Abstract
In the Dlk1-Dio3 imprinted domain, an intergenic differentially methylated region (IG-DMR) regulates the parental allele-specific expression of imprinted genes. The maternally inherited deletion of IG-DMR (IG-DMR(−/+)) results in perinatal lethality because of the overexpression of paternally expressed genes and repression of maternally expressed noncoding RNAs (ncRNAs), including Gtl2. To better understand the possible contribution of paternally expressed genes to the lethality, we attempted to rescue the lethality of IG-DMR(−/+) mutants by restoring the paternally expressed genes. Because the paternally inherited Gtl2 deletion (Gtl2(+/−)) induced a decrease in the expression of paternally expressed genes, we crossed female IG-DMR heterozygous mice and male Gtl2 heterozygous mutant mice. The resultant IG-DMR(−/+)/Gtl2(+/−) double mutant mice had normal expression levels of paternally expressed genes, and none of them showed perinatal lethality; however, most mice showed postnatal lethality with decreased expression of the maternally expressed ncRNAs. Thus, we inferred that paternally expressed genes are necessary for perinatal survivability and that maternally expressed ncRNAs are involved in postnatal lethality.
Keywords: Diseases/Genetic, Gene/Knockout, Genetics/Mouse, RNA/MicroRNA, RNA/Small Nuclear RNA, Transcription/Development, Development, Imprinted Gene
Introduction
Genomic imprinting is an epigenetic mechanism in mammals that results in the functional nonequivalence of parental genomes (1, 2). Imprinted genes are regulated by methylation at differentially methylated regions (DMRs),2 which are dependent on the parent of origin and mainly show monoallelic expression. The loss of imprinting leads to the biallelic expression or silencing of genes, which can result in various developmental defects and diseases (3, 4). Most imprinted genes are regulated by maternally derived methylation, whereas only three domains are reported to be regulated by paternally methylated imprinted domains: Dlk1-Dio3, Igf2-H19, and Rasgrf1 domains (5–7). Studies on mouse embryos containing only the maternal genome have reported that the Dlk1-Dio3 and Igf2-H19 domains are essential for fetal survivability (8, 9).
The Dlk1-Dio3 domain on the distal arm of mouse chromosome 12 contains the paternally expressed genes Dlk1, Rtl1, and Dio3 and the maternally expressed noncoding RNAs (ncRNAs) Gtl2, anti-Rtl1, Rian, and Mirg (see Fig. 1) (10–15). These imprinted genes have been shown to be essential for perinatal survivability in a previous study reporting that embryos with MatD12 and PatD12 (maternal and paternal uniparental disomy for chromosome 12) died late in gestation with biallelic or no expression of these genes (16).
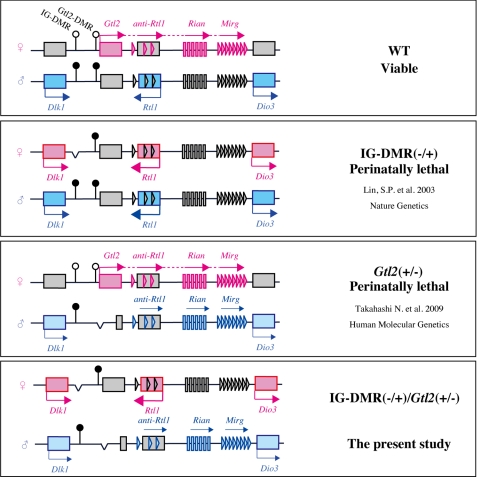
FIGURE 1.
The predicted expression pattern of imprinted genes in the Dlk1-Dio3 domain. The position of the imprinted genes is indicated by squares, vertical bars (small nucleolar RNAs), or triangles (microRNAs). The elements are shown in gray when inactive, in pink when active on the maternal allele, and in blue when active on paternal allele. IG-DMR and Gtl2-DMR are indicated by circles (filled circles, hypermethylated; open circles, hypomethylated).
Imprinting on the Dlk1-Dio3 domain is regulated primarily by an intergenic DMR (IG-DMR) that is methylated during spermatogenesis (5). In mice, when the IG-DMR deletion is inherited from the paternal allele, the mutants have a normal phenotype and gene expression. In contrast, the maternally inherited IG-DMR deletion (IG-DMR(−/+)) results in perinatal lethality as well as biallelic expression of the paternally expressed genes and repression of the maternally expressed genes. The overexpression of Dlk1 and Rtl1 induces partial perinatal lethality in mice; therefore, biallelic expression is a possible cause of the lethality (17, 18). However, this possibility has not been confirmed completely because the expression levels of all of the genes in this domain are significantly different between the IG-DMR(−/+) mutant and wild-type (WT) mice. To clarify the relation between the lethality of the IG-DMR(−/+) mutants and overexpression of Dlk1 and Rtl1, we attempted a rescue experiment by mating female IG-DMR heterozygous mutants with male Gtl2 heterozygous mutants. The paternally inherited Gtl2/Gtl2 DMR deletion (Gtl2(+/−)) leads to a decrease in the expression levels of Dlk1 and Rtl1; consequently, the double mutant mice (IG-DMR(−/+)/ Gtl2(+/−)) would have normal expression levels of Dlk1 and Rtl1 (Fig. 1) (19). The double mutants showed no lethality at the perinatal stage; however, most of them died at the postnatal stage. These results indicate that the restoration of Dlk1 and Rtl1 was necessary but insufficient for normal development in the IG-DMR(−/+) mutants.
EXPERIMENTAL PROCEDURES
Mice
The IG-DMR deletion and Gtl2 knock-out (KO) mutant mice were produced as described previously (5, 19). These mutant lines only could be maintained when the deletion was inherited from the father, as deletion of maternally inherited IG-DMR results in perinatal lethality and maternally inherited Gtl2 deletion results in postnatal lethality. The IG-DMR deletion and Gtl2 KO mutant mice were maintained in a C57BL/6 and B6D2F1 background, respectively. The IG-DMR(−/+)/Gtl2(+/−) double mutant mice were obtained by mating female IG-DMR(+/−) with male Gtl2(+/−) mutant mice. Genotyping of the mutants was conducted by PCR amplification of DNA as described previously. These mutant mice were marked at birth and weighed every 3 days from birth to 7 weeks.
Skeletal Analysis
E18.5 embryos were eviscerated and fixed in 99.5% ethanol for 1 week. For cartilage staining, samples were incubated with 0.015% Alcian blue (Sigma) in 80% ethanol and 20% glacial acetic acid for 1 day. Samples were hydrolyzed overnight in 2% KOH. For bone staining, samples were stained with 0.001% Alizarin red (Sigma) in 1% KOH for 3 h.
Gene Expression Analysis
Total RNA was extracted from the brain, tongue, lungs, and liver of E18.5 embryos using a mirVana miRNA isolation kit (Ambion). The cDNAs were synthesized using the SuperScript III reverse transcriptase (Invitrogen) in a reaction solution (20 μl) containing total RNA (1 μg). Finally, we performed quantitative analysis using real-time PCR (7500 Real-time PCR system; Applied Biosystems) with SYBR Green PCR Master Mix (Applied Biosystems). The primers used for the analysis have been described previously (19).
To conduct the Northern blot hybridization analysis for Rtl1, we isolated total and poly(A)+ RNA from the tongue and placenta using TRIzol (Invitrogen) and the PolyATract(R) mRNA isolation system (Promega). We used pooled samples (n = 3 for each genotype), and the analysis was conducted three times. Signal intensities were calculated using ImageJ software (http://rsb.info.nih.gov/ij/index.html). The probes and methods used for analysis have been described previously (8).
RESULTS
The Mode of Inheritance and Phenotype
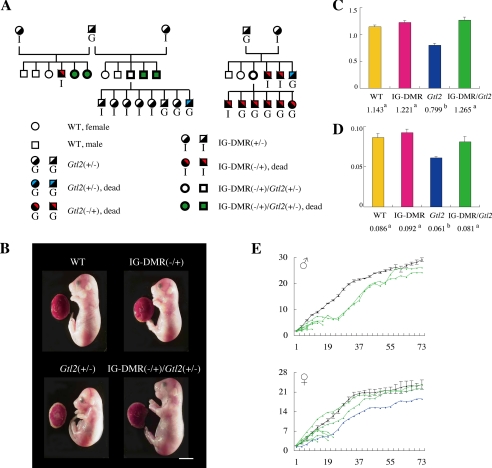
By crossing female IG-DMR heterozygous with male Gtl2 heterozygous mutants, we generated three genotypes of mice: IG-DMR(−/+), Gtl2(+/−), and IG-DMR(−/+)/Gtl2(+/−) (Fig. 2A). The IG-DMR(−/+) mutants died at the perinatal stage showing morphological phenotypes, including a short body length and broad neck, whereas the Gtl2(+/−) mutants showed perinatal lethality with growth retardation of the fetus and placenta (Fig. 2B; Table 1); these findings are consistent with those of previous studies (19, 20). In contrast, the IG-DMR(−/+)/Gtl2(+/−) mutant mice were born in the expected Mendelian ratio and had normal physical characteristics (Fig. 2, B–E; Table 1). Although the double mutants survived at the perinatal stage, 70% of them died within 3 weeks of birth. The remaining double mutants showed weight loss at the weaning stage but grew into normal adults with reproductive ability. This suggests that although perinatal lethality disappears, postnatal abnormalities appear in the double mutants.
FIGURE 2.
Growth phenotype of the mutant mice. A, pedigree of the mutants. B, picture of the fetus and placenta at E18.5. Scale bar, 5 mm. C and D, fetus body weight (C) and placenta weight (D) are represented by the bar chart. Wild-type, n = 9; IG-DMR(−/+), n = 6; Gtl2(+/−), n = 5; and IG-DMR(−/+)/Gtl2(+/−), n = 5. The values are expressed as the means ± S.E. For comparison of the date, statistical analyses were carried out with one-way analysis of variance. The different superscripts denote the statistically differences (p < 0.05) between the genotype. E, the growth curve after birth. Wild-type, n = 11, black; Gtl2(+/−), n = 1, blue; IG-DMR(−/+)/Gtl2(+/−), n = 14, green.
TABLE 1.
The postnatal lethality depending on genotype
| Stage | No. of alive pups/total (%) |
|||
|---|---|---|---|---|
| WT | IG-DMR(−/+) | Gtl2(+/−) | IG-DMR(−/+)/Gtl2(+/−) | |
| ∼1 day | 14/15 (93) | 0/6 (0) | 1/11 (9) | 13/15 (87) |
| ∼1 week | 14/14 (100) | 1/1 (100) | 9/13 (69) | |
| ∼2 weeks | 14/14 (100) | 1/1 (100) | 5/9 (56) | |
| ∼3 weeks | 14/14 (100) | 1/1 (100) | 4/5 (80) | |
| Adult | 14/14 (100) | 1/1 (100) | 4/4 (100) | |
| Total | 14/15 (93) | 0/6 (0) | 1/11 (9) | 4/15 (27) |
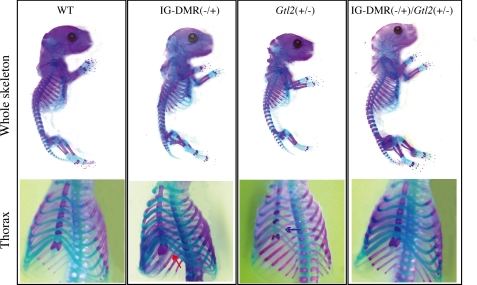
Recovery of the Skeletal Defects in the IG-DMR(−/+)/Gtl2(+/−) Double Mutants
Over- or underexpression of Dlk1 leads to skeletal defects. The IG-DMR(−/+) mutants showed the abnormal attachment of the eighth rib to the sternum (two of four embryos), unlike the WT embryos in whom only seven ribs were attached to the sternum. The Gtl2(+/−) mutants lacked the fifth ossification center (four of five embryos) (Fig. 3). In contrast, these skeletal defects were absent in all the IG-DMR(−/+)/Gtl2(+/−) double mutants (four embryos) (Fig. 3). This indicates that the defect induced by abnormal Dlk1 expression was resolved in the double mutants.
FIGURE 3.
Comparison of skeletal defects among the mutants. The red arrow indicates that the eight ribs were attached the sternum in the IG-DMR(−/+) mutants. The blue arrow points the absence of the fifth ossification center in the Gtl2(+/−) mutants. The skeletal defect was absent in the IG-DMR(−/+)/Gtl2(+/−) double mutants.
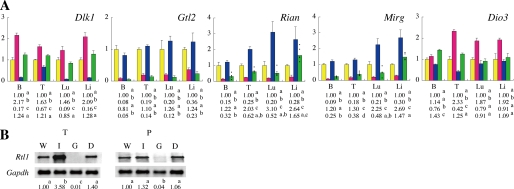
Gene Expression Analysis for the Dlk1-Dio3 Domain in the IG-DMR(−/+)/Gtl2(+/−) Double Mutants
To obtain a better understanding of the IG-DMR(−/+)/Gtl2(+/−) double mutant phenotype, we performed gene expression analysis of the imprinted genes in this domain (Fig. 4, A and B). First, we assayed the expression of Dlk1 and Rtl1 to determine the reason for perinatal survivability in the double mutants. The assay results showed that the IG-DMR(−/+) mutants overexpressed Dlk1 and Rtl1, whereas the Gtl2(+/−) mutants had decreased expression levels of Dlk1 and Rtl1, as expected. In the IG-DMR(−/+)/Gtl2(+/−) double mutants, the expression levels of Dlk1 and Rtl1 were similar to those of the controls in all the tissues examined. This suggests that the restoration of Dlk1 and Rtl1 expression rescued perinatal lethality in the double mutants.
FIGURE 4.
Gene expression in the Dlk1-Dio3 domain. A, graphical representations of the expression levels of the five imprinted genes Dlk1, Gtl2, Rian, Mirg, and Dio3 at E18.5. The values are expressed as the means ± S.E. Wild-type, n = 4, yellow; IG-DMR(−/+), n = 4, red; Gtl2(+/−), n = 3, blue; and IG-DMR(−/+)/Gtl2(+/−), n = 5, green. It is normalized to 1 in each tissue, which represents the average expression level of the WT. The expression value of Rian and Mirg in the IG-DMR(−/+)/Gtl2(+/−) double mutants was individually indicated by dots because the expression was varied among samples. B, Northern blot analysis of Rtl1 expression at E18.5. B, brain; T, tongue; Lu, lungs; Li, liver; and P, placenta. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was used as control. For comparison of the date, statistical analyses were carried out with one-way analysis of variance. The superscripts denote the differences (p < 0.05) between the genotype means. W, wild-type; I, IG-DMR(−/+); G, Gtl2(+/−); D, IG-DMR(−/+)/Gtl2(+/−).
To gain further insight into the mechanism underlying the postnatal lethality of the IG-DMR(−/+)/Gtl2(+/−) double mutant mice, we performed gene expression analysis of Rian and Mirg. The expression of Rian and Mirg was repressed in the IG-DMR(−/+) mutants and enhanced in Gtl2(+/−) mutants, as expected. In the IG-DMR(−/+)/Gtl2(+/−) double mutants, the average expression level of Rian and Mirg in the brain was reduced to nearly one-fourth of that in the controls, and the average expression level of these genes in the tongue and lungs were half that in the controls. This demonstrates that postnatal lethality might be caused by the decrease in the expression levels of Rian and Mirg. The expression levels of these ncRNAs varied among the double mutants. In four of five samples, the expression levels of Rian and Mirg in the lungs decreased to <54% of that in the control levels; however, one sample showed an expression level identical to that of the controls. Additionally, in one of the five samples, the Rian expression level in the brain was two times the average of the other four samples. This might explain why 30% of the double mutants survived. These data suggest that the expression levels of Rian and Mirg determine the viability of the double mutant at the postnatal stage.
DISCUSSION
The correct dosage of Dlk1 and Rtl1 is necessary for normal development, which is clearly evidenced in overexpression and deletion studies on genetically modified mice. One-third of the mutants expressing twice the normal dose of Dlk1 exhibit skeletal defects and die within the first 3 days of life (17). Dlk1 KO mice show dwarfism, and half of them die at the perinatal stage (21). In contrast, mutants overexpressing Rtl1 display partial lethality with placentomegaly, and Rtl1 KO mice show perinatal lethality with growth retardation (18). These reports suggested that over- and underexpression of Dlk1 and Rtl1 induced perinatal lethality.
The IG-DMR regulates the expression of all the imprinted genes in the Dlk1-Dio3 domain, and all of the IG-DMR(−/+) mutants die perinatally and show biallelic expression of Dlk1 and Rtl1 (5). To investigate whether Dlk1 and Rtl1 overexpression caused perinatal lethality, we tried to restore the expression levels of Dlk1 and Rtl1 in the IG-DMR(−/+) mutants by mating IG-DMR(−/+) females with male Gtl2 heterozygous mutants. The paternally inherited Gtl2 deletion led to a lowering of the expression levels of Dlk1 and Rtl1; consequently, the IG-DMR(−/+)/Gtl2(+/−) double mutants showed normal expression levels of Dlk1 and Rtl1. The double mutants were normal at birth and demonstrated no skeletal defects, indicating that the perinatal abnormalities were rescued by the restoration of the Dlk1 and Rtl1 expression dosage. However, all of the double mutants showed growth retardation at the weaning stage, and 70% of them died within 3 weeks of birth. These findings indicate that the restoration of Dlk1 and Rtl1 expression was not sufficient to rescue the lethality of the IG-DMR(−/+) mutants.
Gene expression analysis showed that the levels of maternally expressed ncRNA in the IG-DMR(−/+)/Gtl2(+/−) double mutants did not return to those in the WT. Our previous study showed that the maternal Gtl2 deletion led to postnatal lethality due to hypoplastic pulmonary alveoli and hepatocellular necrosis, probably due to the decreased expression of small nucleolar RNAs and miRNAs derived from Rian and anti-Rtl1/Mirg (19). Histological abnormalities of the liver and lungs also were observed in the IG-DMR(−/+)/Gtl2(+/−) double mutants (two of three; data not shown). Thus, it is highly likely that the postnatal abnormalities observed in the double mutants were caused by the decrease in the expression levels of these ncRNAs. We measured the gene expression levels in five double mutant samples and observed that the expression dosage of Rian and Mirg varied largely among the samples. One sample showed normal expression levels of Rian and Mirg in the lungs, while another sample had expression levels two times the average of the other double-mutant samples in the brain. Four out of the 15 double mutant mice survived; therefore, it can be considered that the sample-specific recovery of gene expression might induce partial survivability of the double mutants. Our finding would be supported by recently published reports indicating that ncRNAs in the Dlk1-Dio3 domain have a great influence on the pluripotency level of induced pluripotent stem cells (22, 23).
In conclusion, the present study suggests that the decrease in the ncRNA expression downstream of Gtl2 might be involved in postnatal development and lethality. Further studies are required to identify the ncRNAs involved in the lethality induced by altered imprinting pattern in the Dlk1-Dio3 domain and to understand rhe mechanism by which these ncRNAs exert their effects.
Acknowledgment
We thank Anne C. Ferguson-Smith for the gift of IG-DMR KO mice.
This work was supported by Grants-in-aid for Scientific Research on Priority Area and for Scientific Research A from the Ministry of Education, Science, Culture and Sports of Japan (to T. K.).
- DMR
- differentially methylated region
- IG-DMR
- intergenic DMR
- ncRNA
- noncoding RNA
- WT
- wild-type
- KO
- knock-out
- E
- embryonic day.
REFERENCES
- 1.Barton S. C., Surani M. A., Norris M. L. (1984) Nature 311, 374–376 [DOI] [PubMed] [Google Scholar]
- 2.McGrath J., Solter D. (1984) Cell 37, 179–183 [DOI] [PubMed] [Google Scholar]
- 3.Allegrucci C., Thurston A., Lucas E., Young L. (2005) Reproduction 129, 137–149 [DOI] [PubMed] [Google Scholar]
- 4.Reik W. (2007) Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 5.Lin S. P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J., Ferguson-Smith A. C. (2003) Nat. Genet. 35, 97–102 [DOI] [PubMed] [Google Scholar]
- 6.Pearsall R. S., Plass C., Romano M. A., Garrick M. D., Shibata H., Hayashizaki Y., Held W. A. (1999) Genomics 55, 194–201 [DOI] [PubMed] [Google Scholar]
- 7.Tremblay K. D., Saam J. R., Ingram R. S., Tilghman S. M., Bartolomei M. S. (1995) Nat. Genet. 9, 407–413 [DOI] [PubMed] [Google Scholar]
- 8.Kawahara M., Wu Q., Takahashi N., Morita S., Yamada K., Ito M., Ferguson-Smith A. C., Kono T. (2007) Nat. Biotechnol. 25, 1045–1050 [DOI] [PubMed] [Google Scholar]
- 9.Kono T., Obata Y., Wu Q., Niwa K., Ono Y., Yamamoto Y., Park E. S., Seo J. S., Ogawa H. (2004) Nature 428, 860–864 [DOI] [PubMed] [Google Scholar]
- 10.Cavaillé J., Seitz H., Paulsen M., Ferguson-Smith A. C., Bachellerie J. P. (2002) Hum. Mol. Genet. 11, 1527–1538 [DOI] [PubMed] [Google Scholar]
- 11.Hatada I., Morita S., Obata Y., Sotomaru Y., Shimoda M., Kono T. (2001) J. Biochem. 130, 187–190 [DOI] [PubMed] [Google Scholar]
- 12.Schmidt J. V., Matteson P. G., Jones B. K., Guan X. J., Tilghman S. M. (2000) Genes. Dev. 14, 1997–2002 [PMC free article] [PubMed] [Google Scholar]
- 13.Seitz H., Youngson N., Lin S. P., Dalbert S., Paulsen M., Bachellerie J. P., Ferguson-Smith A. C., Cavaillé J. (2003) Nat. Genet. 34, 261–262 [DOI] [PubMed] [Google Scholar]
- 14.Tierling S., Dalbert S., Schoppenhorst S., Tsai C. E., Oliger S., Ferguson-Smith A. C., Paulsen M., Walter J. (2006) Genomics 87, 225–235 [DOI] [PubMed] [Google Scholar]
- 15.Tsai C. E., Lin S. P., Ito M., Takagi N., Takada S., Ferguson-Smith A. C. (2002) Curr. Biol. 12, 1221–1226 [DOI] [PubMed] [Google Scholar]
- 16.Georgiades P., Watkins M., Surani M. A., Ferguson-Smith A. C. (2000) Development 127, 4719–4728 [DOI] [PubMed] [Google Scholar]
- 17.da Rocha S. T., Charalambous M., Lin S. P., Gutteridge I., Ito Y., Gray D., Dean W., Ferguson-Smith A. C. (2009) PLoS. Genet. 5, e1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekita Y., Wagatsuma H., Nakamura K., Ono R., Kagami M., Wakisaka N., Hino T., Suzuki-Migishima R., Kohda T., Ogura A., Ogata T., Yokoyama M., Kaneko-Ishino T., Ishino F. (2008) Nat. Genet. 40, 243–248 [DOI] [PubMed] [Google Scholar]
- 19.Takahashi N., Okamoto A., Kobayashi R., Shirai M., Obata Y., Ogawa H., Sotomaru Y., Kono T. (2009) Hum. Mol. Genet. 18, 1879–1888 [DOI] [PubMed] [Google Scholar]
- 20.Lin S. P., Coan P., da Rocha S. T., Seitz H., Cavaille J., Teng P. W., Takada S., Ferguson-Smith A. C. (2007) Development 134, 417–426 [DOI] [PubMed] [Google Scholar]
- 21.Moon Y. S., Smas C. M., Lee K., Villena J. A., Kim K. H., Yun E. J., Sul H. S. (2002) Mol. Cell. Biol. 22, 5585–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Luo G. Z., Yang W., Zhao X., Zheng Q., Lv Z., Li W., Wu H. J., Wang L., Wang X. J., Zhou Q. (2010) J. Biol. Chem. 285, 19483–19490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. (2010) Nature 465, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]