Abstract
The Dnmt3a DNA methyltransferase contains in its N-terminal part a PWWP domain that is involved in chromatin targeting. Here, we have investigated the interaction of the PWWP domain with modified histone tails using peptide arrays and show that it specifically recognizes the histone 3 lysine 36 trimethylation mark. H3K36me3 is known to be a repressive modification correlated with DNA methylation in mammals and heterochromatin in Schizosaccharomyces pombe. These results were confirmed by equilibrium peptide binding studies and pulldown experiments with native histones and purified native nucleosomes. The PWWP-H3K36me3 interaction is important for the subnuclear localization of enhanced yellow fluorescent protein-fused Dnmt3a. Furthermore, the PWWP-H3K36me3 interaction increases the activity of Dnmt3a for methylation of nucleosomal DNA as observed using native nucleosomes isolated from human cells after demethylation of the DNA with 5-aza-2′-deoxycytidine as substrate for methylation with Dnmt3a. These data suggest that the interaction of the PWWP domain with H3K36me3 is involved in targeting of Dnmt3a to chromatin carrying that mark, a model that is in agreement with several studies on the genome-wide distribution of DNA methylation and H3K36me3.
Keywords: Chromatin Histone Modification, DNA Methylation, DNA Methyltransferase, Epigenetics, Histone Methylation
Introduction
In mammals, DNA methylation plays important roles in differentiation, gene regulation, genomic imprinting, X chromosome inactivation, and disease-related processes (1–3). DNA methylation patterns are set during embryogenesis by the Dnmt3a and Dnmt3b DNA methyltransferases and their regulatory factor Dnmt3L (Dnmt3-like) (4–6). However, the mechanisms guiding these enzymes to their target regions are not well understood. Dnmt3a and 3b consist of a C-terminal catalytic domain and an N-terminal part containing a PWWP domain and an ADD2 domain (1, 7, 8). Biochemical studies provide evidence for a direct interaction of Dnmt3a and 3b with native nucleosomes (9), which could be mediated by the ADD domain or the PWWP domain. The ADD domains of Dnmt3L and Dnmt3a have been shown to interact with the histone 3 tail unmethylated at Lys4 (10–12), which can explain the anticorrelation of DNA methylation and the activating H3K4me3 mark as observed in many genome-wide DNA methylation studies (13–16). However, the ADD domain is not directly involved in heterochromatic targeting of Dnmt3a (17, 18).
PWWP domains belong to the Royal domain superfamily, members of which were identified to interact with histone tails in various modification states (19). The PWWP domains of Dnmt3a and 3b are essential for heterochromatic targeting (17, 18). An S333P missense mutation in the Dnmt3a PWWP domain (numbering refers to murine Dnmt3a) led to the loss of chromatin targeting of Dnmt3a (18). This mutation corresponds to the S282P mutation in the PWWP domain of human Dnmt3b, which has been identified in immunodeficiency, centromeric heterochromatin instability, and facial anomalies syndrome patients (20).
Here, we explore the possibility of the interaction of the Dnmt3a PWWP domain with histone peptides using modified histone tail peptide arrays. We discovered its specific interaction with H3K36me3 and show that this interaction is important for the subnuclear localization of Dnmt3a and for its catalytic activity on native chromatin.
EXPERIMENTAL PROCEDURES
Cloning, Site-directed Mutagenesis, Expression, and Purification
The sequence encoding the PWWP domain of murine Dnmt3a (residues 279–420) was subcloned as a GST fusion protein into the pGEX-6P2 vector (GE Healthcare) using BamHI/XhoI sites. The GST-tagged PWWP domain and its variants were expressed and purified as described (21). The full-length murine Dnmt3a2 cloned into pET28a (Novagen) vector was used for the expression and purification of full-length Dnmt3a2 (22), which is the predominant isoform of Dnmt3a in embryonic stem cells (23). The catalytic domain of Dnmt3a (Dnmt3a-CD) was expressed and purified as described (24). To prepare the EYFP-Dnmt3a expression construct used for subnuclear localization studies, Dnmt3a was cloned from a pET expression vector (25) into pcDNA3.1, yielding pcDNA-Dnmt3a. Then EYFP was amplified from pEYFP-C1 (Clontech) and cloned into pcDNA-Dnmt3a in front of the Dnmt3a gene to encode for an EGFP-Dnmt3a fusion protein.
The E290A and D329A mutations in the PWWP domain of Dnmt3a and the D329A mutation in Dnmt3a2 and in EGFP-Dnmt3a were introduced by using a PCR megaprimer mutagenesis method as described previously (26). Successful mutagenesis and cloning was confirmed by restriction marker site analysis and DNA sequencing.
Synthesis of Peptide SPOT Arrays
Peptide arrays were synthesized on cellulose membranes using the SPOT synthesis method (27). Each spot had diameters of 2 mm and contained ∼9 nmol of peptide (Autospot Reference Handbook, Intavis AG). Successful synthesis of each peptide was confirmed by bromphenol blue staining of the membranes. H3 26–44 K36me3 (with N-terminal fluorescein label) and unmodified H3 26–44 peptides were purchased from Intavis AG in purified form.
Binding of Protein Domains to Peptides Arrays
The cellulose membrane was blocked by incubation in TTBS buffer (10 mm Tris/HCl, pH 8.3, 0.05% Tween 20, and 150 mm NaCl) containing 5% nonfat dried milk at 4 °C overnight. The membrane was then washed once with TTBS buffer and incubated with purified GST-tagged PWWP domain of Dnmt3a or mutant domains (10 nm) at room temperature for 1 h in interaction buffer (100 mm KCl, 20 mm Hepes, pH 7.5, 1 mm EDTA, 0.1 mm dithiothreitol, and 10% glycerol). After washing in TTBS buffer, the membrane was incubated with goat anti-GST antibody (GE Healthcare, catalog number 27-4577-01 at 1:5000 dilution) in TTBS buffer for 1 h at room temperature. Then the membrane was washed three times with TTBS and incubated with horseradish peroxidase-conjugated anti-goat antibody (Invitrogen catalog number 81-1620 1:12000) in TTBS for 1 h at room temperature. Finally, the membrane was submerged in ECL developing solution (GE Healthcare), and the image was captured in x-ray film.
Fluorescence Depolarization Peptide Binding Experiments
Fluorescence depolarization experiments were carried out at 25 °C using a Varian Carry Eclipse fluorescence spectrophotometer. For the determination of the binding constant of the PWWP domain to the fluorophore-coupled H3 K36me3 peptide, 0.1 μm of peptide was incubated with increasing concentrations of PWWP domain (0.8–60 μm) in interaction buffer (100 mm NaCl, 50 mm Tris/HCl, pH 8.0, 2 mm EDTA, 0.1% Triton X-100), and fluorescence depolarization was determined (excitation at 494 nm, emission at 524 nm, and excitation and emission slits at 5 nm). The data were fitted to a binary binding equilibrium to determine the equilibrium binding constant. For competition experiments, 30 μm of fluorophore-coupled H3K36me3 was incubated with 30 μm of PWWP domain in interaction buffer, and their interaction was competed by 2- and 5-fold molar excesses of unlabeled unmodified H3 26–44 peptide.
Preparation of Native Histones and Nucleosomes
Native histones were isolated from HEK293 cells by acid extraction as described (28). Native nucleosomes were prepared from HEK293 cells by partial micrococcal nuclease (New England Biolabs) digestion of nuclei as described (29), except that after micrococcal nuclease digestion the nuclei were removed by centrifugation, and the supernatant containing nucleosomes (predominantly mononucleosomes) were used (supplemental Fig. S3). For the preparation of demethylated nucleosomes, HEK293 cells were treated with 2 μm 5-aza-2′-deoxycytidine (Sigma-Aldrich) for 72 h (corresponding to three cell divisions under these conditions), and the cells were allowed to recover for another 48 h in medium without 5-aza-2′-deoxycytidine. From these cells, nucleosomes were prepared as described above.
GST Pulldown Experiments
For GST pulldown experiments, 20 μl of glutathione-Sepharose 4B beads (Amersham Biosciences) were incubated with 20 μg of GST-tagged PWWP domain of Dnmt3a or mutant domain for 1 h at 4 °C in the interaction buffer (50 mm Tris/HCl, pH 8.0, 100 mm NaCl, 2 mm EDTA, and 0.1% Triton X-100). Then the beads were washed once with interaction buffer and blocked with interaction buffer containing 5% bovine serum albumin for 1 h at 4 °C. 35 μg of native histones or 100 μg of native oligonucleosomes were added to the beads in interaction buffer and incubated for 3 h. Finally, the beads were washed three times with interaction buffer, resuspended in 2× SDS-PAGE loading dye, and boiled for 10 min, and the supernatant containing the bound fractions was separated on 16% SDS-PAGE and transferred to nitrocellulose membrane. The membrane was cut in two parts. The lower part (<20 kDa) was probed with anti-H3K36me3 antibody (Abcam ab9050), and the upper part was probed with anti-GST antibody (GE Healthcare) to provide a gel loading control (supplemental Fig. S4, A and B).
For the specificity analysis, the beads were loaded with GST-tagged PWWP domain and incubated with native oligonucleosomes in interaction buffer as described above, washed three times with washing buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 2 mm EDTA, and 0.1% Triton X-100), resuspended in 2× SDS-PAGE loading buffer and boiled for 10 min. The supernatant was split equally and loaded on 16% SDS-PAGE gels. The separated proteins were transferred to nitrocellulose membrane and probed with different modification specific histone antibodies: anti-H3K36me3 (Abcam ab9050), anti-H3K9me3 (Active Motif 39161), anti-H3K4me3 (Active Motif 39159), and anti-H3K27me3 (Active Motif 39535). Equal loading of bound fractions was confirmed by Ponceau S staining of the blot prior to the antibody staining (supplemental Fig. S4C).
Cell Culture, Transfection, and Microscopy
HEK293 and NIH3T3 cells were grown in Dulbecco's modified Eagle's medium with 5% fetal calf serum at 37 °C at 5% CO2. The NIH3T3 cells were seeded in coverslips and transfected with EYFP-Dnmt3a or EYFP-Dnmt3a-D329A mutant constructs using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. Two days after transfection, the cells were fixed with 4% paraformaldehyde and embedded with Mowiol (Carl Roth). Confocal images were taken using a Zeiss LSM 510 Meta instrument (software version 3.0) and 63× oil immersion objective. An argon laser line at 514 nm was used to excite EYFP fluorescence, and a BP530–550 filter was used for image recording. Fluorescence microscopy pictures were taken using an AXIOPLAN2 microscope equipped with AxioCam HRc camera and Achroplan 100× oil immersion objective with filter sets 4′,6′-diamino-2-phenylindole FT395/LP420 and green fluorescent protein BP450–490/FT510 (all from Carl Zeiss).
In Vitro Methylation of Native Chromatin
The methylation reactions were performed with either Dnmt3a2 enzyme, Dnmt3a2-D329A mutant, or Dnmt3a-CD in methylation buffer (20 mm Hepes, pH 7.5, 2 mm EDTA, and 50 mm KCl) containing 0.75 μm radioactively labeled S-adenosyl-l-methionine (PerkinElmer Life Sciences) and 1 μg of demethylated native chromatin in 25 μl of total reaction volume. Additional methylation experiments were conducted in the presence of anti-H3K36me3 antibody (Abcam ab9050), anti-H3K9me3 antibody (Active Motif 39161), anti-H3K4me3 antibody (Active Motif 39159), or anti-H3K27me3 antibody (Active Motif 39535) all in 1:5000 final dilution. The methylation reactions were incubated at 37 °C for 2 h, and the reactions were stopped by the addition of excess of unlabeled S-adenosyl-l-methionine. Then the reaction mixtures were subjected to proteinase K (New England Biolabs) digestion at 42 °C for 2 h, and the nucleic acids in the reaction mixture were purified using Nucleospin PCR purification (Machery Nagel). The amount of radioactivity incorporated into the DNA was measured by liquid scintillation counting and normalized with respect to the total amount of DNA as determined by UV spectroscopy. Methylation of biotinylated oligonucleotide substrates by Dnmt3a2 and its variants was studied by measuring the transfer of titrated methyl groups from labeled S-adenosyl-l-methionine into biotinylated oligonucleotides (Bt-GAG AAG CTG GGA CTT CCG GGA GGA GAG TGC/GCA CTC TCC TCC CGG AAG TCC CAG CTT CTC) using the avidin-biotin methylation assay as described (22, 30).
RESULTS
Peptide Array Binding and Structural Modeling
To investigate whether the Dnmt3a PWWP domain interacts with modified histone peptides, we employed two different modified histone tail peptide arrays containing in total more than 1000 peptides in various combinations of known and hypothetical modification states of the H3, H4, H2A, and H2B tails. We repeatedly observed that the GST-tagged PWWP domain of Dnmt3a interacted with the histone H3 K36 trimethylation mark with high specificity (Fig. 1).
FIGURE 1.
Peptide binding by the Dnmt3a wild type PWWP domain and its variants. A, interaction of the wild type PWWP domain with histone tail arrays containing different combinations of 107 known and hypothetical modifications at the H3, H4, H2A, H2B, and H1b histone tails. The isolated GST domain does not interact with peptide arrays (data not shown). The spot corresponding to the H3 29–46 K36me3 peptide is indicated with an arrow; some other spots that do not interact with the PWWP domain are labeled with numbers and annotated below the image. B, interaction of wild type PWWP domain, D329A, and E290A variants with modified histone tails. The H3K36me3 and H3K36me2 spots on the different arrays used for this study are highlighted with black and gray arrows, respectively. C, binding of the H3K36me3 peptide to PWWP domain studied by fluorescence depolarization (black diamonds). The line represents a fit of the data to a binary binding equilibrium, which revealed a Kd of 64 μm. The gray squares display changes in fluorescence depolarization observed after adding same volumes of buffer. D, weak competition of H3K36me3 peptide binding to the PWWP domain by the addition of unmethylated H3K36 peptide confirms specific binding.
Binding to trimethylated lysine usually involves hydrophobic cages, which often have an acidic residue nearby (19). Because the PWWP domains from Dnmt3a and 3b share high amino acid homology, we used the structure of the Dnmt3b PWWP domain (31) to model the structure of the Dnmt3a PWWP domain and identify such potential binding pockets next to Glu290 and Asp329 (supplemental Fig. S1). Trying to disrupt the histone interaction of PWWP, we mutated Glu290 and Asp329 to alanine. These residues are both exposed on the surface of the Dnmt3a PWWP structural model, suggesting that their exchange will not affect the overall domain structure. We did not mutate the buried hydrophobic residues of the predicted pockets to avoid effects on protein folding and stability. The mutant domain proteins could be expressed and purified with similar yields as the wild type domain (supplemental Fig. S2A), and CD spectroscopy confirmed the wild type-like folding of the D329A mutant domain (supplemental Fig. S2B). Binding experiments on peptide arrays showed that there was no significant change in the peptide interaction of the E290A variant, but the D329A variant had completely lost its ability to bind specifically to the H3K36me3 peptide (Fig. 1B).
Recently, the structure of the Eaf3 chromo barrel domain was solved in complex with an H3K36me2 analog (32). Over 144 backbone atoms comprising the conserved β-barrel and the adjacent α-helix fold, the Eaf3 chromo barrel domain structure could be superimposed with the mouse Dnmt3b PWWP domain structure with a root mean square deviation of 1.37 Å (supplemental Fig. S1). The overlay positioned the methylated Lys36 residue immediately adjacent to Asp329 in the Dnmt3a PWWP domain model. This observation indicates that the biochemical data obtained here are compatible with the structural results obtained with related domains. Furthermore, the Asp329 residue is close to the Ser333 residue (supplemental Fig. S1), mutation of which leads to loss of heterochromatic localization (18).
Equilibrium Peptide Binding Experiments
The interaction of the PWWP domain with H3K36me3 was confirmed by fluorescence depolarization using a fluorescently labeled H3 26–44 K36me3 peptide (Fig. 1C). The binding constant was determined as 64 μm, which is weak but in the range of results observed with other individual chromatin-binding domains, like BHC80 Plant Homeodomain like domain binding to H3K4me0 peptide: 33 μm (33), L3MBTL1 to the H3K9me1 peptide: 26 μm (34), BRD2 bromodomain 1 to the H4K5ac/K12ac peptide: 360 μm (35), and Brd4 bromodomain 1 binding to H3K14ac: 118 μm (36). Peptide binding of the PWWP-D329A variant was at least five times weaker (data not shown). To assess the binding specificity, binding was competed with unmodified H3 26–44 peptide (Fig. 1D). We observed that at a 5-fold excess of competitor only 20% of binding was lost, corresponding to a 25-fold preference for binding to the methylated peptide.
Nucleosome and Histone Pulldown Experiments
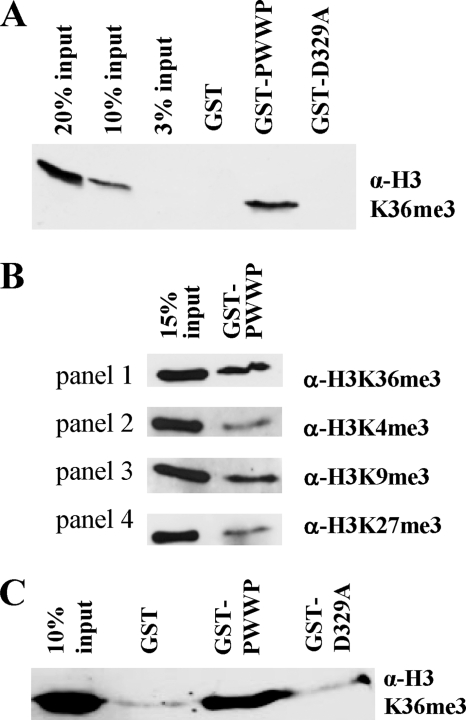
The PWWP-H3K36m3 interaction was also confirmed by pulldown experiments. The GST-PWWP domain of Dnmt3a was able to pull down native nucleosomes isolated from human cells, which contained the H3K36me3 mark (Fig. 2A and supplemental Figs. S3A and 4A). In contrast, the PWWP-D329A mutant completely failed to interact with nucleosomes (Fig. 2A and supplemental Fig. S4A). In addition to H3K36me3, the nucleosome fraction bound by the PWWP domain was also enriched with H3K9me3, but the amounts of H3K4me3 and H3K27me3 were reduced in comparison with H3K36me3 (Fig. 2B). The relative reduction in the amounts of H3K4me3 and K27me3 indicates that the pulldown is specific. The presence of the H3K9me3 mark in the pull down was expected on the basis of the overlapping distribution profiles of the H3K36me3 and H3K9me3 marks across transcribed chromatin (37). Using native histones isolated from human cells (supplemental Fig. S3B), we observed that the GST-PWWP domain of Dnmt3a, but not its D329A mutant, interacted with histone H3K36me3 mark (Fig. 2C and supplemental Fig. S4B), indicating that the interaction does not require DNA.
FIGURE 2.
Native chromatin and histone binding of the Dnmt3a PWWP domain and its variants. A, GST pulldown assay interaction analysis of the PWWP domain and the D329A variant with mononucleosomes purified from human cells. The bound proteins were separated and immunoblotted with anti-H3K36me3 antibody. B, for specificity analysis, the bound fractions were separated and immunoblotted with anti-H3K36me3 antibody, anti-H3K4me3 antibody, anti-H3K9me3 antibody, and H3K27me3 antibody. C, GST pulldown interaction analysis of PWWP domain and D329A mutant domain with native histone proteins, purified from human cells. The bound fractions were separated and immunoblotted with anti-H3K36me3 antibody. In A–C, all of the bands correspond to histone H3.
Subnuclear Localization of Dnmt3a
We investigated the subnuclear distribution of the EYFP-fused Dnmt3a and its D329A mutant in NIH3T3 cells after transient expression of the proteins. Both proteins were located in the nucleus, which was expected because the nuclear localization signals of Dnmt3a are outside of the PWWP domain (17). Within the nucleus, Dnmt3a was found in large spots corresponding to 4′,6′-diamino-2-phenylindole-stained heterochromatin as reported earlier (17, 18) (Fig. 3 and supplemental Fig. S5). In contrast, the mutant protein was found more homogenously distributed in the nucleus, indicating a partial loss of heterochromatic localization. This result suggests that the interaction of PWWP with K36me3 is important for the localization of Dnmt3a to heterochromatin.
FIGURE 3.
Subnuclear distribution of wild type EYFP-Dnmt3a and EYFP-Dnmt3a-D329A mutant in NIH3T3 cells. The subnuclear localization patterns were analyzed by laser scanning microscopy and assigned into one of the following categories: localization mainly homogenous, homogenous localization with some large spots, and localization almost exclusively in large spots. The figure shows examples of each type of localization patterns and gives the distribution of occurrence of the different patterns in both experiments in 100 arbitrarily chosen cells.
DNA Methylation Experiments
The influence of the PWWP-H3K36me3 interaction on the methylation of nucleosomal DNA by Dnmt3a was investigated using native nucleosomes from human cells prepared after treatment of the cells with 5-aza-2′-deoxycytidine. This treatment reduces DNA methylation at unique sequences and repeats by more than 50% (15, 38). The native nucleosomes carry their endogenous histone modification pattern and partially demethylated DNA. This experimental design allowed us to study the methylation activity of Dnmt3a using a substrate that mimics the in vivo situation. Demethylated nucleosomes were methylated with Dnmt3a2, a shorter isoform of Dnmt3a lacking the N-terminal variable region (23), the Dnmt3a2 D329A mutant (Dnmt3a2-D329A), and the catalytic domain of Dnmt3a (Dnmt3a-CD) using radioactively labeled S-adenosyl-l-methionine as cofactor. The amounts of Dnmt3a2, Dnmt3a2-D329A, and Dnmt3a-CD enzymes used in the nucleosome methylation experiments were normalized to have similar activity on oligonucleotide substrates (supplemental Fig. S6). Using the native chromatin as substrate, we observed that Dnmt3a2 is ∼2.5-fold more active than Dnmt3a-CD (Fig. 4 and supplemental Fig. S6). The Dnmt3a2 enzyme containing the mutated D329A-PWWP domain, which fails to interact with the H3K36me3 mark, only showed a basal level of activity similar to Dnmt3a-CD (Fig. 4).
FIGURE 4.
A, methylation of demethylated native nucleosomes by Dnmt3a enzymes. Relative methylation activity of Dnmt3a2, Dnmt3a2-D329A, and Dnmt3a-CD on demethylated native nucleosomes in the absence (dark gray bars) and the presence (light gray bars) of anti-H3K36me3 antibody. The error bars indicate standard deviations derived from at least three independent experiments. B, schematic picture showing how the H3 tail interactions of the Dnmt3a ADD domain (dark grey rectangle) binding to the end of the H3 tail (12) and the PWWP domain (light grey hexagon) binding to Lys36 (black triangle) may anchor the catalytic domain of the enzyme (dark grey oval) to methylate DNA preferentially in linker region (12).
To confirm that the increase in activity of the wild type Dnmt3a2 is due to the PWWP-H3K36me3 interaction, we performed the chromatin methylation experiments in the presence of an anti-H3K36me3 antibody. Although the addition of the anti-H3K36me3 antibody had no effect on the activity of Dnmt3a2-D329A or Dnmt3a-CD, it decreased the activity of Dnmt3a2 to a basal level, similar to the level of activity observed with the Dnmt3a2-D329A or Dnmt3a-CD (Fig. 4). Additional unrelated antibodies used as controls had no effect or a very weak effect on the catalytic activity of Dnmt3a2 (supplemental Fig. S6). Taken together, these results demonstrate that the PWWP domain-H3K36me3 interaction increases the activity of Dnmt3a2 for methylation of native nucleosomal DNA.
DISCUSSION
We show here that the Dnmt3a PWWP domain specifically interacts with H3 tails containing the K36me3 mark. Peptide binding is weak but in the range of binding constants seen with other individual domains interacting with histone peptides. In the cell binding may be further supported by an interaction of the PWWP domain with DNA, because the Lys36 position of the H3 tail is very close to the DNA, such that the PWWP domain could interact with both epitopes simultaneously. Interestingly, the DNA binding and peptide binding properties of the PWWP domains from Dnmt3a and 3b seem to be inverse; we detected peptide binding of the Dnmt3a-PWWP domain but not for Dnmt3b (data not shown), whereas DNA binding of the Dnmt3b-PWWP domain has been reported to be stronger than that of Dnmt3a (17).
We show that the H3K36me3-PWWP interaction mediates chromatin binding, because the heterochromatic localization of Dnmt3a was disturbed in the D329A mutant, which no longer binds to H3K36me3. Targeting of Dnmt3a by an H3K36me3-PWWP interaction is in agreement with previous results showing that the Dnmt3a PWWP domain is necessary for heterochromatic localization of Dnmt3a (17, 18). In contrast, the ADD domain of Dnmt3a, which binds to H3 tails unmethylated at Lys4 (11, 12), does not have a role in heterochromatic localization (17, 18).
We show that the PWWP-H3K36me3 interaction stimulates the methylation activity of Dnmt3a on chromatin-bound DNA isolated from human cells, because the catalytic activity of the D329A mutant dropped to levels similar to the isolated catalytic domain. This result suggests an important role for the PWWP domain in guiding DNA methylation to chromatin carrying the H3K36me3 mark, which is in agreement with the observation that the PWWP domain is required for the activity of Dnmt3a in the cell (17, 39). This model is further supported by the observation that the genome-wide distribution of DNA methylation is very similar to that of K36me3 methylation. Many studies have found that H3K36me3 accumulates in euchromatin in the body of active genes, and its distribution is anticorrelated with H3K4me3 (37, 40–43). Similarly, active genes showed high DNA methylation in the gene bodies, whereas inactive genes did not (44, 45). The strong correlation of DNA methylation, absence of H3K4me3, and presence of H3K36me3 were also observed in two recent genome-wide studies (14, 16). Additionally, a correlation of H3K36me3 and DNA methylation was observed at exon-intron boundaries where exons were shown to have increased levels of both H3K36me3 (46) and DNA methylation (16).
Like DNA methylation, H3K36me3 is a silencing mark that recruits histone deacetylation and represses intragenic transcriptional initiation (47). Consequently, the Smyd2 histone methlyltransferase (which generates K36me2) has a repressive effect on reporter gene expression (48). In Schizosaccharomyces pombe the Set2 histone methyltransferase methylates histone H3 lysine 36 at repeat loci during the S phase (49). Consequently, heterochromatic H3K36me3 transiently peaks in the S phase and has an important role in the re-establishment of heterochromatin after DNA replication that acts in a pathway parallel to Clr4. It has been proposed that in mammals Dnmt3a contributes to the maintenance of the DNA methylation at highly methylated regions (like repeats) (50). H3K36 methylation is an evolutionarily conserved mark, suggesting that a transient methylation of H3K36 might occur in mammalian genomes as well and that transient H3K36 methylation might be one of the modifications that recruit Dnmt3a to heterochromatin.
Dnmt3a has been shown to bind tightly to chromatin (9). When combined with the previous findings, our data indicate that there are several mechanisms that contribute to the interaction of Dnmt3a with chromatin; Dnmt3a is engaged in two interactions with the H3 tail, mediated by its ADD domain, which binds to the end of the H3 tail unmodified at Lys4 (11, 12), and by its PWWP domain, which interacts with H3K36me3 located at the more basal region of the H3 tail (Fig. 4B). The anticorrelation of the H3K4me3 and H3K36me3 marks suggests that in the cell most H3 tails will either bind to both ADD and PWWP or neither of them, such that the targeting effects of both domains are synergistic. In addition, the catalytic domain of Dnmt3a binds to DNA and polymerizes on the DNA (22, 51), and it interacts with many other proteins including Dnmt3L and Dnmt3b (52), which provide additional contact points to chromatin (10, 53), suggesting that a complex network of interactions is involved in targeting DNA methylation.
Supplementary Material
Acknowledgment
Many thanks are due to Dr. X. Cheng for insightful comments and valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM068680 and DK082678. This work was also supported by Deutsche Forschungsgemeinschaft Grant JE 252/6-1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- ADD
- ATRX-DNMT3-DNMT3L
- GST
- glutathione S-transferase
- EYFP
- enhanced yellow fluorescent protein.
REFERENCES
- 1.Goll M. G., Bestor T. H. (2005) Annu. Rev. Biochem. 74, 481–514 [DOI] [PubMed] [Google Scholar]
- 2.Klose R. J., Bird A. P. (2006) Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 3.Reik W. (2007) Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 4.Okano M., Bell D. W., Haber D. A., Li E. (1999) Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 5.Bourc'his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H. (2001) Science 294, 2536–2539 [DOI] [PubMed] [Google Scholar]
- 6.Hata K., Okano M., Lei H., Li E. (2002) Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
- 7.Hermann A., Gowher H., Jeltsch A. (2004) Cell Mol. Life Sci. 61, 2571–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng X., Blumenthal R. M. (2008) Structure 16, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeong S., Liang G., Sharma S., Lin J. C., Choi S. H., Han H., Yoo C. B., Egger G., Yang A. S., Jones P. A. (2009) Mol. Cell. Biol. 29, 5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ooi S. K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S. P., Allis C. D., Cheng X., Bestor T. H. (2007) Nature 448, 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otani J., Nankumo T., Arita K., Inamoto S., Ariyoshi M., Shirakawa M. (2009) EMBO Rep. 10, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., Jeltsch A. (2010) Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber M., Hellmann I., Stadler M. B., Ramos L., Pääbo S., Rebhan M., Schübeler D. (2007) Nat. Genet. 39, 457–466 [DOI] [PubMed] [Google Scholar]
- 14.Meissner A., Mikkelsen T. S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B. E., Nusbaum C., Jaffe D. B., Gnirke A., Jaenisch R., Lander E. S. (2008) Nature 454, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Rohde C., Tierling S., Jurkowski T. P., Bock C., Santacruz D., Ragozin S., Reinhardt R., Groth M., Walter J., Jeltsch A. (2009) PLoS Genet 5, e1000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodges E., Smith A. D., Kendall J., Xuan Z., Ravi K., Rooks M., Zhang M. Q., Ye K., Bhattacharjee A., Brizuela L., McCombie W. R., Wigler M., Hannon G. J., Hicks J. B. (2009) Genome Res. 19, 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T., Tsujimoto N., Li E. (2004) Mol. Cell. Biol. 24, 9048–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ge Y. Z., Pu M. T., Gowher H., Wu H. P., Ding J. P., Jeltsch A., Xu G. L. (2004) J. Biol. Chem. 279, 25447–25454 [DOI] [PubMed] [Google Scholar]
- 19.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirohzu H., Kubota T., Kumazawa A., Sado T., Chijiwa T., Inagaki K., Suetake I., Tajima S., Wakui K., Miki Y., Hayashi M., Fukushima Y., Sasaki H. (2002) Am. J. Med. Genet. 112, 31–37 [DOI] [PubMed] [Google Scholar]
- 21.Rathert P., Dhayalan A., Murakami M., Zhang X., Tamas R., Jurkowska R., Komatsu Y., Shinkai Y., Cheng X., Jeltsch A. (2008) Nat. Chem. Biol. 4, 344–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurkowska R. Z., Anspach N., Urbanke C., Jia D., Reinhardt R., Nellen W., Cheng X., Jeltsch A. (2008) Nucleic Acids Res. 36, 6656–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T., Ueda Y., Xie S., Li E. (2002) J. Biol. Chem. 277, 38746–38754 [DOI] [PubMed] [Google Scholar]
- 24.Gowher H., Jeltsch A. (2002) J. Biol. Chem. 277, 20409–20414 [DOI] [PubMed] [Google Scholar]
- 25.Gowher H., Jeltsch A. (2001) J. Mol. Biol. 309, 1201–1208 [DOI] [PubMed] [Google Scholar]
- 26.Jeltsch A., Lanio T. (2002) Methods Mol. Biol. 182, 85–94 [DOI] [PubMed] [Google Scholar]
- 27.Frank R. (2002) J. Immunol. Methods 267, 13–26 [DOI] [PubMed] [Google Scholar]
- 28.Shechter D., Dormann H. L., Allis C. D., Hake S. B. (2007) Nat. Protoc. 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 29.Brand M., Rampalli S., Chaturvedi C. P., Dilworth F. J. (2008) Nat. Protoc. 3, 398–409 [DOI] [PubMed] [Google Scholar]
- 30.Roth M., Jeltsch A. (2000) Biol. Chem. 381, 269–272 [DOI] [PubMed] [Google Scholar]
- 31.Qiu C., Sawada K., Zhang X., Cheng X. (2002) Nat. Struct. Biol. 9, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu C., Cui G., Botuyan M. V., Mer G. (2008) Structure 16, 1740–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lan F., Collins R. E., De Cegli R., Alpatov R., Horton J. R., Shi X., Gozani O., Cheng X., Shi Y. (2007) Nature 448, 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalakonda N., Fischle W., Boccuni P., Gurvich N., Hoya-Arias R., Zhao X., Miyata Y., Macgrogan D., Zhang J., Sims J. K., Rice J. C., Nimer S. D. (2008) Oncogene 27, 4293–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umehara T., Nakamura Y., Jang M. K., Nakano K., Tanaka A., Ozato K., Padmanabhan B., Yokoyama S. (2010) J. Biol. Chem. 285, 7610–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vollmuth F., Blankenfeldt W., Geyer M. (2009) J. Biol. Chem. 284, 36547–36556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vakoc C. R., Sachdeva M. M., Wang H., Blobel G. A. (2006) Mol. Cell. Biol. 26, 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohde C., Zhang Y., Stamerjohanns H., Hecher K., Reinhardt R., Jeltsch A. (2009) BioTechniques 47, 781–783 [DOI] [PubMed] [Google Scholar]
- 39.Shikauchi Y., Saiura A., Kubo T., Niwa Y., Yamamoto J., Murase Y., Yoshikawa H. (2009) Mol. Cell. Biol. 29, 1944–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 41.Larschan E., Alekseyenko A. A., Gortchakov A. A., Peng S., Li B., Yang P., Workman J. L., Park P. J., Kuroda M. I. (2007) Mol. Cell 28, 121–133 [DOI] [PubMed] [Google Scholar]
- 42.Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007) Cell 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edmunds J. W., Mahadevan L. C., Clayton A. L. (2008) EMBO J. 27, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellman A., Chess A. (2007) Science 315, 1141–1143 [DOI] [PubMed] [Google Scholar]
- 45.Ball M. P., Li J. B., Gao Y., Lee J. H., LeProust E. M., Park I. H., Xie B., Daley G. Q., Church G. M. (2009) Nat. Biotechnol. 27, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X. S., Ahringer J. (2009) Nat. Genet. 41, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J. S., Shilatifard A. (2007) Mutat Res. 618, 130–134 [DOI] [PubMed] [Google Scholar]
- 48.Brown M. A., Sims R. J., 3rd, Gottlieb P. D., Tucker P. W. (2006) Mol. Cancer 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen E. S., Zhang K., Nicolas E., Cam H. P., Zofall M., Grewal S. I. (2008) Nature 451, 734–737 [DOI] [PubMed] [Google Scholar]
- 50.Jones P. A., Liang G. (2009) Nat. Rev. Genet. 10, 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X. (2007) Nature 449, 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J. Y., Pu M. T., Hirasawa R., Li B. Z., Huang Y. N., Zeng R., Jing N. H., Chen T., Li E., Sasaki H., Xu G. L. (2007) Mol. Cell. Biol. 27, 8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gopalakrishnan S., Sullivan B. A., Trazzi S., Della Valle G., Robertson K. D. (2009) Hum. Mol. Genet. 18, 3178–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.