reduced. 293 cells were co-transfected with varying amounts of α5 cDNA (as indicated) with constant cDNA amounts for either α4 and β2 or α4 and β4, respectively. A, immunoprecipitation with either anti-α5 or the appropriate anti-β subunit was followed by Western blot analysis to determine the amount of associated α4. Quantitation of the blots in this panel shows that as the amount of α5 cDNA co-transfected with α4+β2 subunit is increased, associations between α4 and both β2 and α5, respectively, also increase. In contrast, when β4 is substituted for β2, the amount of α4 associated with β4 diminishes, whereas α5 is enhanced. All of the values were normalized to the no α5 input values, and the error bars were calculated and are shown as ± S.E. for the results from three to seven independent experiments. B, [3H]EB ligand binding to membranes from cells transfected as in A and membranes from cells treated with 1 μm nicotine (gray bars). Increased α5 corresponds with decreased up-regulation induced by nicotine (fold increase is noted above each treatment set). Each experiment was conducted three to eight times, and the error bars reflect ± S.E. for all experiments.
To assess the importance of the β subunit in this process, mixtures where β2 was replaced by β4 were examined. Again, as the α5 concentration increases so do α5 associations with α4; however, associations between β4 and α4 are notably decreased (Fig. 2A). The impact by α5 occurs as the input cDNA concentration is equivalent to or exceeds that of β4 cDNA. The decreased α4+β4 association is accompanied by a decrease in [3H]EB binding, suggesting disruption of ligand-binding complexes by α5 (Fig. 2B).
The α4β4 receptors also exhibit up-regulation to chronic nicotine (1.5-fold), but this is much less than for α4β2 receptors (9, 10, 32). However, upon increasing the α5 contribution, up-regulation is completely abolished. Therefore, although α5 readily forms subunit associations in cells transfected with α4+β4, the net result is reduced [3H]EB binding, opposite that seen for α4+β2 transfected cells. In both cases, inclusion of α5 corresponds with the overall reduced up-regulation in response to nicotine.
Cells Co-expressing α4+α5+β2+β4
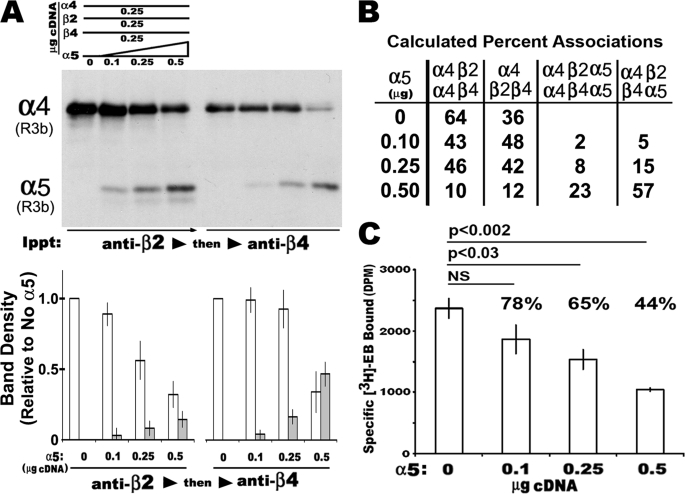
As reported previously (13) when equal amounts of α4, β2, and β4 cDNA are co-transfected into 293 cells, a characteristic distribution of mature receptors of three different subunit composition results (α4+β2, 15%; α4+β4, 55%; and α4+β2+β4, 30%). We examined how α5 impacts upon this distribution. For these experiments 293 cells were co-transfected with a constant amount of cDNA encoding α4+β2+β4 (0.25 μg each) and varied amounts of α5 (0, 0.1, 0.25, and 0.5 μg, respectively). The transfected cells were harvested and subjected to successive immunoprecipitation (4, 13, 14) where β2-containing complexes were removed first followed by a second immunoprecipitation with anti-β4 (Fig. 3). The products of each immunoprecipitation were resolved by SDS-PAGE and the amount of α4 and α5 (when present) in association with the subunit being precipitated was measured by Western blot (Fig. 3A). Because both α4 and α5 are identified with the same epitope tag, both subunits can be resolved simultaneously on the blot by the difference in molecular weights (α4, ∼72 kDa; α5, ∼48 kDa). In some experiments, the anti-α5 antibody was used first to remove complexes containing α5 (not shown). The relative intensity of each band from the respective immunoprecipitation was then quantitated as shown in Fig. 3A. A summary of the results derived in percentages of complexes is shown (Fig. 3B). The results were similar to those obtained before (13) where receptors of α4+β2, α4+β4, and α4+β2+β4 were generated in different but distinct ratios. Associations between α4 and β4 are favored over α4 and β2 in the absence of α5 (not shown). As α5 input cDNA increased so did the proportion of α4 in association with α5. This included a dramatic increase in α4+α5+β2+β4 complexes that reached on average 57% of the total associations in mixtures containing 0.5 μg of α5 cDNA. However, there was a progressive decrease in [3H]EB binding (Fig. 3C). The reduction to 65% is significant when equal cDNA input is used (0.25 μg of each subunit; p < 0.03), and ligand binding decreases further to 44% when α5 amounts exceed those of other subunits. This decrease was not due to cell death because trypan blue exclusion was not increased relative to the other transfections (not shown). Notable is the convergence of values between the 57% value found for the production of α4+α5+β2+β4 complexes and the 56% decrease in ligand binding for cells receiving 0.5 μg of α5 cDNA. Finally, complexes such as α4+α5+β4 were strongly disfavored (see the gel Fig. 3A where anti-β4 immunoprecipitation follows anti-β2). Collectively these results indicate that α5 is a favored assembly partner with α4 and β2. However, other subunits such as β4 alter this preferred association pattern, presumably leading to nonproductive subunit interactions.
FIGURE 3.
The α5 subunit alters the relative subunit association among α4+β2+β4. Increasing amounts of α5 were co-transfected with fixed amounts of α4, β2, and β4 cDNA (0.25 μg of each). Crude membranes were solubilized in detergent and subjected to progressive immunoprecipitation with either anti-β2 followed by anti-β4 or reciprocally anti-β4 followed by anti-β2 as described (see “Experimental Procedures”). A, representative results from cells transfected as described above. A Western blot reveals the relative amount of R3b epitope-tagged α4 or α5 as indicated in association with β2 or β4 after removal of all β2 complexes. Quantitation of these blots is shown where all of the values are normalized to samples with no α5 co-transfected. The error bars reflect plus or minus the standard error of the mean as calculated from three independent experiments done in the same manner. B, average percentages of the relative amounts of mixed subunit complexes that were either directly measured or derived from measuring the relative amount of α4 remaining after successive anti-β subunit immunoprecipitations as in A. The averages are from three independent experiments. C, ligand binding ([3H]EB) to total membranes decreases as α5 input is increased. The loss of ∼56% of the total radioligand binding relative to non-α5 transfection mixtures corresponds well with the increase of α4α5β2β4 complexes (B).
To clarify further the receptor subunit distribution and ligand binding from emerging from mixtures of α4+α5+β2+β4, a combination of sucrose density centrifugation, ligand binding, and two-dimensional blue native gel electrophoresis (BNG) was used. Detergent-solubilized lysates from cells co-transfected with equal amounts of each cDNA (α4+β2+β4 with and without α5; 0.25 μg/subunit) were subjected to sedimentation velocity centrifugation through sucrose gradients. Individual fractions were subsequently analyzed by SDS-PAGE. Because each subunit migrates on SDS-PAGE at different molecular weights, they could each be detected simultaneously on Western blots, and specific [3H]EB binding to corresponding fractions of gradients prepared in parallel could be measured.
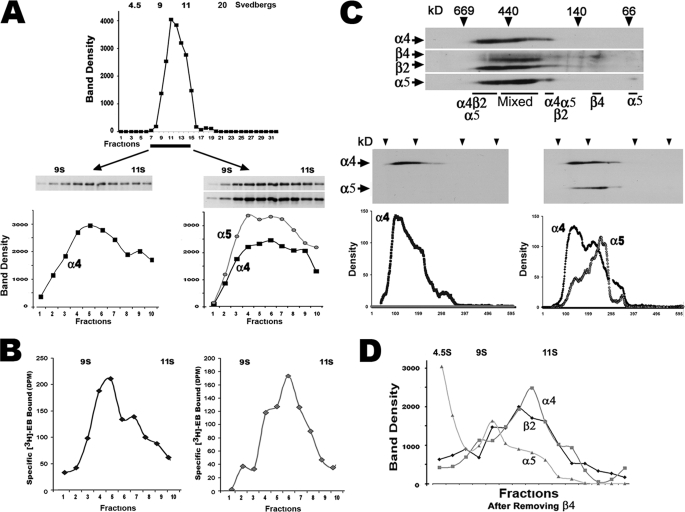
The majority of α4-containing receptor complexes in the absence of α5 migrate as a peak fraction of ∼10 S (Fig. 4A). To examine this region of the gradient in greater detail, smaller fractions (100 μl) were collected between the 9 and 11 S marker fractions to increase the resolution of receptor distribution as reflected by the distribution of α4, which localized mostly to a peak consistent with the 10 S fraction (Fig. 4A). When α5 was included in the transfection mixture, the sucrose gradient profile was altered. Although there was an apparent increase in the amount of α4 and α5 in the 10 S peak, the presence of a prominent 10.5 S shoulder was also detected, indicating that α5 shifts, at least in part, the receptor shape.
FIGURE 4.
Sucrose density gradients and two-dimensional BNG reveals ligand-binding sites of the α4+α5+β2 subunit composition. A, cells were transfected with equal amounts of cDNA (0.25 μg of each) encoding α4R3b and β2-HA with or without α5R3b. Solubilized membranes from these cells were subjected to sucrose gradient sedimentation, and gradient fractions were collected for analysis of the epitope marked α4 and α5 subunits (when present) using SDS-PAGE gel electrophoresis. As shown, the majority of α4 is localized to fractions consistent with a 10 S complex. The profile revealed by α4 immunoreactivity results when gradient resolution between the 9–11 S markers is increased. When α5 is included in the transfection mixtures, there is a broadening of the sedimentation profile curve to include a new distribution covering the 10–10.5 S range. B, the corresponding [3H]EB binding to similar gradient fractions is shown. Inclusion of α5 increases the amount of ligand bound to the 10.5 S peak over the 10 S peak similar to the distribution of the α4 or α4+α5, respectively, measured by Western blots. C, mixtures of α4+α5+β2+β4 were co-transfected, and the cell membranes were prepared for two-dimensional BNG analysis. All of the subunits were detected using Western blot analysis for epitope-tagged β2 and β4 followed by stripping and reprobing to reveal both epitope-tagged α subunits. The gel was then reassembled electronically to show all of the subunits as identified. Most immunoreactivity is of various (mixed) subunit composition, although two major complexes (designated as α4α5β2) do not appear to include β4. Possible β4 dimers and α5 monomers are marked. In the lower left gel, α5 was omitted from the subunit co-transfection mixture (only α4 is shown for clarity); the majority of α4 resides in a smear of complexes, although a prominent peak of immunoreactivity as measured in the gel densitometry tracing below the blot is observed. However, when α5 is included in the transfection mixture, the molecular weight of the overall complex is shifted to add a “lower” molecular weight component that includes both α4 and α5 species. These are the same complexes that are dominated by the presence of α4+α5+β2 in the reference gel above. D, to determine whether α4+α5+β2 subunits are present, α4+α5+β2+β4 were co-transfected (equivalent amounts of cDNA as above) only before separating the complexes on sucrose gradients, complexes harboring β4 were removed using anti-β4 immunoprecipitation. The remaining complexes were then separated on a sucrose gradients, the fractions were collected, the Western blots were prepared, and the density traces of the two-dimensional analysis were done as in C. As shown in the line profiles, two prominent peaks of the remaining α4, α5, and β2 were present that include one at the 10.5 S peak and another near 10 S. A third and smaller peak found in immunoprecipitated samples was composed of mixed subunits of near equivalent amounts at ∼9.5 S and presumed monomers of α5 near the top of the gradient. The results reflect data collected from between two and six independently performed experiments.
The binding of [3H]EB to protein within the gradient fractions collected in parallel are also consistent with a 10 and 10.5 S distribution that is largely dependent upon the presence of α5 (Fig. 4B). As shown there, without co-expression of α5, the majority of ligand binding is associated with the 10 S fraction. However, a persistent 10.5 S shoulder is distinguished by ligand binding that is not resolved well using Western blot analyses. Possible explanations for a shift to a 10.5 S shoulder include altered binding conformation, changes in glycosylation, binding of other intracellular proteins or altered receptor subunit stoichiometry. What is clear is that when α5 is present, ligand binding in the 10.5 S peak is increased substantially, although binding in the 10 S peak remains the same. Thus, ligand binding and Western blot profiles of α4 containing fractions between the 9 and 11 S markers correspond well with at least two major binding components of 10 and 10.5 S, but the 10.5 S fraction is enhanced by the presence of α5, suggesting an altered receptor shape dependent upon inclusion of α5.
To investigate further the receptor subunit differences on receptor shape suggested by sucrose gradient analysis of α4+α5+β2+β4 mixtures, we used two-dimensional BNG (see “Experimental Procedures”). This method is particularly useful for resolving the subunit composition of intact protein complexes including ligand gated ion channels, and we introduced this method to examine the relative subunit composition of nAChRs assembled from α4+β2+β4 subunit mixtures (13). A gel typical of the two-dimensional BNG method produced from cells co-transfected with α4+α5+β2+β4 is shown in Fig. 4C. In this system α5 is present in large complexes consistent with mature nAChRs as well as in a single spot at ∼55 kDa that is likely to be monomeric subunits. Similarly, β4 is present in a ∼100-kDa spot that is of dimer size. Although the majority of nAChR immunoreactivity co-localizes in the complex mixtures of different α4+α5+β2+β4 combinations, three prominent components are resolved (Fig. 4C). First, the largest complex resolved by two-dimensional BNG is composed of α4+α5+β2. Because this peak appears only in α5 transfected cells, it is likely to be related to the 10.5 S peak seen in sucrose gradients reported above. The second complex, the most prominent one in terms of protein content, is composed of a mixture of α4+α5+β2+β4 subunits, which are likely to reflect the relatively complex mixture of α4-containing assemblages suggested by the results of Fig. 3. Finally there is a smaller sized fraction that is also consistent with α4+α5+β2 composition. The presence of this additional peak suggests that α4+α5+β2 receptors are present in two conformations or possibly bound to other intracellular proteins whose identity will require further investigation.
If indeed a receptor of α4+α5+β2 is generated from a subunit mixture of α4+α5+β2+β4, then this complex should be evident in samples where all β4-containing complexes are removed before gradient analysis. To test this, 293 cells were transfected with α4+α5+β2+β4 (0.25 μg of cDNA/subunit), the solubilized membranes were cleared by immunoprecipitation with anti-β4, and the remaining complexes were analyzed using sucrose gradient density centrifugation. The results in Fig. 4D show the band density profiles of a Western blot from these gradients. As predicted, there was essentially no detectable β4 (not shown), although α4+α5+β2 proteins remained. Of these, greater than 80% of all receptor-related protein signals were in the 10–10.5 S fractions, although the 10.5 S peak dominated. A smaller fraction closer to 9.5 S in these gradients harbored almost equivalent amounts of α4+α5+β2 complexes (Fig. 4D), as did a large amount of α5 immunoreactivity near the gradient top, which is consistent with the monomers suggested by two-dimensional BNG analysis (Fig. 4C).
These results show receptor complexes harboring α4 in combination with β2 and/or β4 that bind [3H]EB as reported previously (13, 30, 33). The addition of α5 produces both α4+α5+β2 complexes consistent with a 10.5 S peak revealed by Western blot and ligand binding. Other complexes of mixed α4+α5+β2+β4 subunits are also present as predicted from the results in Fig. 3.
IL-β and TNFα Do Not Impact upon Assembly of α4-containing nAChRs when α5 Is Co-expressed
Pro-inflammatory cytokines also impact upon the expression of receptors assembled from α4 and various mixtures of β2 and/or β4 subunits (13). The α4-based subunit associations measured above were repeated in the presence of α5. For these experiments, the conditions described above were repeated, but cells were also treated with either IL-1β or TNFα. Crude membranes were then prepared from cells of each treatment group and analyzed for the relative amount of α4 co-immunoprecipitated with either of the respective β subunits or α5. As shown in Fig. 5A, the results from these experiments reveal no impact by cytokines on subunit associations in the presence of α5 (compare results with those in Fig. 2A and Ref. 13). When complex mixtures of subunits were co-transfected, and subunit profiles of the receptors were examined using two-dimensional BNG (Fig. 5B), the receptor profiles emerging, regardless of IL-1β or TNFα treatment, exhibited equivalent assembly to the control. Thus, the assembly of α4 when α5 is present is unaffected by the presence of these pro-inflammatory cytokines.
FIGURE 5.
The presence of α5 moderates the influence of the pro-inflammatory cytokines IL-1β and TNFα on nAChR receptor assembly. A, similar to experiments in Fig. 2 only IL-β or TNFα was added for the duration of the transfection (control gels are the same as in Fig. 2). Western blot analysis of cells co-transfected with fixed amounts of α4 and the indicated β subunit and increasing amounts of α5 cDNA. The relative amount of association of α4 with the indicated β subunit is detected using Western blot, and the relative band density is plotted. B, in similar experiments cell membranes were examined by two-dimensional BNG analysis, and the results for just α4 and α5 are shown for clarity. Plots of α4 immunoreactivity in cells treated with either IL-β or TNFα and co-transfected with α5 or not co-transfected (as labeled) are placed below the blots. Control experiments were conducted between three and eight times as in Fig. 2. Cytokine experiments are based upon an n value of 3.
DISCUSSION
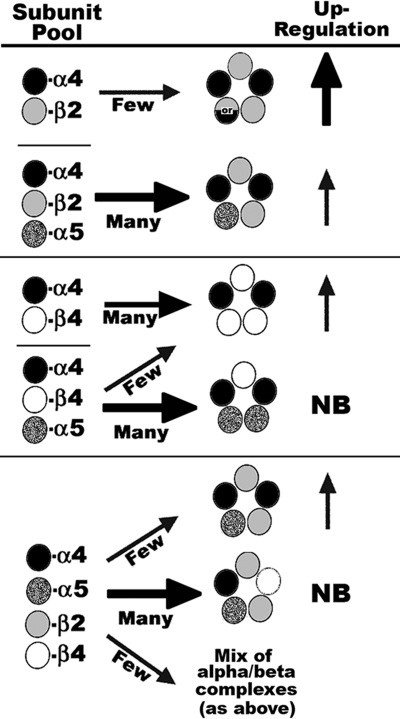
The results of this study suggest that α5 promotes α4+β2 associations but interferes with α4+β4 as detailed in Fig. 6. A possible explanation for this result is that in the starting subunit pools of α4+β2, α5 provides an “extra” subunit to promote direct assembly into mature receptors as defined by ligand binding. However, if the starting pool is α4+β4, then α5 binding to α4 is favored but occurs at the expense of α4+β4 interactions, resulting in less efficient production of mature receptors. This basic priority in subunit association is retained in more complex subunit mixtures such as α4+α5+β2+β4, where again α5 competes with β4 for association with α4, leading to complexes that fail to bind ligand as well as promote the final outcome of favoring α4+α5+β2 complexes.
FIGURE 6.
Model of α5 impact upon assembly of nAChRs. Depending upon the β subunit (either β2 or β4), the relative outcome of assembly with α4 varies dramatically when α5 is included in the mixture. Although α4+β2 does assemble to bind ligand with high affinity (as indicated by upward arrows), α5 favors this assembly through possibly filling the fifth position in nAChR assembly. The relative level of up-regulation is indicated by the size and thickness of the arrow. Two possible stoichiometries for α4+β2 receptors that were not investigated in this study are designated by the mixed circle containing or (31). In α4+β4 mixtures, α5 competes with β4 for binding to α4, resulting in reduced complexes detected by ligand binding. Although assembly of α4+β2 containing nAChRs is favored by α5, this decreases the relative amount of up-regulation promoted by nicotine because this subunit takes the place of nicotine in stabilizing or promoting expression of maturing nAChR complexes. The majority of complexes formed from co-transfection of α4+α5+β2+β4 result in mostly subunit associations that are not detected by ligand binding. NB, no binding. However, these mixtures also give rise to α4+β4±α5 complexes and α4+α5+β2 complexes, which dominate ligand binding measurements.
In terms of up-regulation, one of several mechanisms suggested to regulate this process (for review and discussion see Ref. 8) is improved assembly caused by stabilization of associations between α and β subunits upon binding of ligands such as nicotine (45). However, because α5 strongly favors α4+β2 assembly, binding of ligand to α4+α5+β2 receptors would have only a minimal effect on subsequent events related to up-regulation. In turn, this also reveals the importance of the β subunit to this process. Because the presence of β4 favors α4+α5 associations, which do not form a ligand-binding site, both up-regulation and overall assembly of receptors from these subunit combinations would be discouraged. This is consistent with other reports where the role of α5 in receptor formation in the presence of different β subunits has been examined. For example, Lindstrom and colleagues (34, 35) found that α5 promoted receptor assembly of α3+β2 receptors, but assembly was diminished if β4 was substituted for β2. Also similar to the present study, ligand binding was present only in regions of sucrose gradients harboring receptors of pentameric structure as measured by centrifugation and inclusion of the α5 subunit.
These studies also appear to be relevant to nAChRs identified in the animal. Chick ciliary ganglia that express predominantly α3 subtype receptors are most prevalent in ganglia, where 80% of these expressed were co-assembled with β4 and α5, whereas others harbor α3+α5+β2+β4 (36). Similar detailed examinations of the coincident expression of nAChR subunit expression by a subset of inhibitory interneurons in the CA1 hippocampal region suggest that although ∼85% of these express α4 also co-express α5, there are those that do not (8, 21, 24). Thus, in a model of local cellular heterogeneity of nAChR expression, our findings would predict that the variability in the co-expression of α5 with varying β subunits would render some cells highly resistant to change promoted by exogenous conditions, whereas others could modify their receptor expression. This could also address the issue of the relatively limited evidence for the presence of functional α4+β4 receptors in the brain despite multiple lines of evidence that support the expression of this subunit (46–48). In this case the production of α4+α5+β2 receptors or possibly α3+β4(±α5) receptors (37, 38) would be favored, whereas those of α4+β4 composition would fail to be detected.
Finally, nAChRs can modulate and be modulated by endogenous agents controlling the pro-inflammatory environment (32, 39–43). As noted earlier α7 nAChRs suppress inflammatory responses in neuronal and non-neuronal systems including the digestive system, lung, and skin (39–44). However, in turn IL-1β or TNFα significantly modifies certain nAChR receptor assembly where IL-1β favors α4+β2 assembly, and TNFα favors the formation of effectively any transfected α4+β2+β4 receptor subunit combination (13). TNFα dramatically enhances nicotine-mediated up-regulation of α4+β2 receptors through a mechanism that requires participation of the p38 MAPK pathway (32). However, in the current study the presence of α5 stabilizes assembly from these agents. Thus, in addition to ligand, the inflammatory environment may also impact the expression of certain nAChRs. However, these results also suggest that the occurrence of reciprocal regulatory interactions between the pro-inflammatory and nicotinic cholinergic receptors systems would be facilitated by the presence or absence of α5 and stabilization of key receptor expression.
Acknowledgments
Many thanks to Emily Days, Tuesday Kaasch, Karina Persiyanov, and Gustavo Vasquez-Opazo for excellent technical contributions during the course of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants AG017517 (to S. W. R.), AG029838 (to L. C. G.), and DA025057 (to L. C. G. and S. W. R.). This work was also supported by a Veterans Affairs-Merit award (to L. C. G.).
- nAChR
- nicotinic acetylcholine receptor
- BNG
- blue native gel
- EB
- epibatidine.
REFERENCES
- 1.Schwartz R. D., Kellar K. J. (1985) J. Neurochem. 45, 427–433 [DOI] [PubMed] [Google Scholar]
- 2.Marks M. J., Stitzel J. A., Collins A. C. (1985) J. Pharmacol. Exp. Ther. 235, 619–628 [PubMed] [Google Scholar]
- 3.Whiting P., Lindstrom J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 595–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores C. M., Rogers S. W., Pabreza L. A., Wolfe B. B., Kellar K. J. (1992) Mol. Pharmacol. 41, 31–37 [PubMed] [Google Scholar]
- 5.Picciotto M. R., Zoli M., Changeux J. P. (1999) Nicotine Tob. Res. Suppl. 2, S121–S125 [DOI] [PubMed] [Google Scholar]
- 6.Cordero-Erausquin M., Marubio L. M., Klink R., Changeux J. P. (2000) Trends Pharmacol. Sci. 21, 211–217 [DOI] [PubMed] [Google Scholar]
- 7.Govind A. P., Vezina P., Green W. N. (2009) Biochem. Pharmacol 78, 756–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albuquerque E. X., Pereira E. F., Alkondon M., Rogers S. W. (2009) Physiol. Rev. 89, 73–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao Y., Kellar K. J. (2004) J. Pharmacol. Exp. Ther. 310, 98–107 [DOI] [PubMed] [Google Scholar]
- 10.Xiao Y., Baydyuk M., Wang H. P., Davis H. E., Kellar K. J. (2004) Bioorg. Med. Chem. Lett. 14, 1845–1848 [DOI] [PubMed] [Google Scholar]
- 11.Walsh H., Govind A. P., Mastro R., Hoda J. C., Bertrand D., Vallejo Y., Green W. N. (2008) J. Biol. Chem. 283, 6022–6032 [DOI] [PubMed] [Google Scholar]
- 12.Vallejo Y. F., Buisson B., Bertrand D., Green W. N. (2005) J. Neurosci. 25, 5563–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gahring L. C., Days E. L., Kaasch T., González de Mendoza M., Owen L., Persiyanov K., Rogers S. W. (2005) J. Neuroimmunol. 166, 88–101 [DOI] [PubMed] [Google Scholar]
- 14.Mao D., Perry D. C., Yasuda R. P., Wolfe B. B., Kellar K. J. (2008) J. Neurochem. 104, 446–456 [DOI] [PubMed] [Google Scholar]
- 15.Albuquerque E. X., Alkondon M., Pereira E. F., Castro N. G., Schrattenholz A., Barbosa C. T., Bonfante-Cabarcas R., Aracava Y., Eisenberg H. M., Maelicke A. (1997) J. Pharmacol. Exp. Ther. 280, 1117–1136 [PubMed] [Google Scholar]
- 16.Gerzanich V., Wang F., Kuryatov A., Lindstrom J. (1998) J. Pharmacol. Exp. Ther. 286, 311–320 [PubMed] [Google Scholar]
- 17.Salas R., Orr-Urtreger A., Broide R. S., Beaudet A., Paylor R., De Biasi M. (2003) Mol. Pharmacol. 63, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 18.Orr-Urtreger A., Kedmi M., Rosner S., Karmeli F., Rachmilewitz D. (2005) Neuroreport 16, 1123–1127 [DOI] [PubMed] [Google Scholar]
- 19.Stevens V. L., Bierut L. J., Talbot J. T., Wang J. C., Sun J., Hinrichs A. L., Thun M. J., Goate A., Calle E. E. (2008) Cancer Epidemiol. Biomarkers Prev. 17, 3517–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss R. B., Baker T. B., Cannon D. S., von Niederhausern A., Dunn D. M., Matsunami N., Singh N. A., Baird L., Coon H., McMahon W. M., Piper M. E., Fiore M. C., Scholand M. B., Connett J. E., Kanner R. E., Gahring L. C., Rogers S. W., Hoidal J. R., Leppert M. F. (2008) PLoS Genet. 4, e1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudweeks S. N., Yakel J. L. (2000) J. Physiol. 527, 515–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoli M., Moretti M., Zanardi A., McIntosh J. M., Clementi F., Gotti C. (2002) J. Neurosci. 22, 8785–8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salminen O., Murphy K. L., McIntosh J. M., Drago J., Marks M. J., Collins A. C., Grady S. R. (2004) Mol. Pharmacol. 65, 1526–1535 [DOI] [PubMed] [Google Scholar]
- 24.Gahring L. C., Persiyanov K., Rogers S. W. (2005) Neurobiol. Aging 26, 973–980 [DOI] [PubMed] [Google Scholar]
- 25.Rogers S. W., Mandelzys A., Deneris E. S., Cooper E., Heinemann S. (1992) J. Neurosci. 12, 4611–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cauley K., Marks M., Gahring L. C., Rogers S. W. (1996) J. Neurobiol. 30, 303–314 [DOI] [PubMed] [Google Scholar]
- 27.Rogers S. W., Gahring L. C., Collins A. C., Marks M. (1998) J. Neurosci. 18, 4825–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores C. M., DeCamp R. M., Kilo S., Rogers S. W., Hargreaves K. M. (1996) J. Neurosci. 16, 7892–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Hara B. F., Macdonald E., Clegg D., Wiler S. W., Andretic R., Cao V. H., Miller J. D., Heller H. C., Kilduff T. S. (1999) Brain Res. Mol. Brain Res. 66, 71–82 [DOI] [PubMed] [Google Scholar]
- 30.Paulson H. L., Ross A. F., Green W. N., Claudio T. (1991) J. Cell Biol. 113, 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson M. E., Kuryatov A., Choi C. H., Zhou Y., Lindstrom J. (2003) Mol. Pharmacol. 63, 332–341 [DOI] [PubMed] [Google Scholar]
- 32.Gahring L. C., Osborne-Hereford A. V., Vasquez-Opazo G. A., Rogers S. W. (2008) J. Biol. Chem. 283, 693–699 [DOI] [PubMed] [Google Scholar]
- 33.Anand R., Conroy W. G., Schoepfer R., Whiting P., Lindstrom J. (1991) J. Biol. Chem. 266, 11192–11198 [PubMed] [Google Scholar]
- 34.Wang F., Gerzanich V., Wells G. B., Anand R., Peng X., Keyser K., Lindstrom J. (1996) J. Biol. Chem. 271, 17656–17665 [DOI] [PubMed] [Google Scholar]
- 35.Wang F., Nelson M. E., Kuryatov A., Olale F., Cooper J., Keyser K., Lindstrom J. (1998) J. Biol. Chem. 273, 28721–28732 [DOI] [PubMed] [Google Scholar]
- 36.Conroy W. G., Berg D. K. (1995) J. Biol. Chem. 270, 4424–4431 [DOI] [PubMed] [Google Scholar]
- 37.Alkondon M., Pereira E. F., Albuquerque E. X. (1996) J. Pharmacol. Exp. Ther. 279, 1491–1506 [PubMed] [Google Scholar]
- 38.Alkondon M., Albuquerque E. X. (2001) J. Neurophysiol. 86, 3043–3055 [DOI] [PubMed] [Google Scholar]
- 39.Arredondo J., Hall L. L., Ndoye A., Nguyen V. T., Chernyavsky A. I., Bercovich D., Orr-Urtreger A., Beaudet A. L., Grando S. A. (2003) Lab. Invest. 83, 207–225 [DOI] [PubMed] [Google Scholar]
- 40.de Jonge W. J., Ulloa L. (2007) Br. J. Pharmacol. 151, 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gahring L. C., Rogers S. W. (2005) AAPS J. 7, E885–E894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborne-Hereford A. V., Rogers S. W., Gahring L. C. (2008) J. Neuroimmunol. 193, 130–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang D. W., Zhou R. B., Yao Y. M. (2009) Chin. J. Traumatol. 12, 355–364 [PubMed] [Google Scholar]
- 44.Carlson N. G., Bacchi A., Rogers S. W., Gahring L. C. (1998) J. Neurobiol. 35, 29–36 [DOI] [PubMed] [Google Scholar]
- 45.Kuryatov A., Luo J., Cooper J., Lindstrom J. (2005) Mol. Pharmacol. 68, 1839–1851 [DOI] [PubMed] [Google Scholar]
- 46.Dineley-Miller K., Patrick J. (1992) Brain Res. Mol. Brain Res. 16, 339–344 [DOI] [PubMed] [Google Scholar]
- 47.Gahring L. C., Persiyanov K., Rogers S. W. (2004) J. Comp. Neurol. 468, 322–333 [DOI] [PubMed] [Google Scholar]
- 48.Bruschweiler-Li L., Fuentes Medel Y. F., Scofield M. D., Trang E. B., Binke S. A., Gardner P. D. (2010) Neuroscience 166, 864–877 [DOI] [PMC free article] [PubMed] [Google Scholar]