Abstract
CUB-domain-containing protein 1 (CDCP1) is an integral membrane glycoprotein with potential as a marker and therapeutic target for a number of cancers. Here we examine mechanisms regulating cellular processing of CDCP1. By analyzing cell lines exclusively passaged non-enzymatically and through use of a panel of protease inhibitors, we demonstrate that full-length 135 kDa CDCP1 is post-translationally processed in a range of cell lines by a mechanism involving serine protease activity, generating a C-terminal 70-kDa fragment. Immunopurification and N-terminal sequencing of this cell-retained fragment and detailed mutagenesis, show that proteolytic processing of CDCP1 occurs at two sites, Arg-368 and Lys-369. We show that the serine protease matriptase is an efficient, but not essential, cellular processor of CDCP1 at Arg-368. Importantly, we also demonstrate that proteolysis induces tyrosine phosphorylation of 70-kDa CDCP1 and recruitment of Src and PKCδ to this fragment. In addition, Western blot and mass spectroscopy analyses show that an N-terminal 65-kDa CDCP1 ectodomain is shed intact from the cell surface. These data provide new insights into mechanisms regulating CDCP1 and suggest that the biological role of this protein and, potentially, its function in cancer, may be mediated by both 70-kDa cell retained and 65-kDa shed fragments, as well as the full-length 135-kDa protein.
Keywords: Membrane Proteins, Phosphotyrosine, Protease, Receptors, Shedding, CDCP1, PKCδ, Src, Serine Protease, Tyrosine Phosphorylation
Introduction
CUB-domain-containing protein 1 (CDCP1)3 is an 836 amino acid integral membrane glycoprotein with a type I orientation at the cell surface (1–4), that is up-regulated in a number of malignancies including breast (1, 5, 6), colon (1, 2, 7), and lung (1) cancers. Of potential clinical significance, CDCP1 expression correlates with recurrence and patient survival rate in renal cell carcinomas (8) and lung adenocarcinomas (9), indicating that it may be suitable as a prognostic marker. Consistent with a role in cancer progression, silencing of CDCP1 reduced the metastatic ability of lung cancer A549 cells (10) and the peritoneal dissemination of gastric cancer 44As3 cells (11) in mice. Although its biological function is not known, the potential of CDCP1 as a therapeutic target for cancer treatment has been highlighted by studies showing that antibody-mediated inhibition of CDCP1 reduced metastasis of prostate cancer PC3 cells in mice (12, 13) and chicken embryos (13). Currently the mechanisms regulating CDCP1 in cancer and normal physiology are not well defined (14).
During cellular processing, the 29-residue CDCP1 N-terminal signal peptide is removed generating a protein with molecular mass identified as either 135 kDa (2, 10, 15) or 140 kDa (3, 4) that contains 30–40 kDa of N-linked glycans (2). In addition to this full-length form, there is evidence that a shorter CDCP1 species is expressed endogenously by a range of cell lines or is generated through the action of exogenous serine proteases. For example, lung cancer A549, PC14, H520, H322, and H157 cells (10) and gastric cancer 44As3 and 58As9 cells (11) resuspended non-enzymatically using a metal ion chelator (EDTA) to disrupt cell/matrix interactions, express CDCP1 as the full-length protein as well as a shorter species with a reported molecular mass of 70 kDa. Similarly, colon HCT116 and DLD-1, pancreatic MiaPaCa2 and prostate cancer PC3 cells resuspended with EDTA express a shorter form of CDCP1 with a reported molecular mass of 85 kDa (16). Interestingly, exogenous serine proteases also generate short CDCP1 species that have different reported molecular weights. For example, the serine protease trypsin generates a CDCP1 fragment in human foreskin keratinoytes (3) and breast MCF10A cells (17) with reported molecular masses of 80 kDa and 85 kDa, respectively, while another serine protease, plasmin, generates a reported 80-kDa CDCP1 protein in cultured keratinocytes (3). Significantly, the observation that 80 kDa and full-length CDCP1 are expressed in untreated preparations of neonatal mouse epidermis (3), suggests that mechanisms generating short forms of CDCP1 occur in vivo and that it will be important to understand the mechanisms controlling the generation of these species.
Here we examine the expression of full-length and lower molecular weight CDCP1 in cell lines originating from five different tissues focusing on prostate-derived cells to demonstrate that endogenous lower molecular weight CDCP1 is generated though the action of serine proteases. We also examine the ability of the type II transmembrane serine protease (TTSP) matriptase (18) to proteolytically process and induce tyrosine phosphorylation of CDCP1. Importantly, we analyze downstream consequences of CDCP1 cleavage showing that it results in shedding of a 65-kDa CDCP1 ectodomain and tyrosine phosphorylation of cell-retained 70-kDa CDCP1 and recruitment of Src and PKCδ to this fragment. Our data indicate that it will be important to better understand the molecular regulators and downstream signaling events coupled to normal and dysregulated CDCP1 processing.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies were from the following suppliers: goat polyclonal antibody against the last 13 C-terminal residues of CDCP1 from Abcam (Cambridge, MA; ab1377); rabbit polyclonal antibody against unspecified C-terminal residues of CDCP1 from Cell Signaling Technology (CST; Danvers, MA; 4115); goat antibody against the extracellular domain of CDCP1 from R&D Systems (Bio-Scientific Pty Ltd, Gymea, Australia; AF2666); rabbit anti-matriptase antibody from Bethyl Laboratories (Montgomery, TX); rabbit anti-Src antibody from CST (2108); rabbit anti-PKCδ antibody from Santa Cruz Biotechnology (Santa Cruz, CA; SC-937); rabbit anti-p-FAK-Y861 antibody that detects both p-CDCP1-Y734 and p-FAK-Y861 (3), from Invitrogen (Mulgrave, Australia); rabbit and mouse monoclonal anti-Flag epitope (DYKDDDDK) antibodies from Sigma; monoclonal anti-phosphotyrosine antibody PY20 from Calbiochem (La Jolla, CA); monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody from Chemicon International (Boronia, Australia); and HRP-conjugated secondary antibodies from Thermo Scientific (Murarrie, Australia). Control immunoglobulins (IgGs) were from Sigma and Invitrogen. The protease inhibitors aprotinin, phenylmethylsulfonyl fluoride (PMSF), tosyl-l-phenylalanine chloromethyl ketone (TPCK), tosyl-l-lysine chloromethyl ketone (TLCK), and (R)-N4-hydroxy-N1-[(S)-2-(1H-indol-3-yl)-1-methylcarbamoyl-ethyl]-2-isobutyl-succinamide (GM6001), leupeptin, pepstatin, and trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane (E-64) were from Sigma. Protein A/G-agarose and Complete EDTA-free protease inhibitor were from Roche Applied Sciences (Castle Hill, Australia). Blasticidin was from InvivoGen (San Diego, CA). Precision Plus Protein Dual Color Standard (catalogue number 161-0374) was from Bio-Rad. All other chemicals were from Sigma.
Expression Constructs
CDCP1-encoding cDNA was reverse transcribed from total RNA extracted from PC3 cells and amplified using Expand high fidelity polymerase (Roche Applied Science), and inserted into pcDNA3.1 vector (Invitrogen) generating an expression construct encoding full-length CDCP1 with a cytoplasmic, C-terminal Flag epitope. A matriptase-encoding expression construct has been described previously (19). Site-directed mutagenesis, to introduce CDCP1 mutations R362A, K365A, R368A, K369A, and F370W, and a stop codon replacing Lys-369 or Phe-370, was performed using Pfu Ultra polymerase (Stratagene, La Jolla, CA). The sequence of all constructs was confirmed by DNA sequencing at the Australian Genome Research Facility (St. Lucia, Australia).
Cell Culture and Transfections
Cells used in this study were purchased from the American Type Culture Collection. HeLa cells stably transfected with either pcDNA3.1 (vector) or the CDCP1-Flag expression construct were described previously (13). Prostate cancer lines PC3, LNCaP, DU145, 22Rv1, and immortalized prostate cell lines RWPE-1 and RWPE-2 and lymphoid K562, U937, Jurkat, and YT cells were grown in RPMI1640 medium, and cervical Ca Ski and HeLa cells, breast cell lines MDA-MB-231, MDA-MB-468, and Hs578t in Dulbecco's modified Eagle's medium. Breast MCF7 and MCF10A cells were grown in MEMα and DMEM/F12 media, respectively, containing insulin (10 μg/ml). Cultures were supplemented with 10% fetal calf serum, 100 units/ml of penicillin, and 100 units/ml of streptomycin unless otherwise specified and incubated at 37 °C in 5% CO2. Unless otherwise specified, all cells were passaged using 0.5 mm EDTA in PBS. In medium-exchanging experiments, donor cells were cultured in serum-containing medium for 3 days before the medium was collected, spun at 800 × g for 5 min, and the cell-free supernatant applied to acceptor cells, which had been cultured for 1–2 days to ∼50% confluence. In experiments to assess the class of protease mediating CDCP1 processing, immediately before transfer to CDCP1-expressing cells, serum containing conditioned medium was supplemented with Complete (EDTA-free) inhibitor mixture, PMSF, aprotinin, TLCK, TPCK, leupeptin, GM6001, pepstatin, or E-64 at the concentrations specified in the relevant figure legend. Cell transfections were performed using Lipofectamine 2000 (Invitrogen), following the instructions of the manufacturer.
Immunoprecipitation
In experiments to detect interacting proteins, cell lysates were collected in either PBS (pH 7.4) containing 1% CHAPS (Sigma) or 10 mm Tris-HCl (pH 7.4) containing 150 mm NaCl, 1% Triton X-100, in the presence of 1× protease inhibitor mixture, 2 mm sodium vanadate and 10 mm sodium fluoride. In experiments to purify low and high molecular weight CDCP1, cells were lysed in a buffer containing 1% SDS, 2 mm DTT, and protease inhibitor mixture followed by denaturation at 100 °C for 5 min. This mixture was then diluted 10-fold in buffer containing 0.55% IGEPAL, 55.55 mm Tris (pH 8.0), 11.1 mm MgCl2, and protease inhibitor mixture. In experiments to characterize the shed ectodomain of CDCP1, immunoprecipitation was performed from conditioned medium. DU145 cells were cultured in serum-free medium for 3 days. The medium was then centrifuged briefly to remove intact cells, and proteins were recovered by acetone precipitation. Lysates and media were pre-cleared with protein A/G-agarose for 1 h at 4 °C on a rolling platform. After centrifugation, the supernatant was mixed with appropriate antibodies or isotype IgGs and incubated overnight at 4 °C. Fresh aliquots of protein A/G-agarose beads were then added, and the mixture was incubated for 4 h at 4 °C with gentle agitation. The beads were then washed three times in cell lysis buffer. Associated proteins were eluted into Laemmli sample buffer and subjected to SDS-PAGE followed by either Western blot analysis, Edman degradation sequencing, or mass spectroscopy analysis.
Western Blot Analysis
Whole cell lysates were collected in a buffer containing protease inhibitor mixture, 2 mm sodium vanadate, and 10 mm sodium fluoride and either Triton X-100 (1%), 50 mm Tris-HCl (pH 7.4), and NaCl (150 mm), or CHAPS (1%), PBS (pH 7.4), and MgCl2 (2 mm). Protein concentrations were determined by a microbicinchoninic acid assay (Thermo Scientific). Proteins from serum-free conditioned medium from cells cultured for 3 days (300 μl) were concentrated by acetone precipitation then resuspended in lysis buffer (20 μl). Cell lysates, acetone-precipitated proteins from conditioned medium and immunoprecipitated proteins were separated by SDS-PAGE under reducing conditions and transferred to nitrocellulose membranes, which were blocked in 5% skim milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T). Membranes were incubated with appropriate antibodies diluted in blocking buffer for 1 h at room temperature, washed with TBS-T, and then incubated with species-appropriate HRP-conjugated secondary antibodies for 45 min. Following washes, membranes were incubated with SuperSignal West Pico Substrate (Pierce) and then exposed to film. Consistent protein loading and transfer was determined by reprobing membranes stripped in Restore Western blot stripping buffer (Pierce) with anti-matriptase, anti-GAPDH, or anti-Flag antibodies as appropriate.
Edman Degradation Protein Sequencing
N-terminal sequencing of immunopurified CDCP1 electrophoretically transferred to PVDF membranes was carried out on an Applied Biosystems 494 Procise Protein Sequencing System at the Australian Proteome Analysis Facility (Macquarie University, Sydney, Australia).
Mass Spectroscopy Analysis
The protein band of interest from a Coomassie-stained gel was excised and subjected to reduction, acetylation, and trypsin digestion. Fragments were resolved by liquid chromatography and analyzed on a QStar Elite quadrupole-time-of-flight mass spectrometer (Applied Biosystems). Data were analyzed using the Paragon algorithm as part of the ProteinPilot software package (Applied Biosystems) and searched against the current version of the SwisProt/UniProt database.
Matriptase Treatment of Cells
Generation and characterization of recombinant matriptase serine protease domain used in this study has been described previously (20). Cells were allowed to attach for 12 h in serum-containing medium, then starved of serum for 24 h before being washed with fresh serum-free medium. Another aliquot of serum-free medium containing matriptase (1, 5, or 10 nm) was then added, and the cells were incubated for 1, 5, 10, or 30 min before being lysed for Western blot analysis as described above. The concentration to stimulate half-maximal processing (EC50) was determined from anti-CDCP1 Western blot analysis of three independent experiments by plotting percentage CDCP1 conversion versus matriptase concentration for the 30 min time point. In experiments to assess changes in phosphorylation of CDCP1 due to matriptase treatment, 22Rv1 cells were either untreated or treated with matriptase (20 nm) for 0.5, 1, or 2 h. Immunoprecipitation was then performed on the collected lysates using either a rabbit anti-CDCP1 antibody or control rabbit IgGs followed by mouse anti-phosphotyrosine Western blot analysis. The blot was reprobed with anti-rabbit secondary antibody to confirm consistency in the amount of antibody used in the immunoprecipitation.
Reduction of Matriptase Expression
Matriptase activity was reduced by silencing of expression. Polyclonal populations of PC3 and DU145 cells with stable suppression of matriptase expression were generated using four constructs generated in a BLOCK-iT Pol II miR RNAi expression vector backbone and containing sequences selected using the Invitrogen Block-iT miR RNAi Select tool (CCAACATTGACTGCACATGGA, GCCCAACAACCAGCATGTGAA, GTACACAAGGCTCCCTCTGTT, and TCCTGCCAGTCAACAACGTCA). Constructs containing either scrambled (GTCTCCACGCGCAGTACATTT) or LacZ-targeting (GACTACACAAATCAGCGATTT) sequences from Invitrogen were used as controls. Cells were selected in medium containing blasticidin (InvivoGen, San Diego, CA) at a concentration of 3 and 10 μg/ml for PC3 and DU145 cells, respectively, for 4 weeks. Suppression of matriptase expression and effect on CDCP1 processing was assessed by Western blot analysis.
RESULTS
Post-translational Processing of 135-kDa CDCP1 Generates a 70 kDa protein
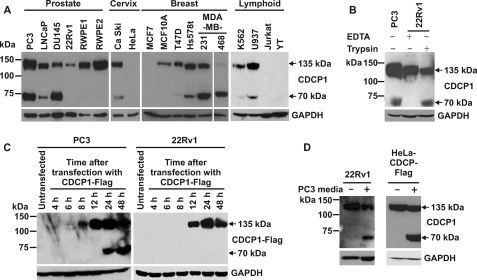
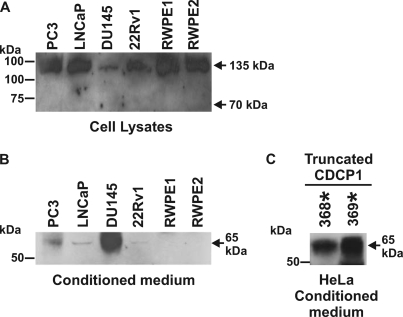
To examine the expression of CDCP1 in human cell lines, Western blot analysis was performed using an antibody generated against a peptide encompassing the last 13 amino acids of CDCP1 (residues 824–836 of GenBankTM entry AAO33397). To ensure that passaging of cells using trypsin did not contribute to generation of lower molecular weight CDCP1 species, analysis was performed on cells that had been passaged exclusively using EDTA (0.5 mm). As shown in Fig. 1A, CDCP1 was detected in 14 of the 19 analyzed cell lines. Of these 14 CDCP1-expressing lines, 9 clearly expressed two CDCP1 forms of 70 and 135 kDa, PC3, LNCaP, and DU145 prostate cells, Ca Ski cervical cells, Hs578t, MDA-MB-231, and MDA-MB-468 breast cells, and K562 and U937 lymphoid cells. In addition, on longer exposure prostate cancer 22Rv1 and breast cancer T47D cells showed low level expression of the 70-kDa band. CDCP1 was not detected in cervical HeLa, breast MCF7, or lymphoid Jurkat and YT cells.
FIGURE 1.
Post-translational processing generates 70 kDa CDCP1 from the full-length 135 kDa protein. A, Western blot analysis using a goat antibody generated against the last 13 C-terminal residues of CDCP1 (Abcam ab1377) was performed on lysates from the indicated cell lines passaged with EDTA. B, trypsin induces the generation of 70 kDa CDCP1. Lysates from prostate cancer PC3 cells passaged with EDTA and 22Rv1 cells treated once with either EDTA or trypsin (5 min) were examined by Western blot analysis using a goat anti-CDCP1 antibody (Abcam ab1377). C, prostate cancer PC3 and 22Rv1 cells were transiently transfected with a CDCP1-Flag expression construct, and lysates were collected at the indicated time points for anti-Flag Western blot analysis. D, Western blot analysis of lysates from 22Rv1 (left panel) and HeLa-CDCP1-Flag (right panel) cells either untreated or cultured for 24 h in PC3 cell-conditioned medium. Lysates from 22Rv1 cells were probed with a goat anti-CDCP1 antibody (Abcam ab1377) and lysates from HeLa-CDCP1-Flag cells were probed with a rabbit anti-Flag antibody. Anti-GAPDH Western blot analysis was performed to examine protein loading.
Previous studies have reported lower molecular mass CDCP1 of 70, 80, or 85 kDa, generated either through the action of exogenous trypsin and plasmin (3, 17) or unknown mechanisms (11, 16). To examine the relationship between these species and endogenous 70 kDa CDCP1 detected in Fig. 1A, we used anti-CDCP1 Western blot analysis to compare lysates from cells that predominantly express only 135 kDa CDCP1 (prostate 22Rv1 cells) with lysates obtained from the same line after a single treatment with trypsin (0.25% solution). Lysates from a cell line that expresses both 70 and 135 kDa CDCP1 (PC3 cells) were used to compare endogenous 70 kDa CDCP1 and the trypsin-induced species. As shown in Fig. 1B, lower molecular weight CDCP1 produced in 22Rv1 cells by trypsin treatment migrated at the same molecular mass as the 70-kDa CDCP1 band expressed endogenously by PC3 cells. These data suggest that cleavage by the serine protease trypsin generates a CDCP1 species of the same molecular weight as produced endogenously by a range of cell lines (Fig. 1A).
To examine the mechanism by which 70 kDa CDCP1 is generated, we transiently transfected prostate PC3 and 22Rv1 cells with a construct encoding full-length CDCP1 with a cytoplasmic, C-terminal Flag epitope (CDCP1-Flag). We have shown that PC3 cells endogenously express both 70 and 135 kDa CDCP1 whereas 22Rv1 cells predominantly express only high molecular weight CDCP1 (Fig. 1A). As shown in Fig. 1C (left panel), Western blot analysis using an anti-Flag antibody indicated that PC3 cells began to express 135 and 70 kDa CDCP1-Flag, 8 and 24 h after transfection, respectively. As CDCP1-Flag expressed by these cells is generated from plasmid DNA encoding only the full-length protein, 70 kDa CDCP1-Flag must have been produced through a post-translational processing event rather than via alternate processing of a pre-mRNA transcript. Consistent with this proposal, anti-Flag Western blot analysis of lysates from 22Rv1 cells transiently transfected with the CDCP1-Flag construct detected only 135 kDa CDCP1-Flag (Fig. 1C, right panel).
To assess whether CDCP1 processing is mediated by a cell secreted or shed factor or factors, 22Rv1 and HeLa cells stably expressing CDCP1-Flag (designated HeLa-CDCP1-Flag (13)), which are low and non-CDCP1-processing cell lines, respectively, were cultured either in standard medium or conditioned medium from CDCP1-processing PC3 cells. As shown in Fig. 1D (left panel), anti-CDCP1 Western blot analysis indicated that PC3-conditioned medium contains a factor that enables processing of 135 kDa CDCP1 to 70 kDa in 22Rv1 cells. The same result was obtained from anti-Flag Western blot analysis of lysates from HeLa-CDCP1-Flag cells treated in the same way (Fig. 1D, right panel). These data indicate that CDCP1 processing is mediated by a cell-secreted or shed factor or factors.
Serine Protease Activity Is Required for Cellular Conversion of 135 kDa CDCP1 to 70 kDa
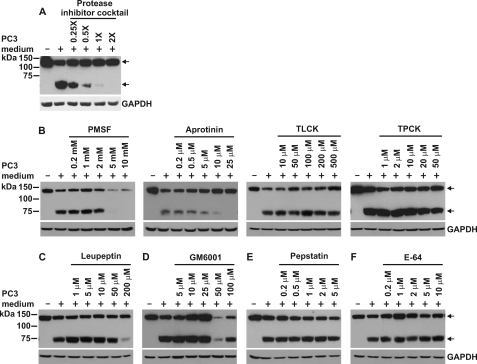
To examine whether endogenous CDCP1 processing is mediated by a protease, we performed experiments in which PC3 cell-conditioned medium was treated with various protease inhibitors including a broad range mixture targeting serine, cysteine, and metalloproteases as well as inhibitors of narrower specificity: phenylmethylsulfonyl fluoride (21), aprotinin (22–24), TLCK (25), and TPCK (26) (serine protease inhibitors); leupeptin (mixed serine/cysteine protease inhibitor (27)); GM6001 (matrix metalloprotease inhibitor (28, 29)); pepstatin (aspartyl protease inhibitor (30)); and E-64 (cysteine protease inhibitor (31)). We incubated HeLa-CDCP1-Flag cells with either untreated or protease inhibitor-treated PC3-conditioned medium for 24 h and analyzed cell lysates by Western blot analysis using an anti-Flag antibody. The protease inhibitor mixture was effective at completely blocking PC3 media-induced proteolysis of CDCP1 suggesting the involvement of serine, cysteine and/or metalloproteases in processing (Fig. 2A). Use of protease inhibitors of narrower specificity indicated that CDCP1 processing is blocked by the serine protease inhibitors PMSF (at intermediate to high concentration) and aprotinin (at low to intermediate concentration) (Fig. 2B), and the mixed serine/cysteine protease inhibitor leupeptin (at high concentration) (Fig. 2C), but not by the serine protease inhibitors TPCK and TLCK (Fig. 2B), the matrix metalloprotease inhibitor GM6001 (Fig. 2D), the aspartyl protease inhibitor pepstatin (Fig. 2E), or the cysteine protease inhibitor E-64 (Fig. 2F). Although at concentrations greater than 25 μm, GM6001 reduced the level of 70 kDa CDCP1, this was accompanied by a similar reduction in expression of 135 kDa CDCP1, likely indicating its effect was to down-regulate expression rather than proteolytic processing of CDCP1. These data suggest that cellular processing of CDCP1 is mediated by secreted or shed serine protease activity. This is supported by a recent report showing that 7 μm ecotin, a trypsin-fold-specific macromolecular serine protease inhibitor, reduces processing of CDCP1 in MDA-MB-468 cells stably overexpressing this protein (4).
FIGURE 2.
Serine protease activity is required for conversion of 135 kDa CDCP1 to 70 kDa. Anti-Flag Western blot analysis of lysates from HeLa-CDCP1-Flag cells untreated or cultured for 24 h in PC3 cell-conditioned medium that was either untreated or had been supplemented with (A) Complete EDTA-free protease inhibitor mixture or (B) PMSF, aprotinin, TLCK, or TPCK, (C) leupeptinin, (D) GM6001, (E) pepstatin, or (F) E-64 at the indicated concentrations. Arrows indicate 70 and 135 kDa CDCP1. Anti-GAPDH Western blot analysis was performed to examine protein loading.
Cellular Proteolytic Processing of CDCP1 Occurs at Arg-368 and Lys-369
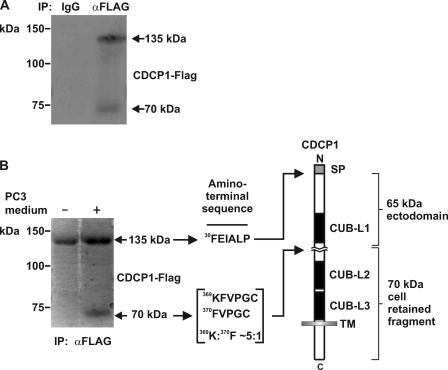
To identify the sites at which CDCP1 processing occurs, we performed immunopurification from HeLa-CDCP1-Flag cells treated with PC3 cell-conditioned medium using a mouse anti-Flag antibody. As shown in Fig. 3A, Western blot analysis indicated that this approach immunoprecipitated both 70 and 135 kDa CDCP1. When performed on a larger scale, quantities sufficient for amino-terminal sequencing of both CDCP1 forms were obtained (Fig. 3B). Edman degradation sequence analysis identified three N-terminal sequences: 30FEIALP, 369KFVPGC, and 370FVPGC. The first of these has previously been reported by us as the amino-terminal of 135 kDa CDCP1 (2), generated through removal of the 29 residue signal peptide (Fig. 3B). The other two N-terminal sequences indicated proteolytic processing of CDCP1 occurs following Arg-368 and Lys-369. The ratio of the signals obtained from the N termini commencing at Lys-369 and Phe-370 was 5:1 (Fig. 3B).
FIGURE 3.
Identification of the sites at which CDCP1 is proteolytically processed. A, anti-Flag Western blot analysis of proteins obtained from mouse IgG and anti-Flag antibody immunoprecipitations from lysates of HeLa-CDCP1-Flag cells treated with PC3 cell-conditioned medium. B, Coomassie-stained PVDF membrane of anti-Flag immunoprecipitates obtained from HeLa-CDCP1-Flag cells either untreated or treated with PC3 cell-conditioned medium. To the right is shown the N-terminal sequence obtained by Edman degradation sequencing of 70 and 135 kDa CDCP1 and a diagram showing the location of the identified N termini within the CDCP1 structure. The data indicated that 135 kDa CDCP1 is processed after Arg-368 and Lys-369 in the ratio 5:1 in a region located between CUB-like domains 1 and 2 (CUB-L1 and CUB-L2). SP, signal peptide; TM, transmembrane domain.
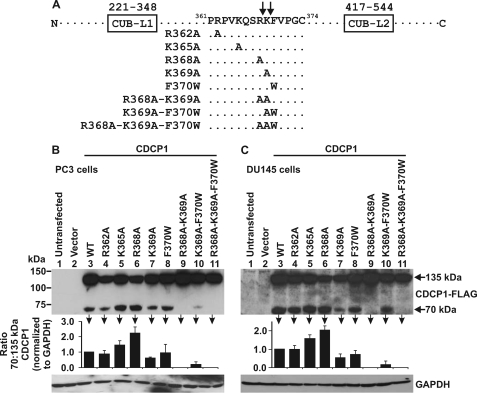
To examine whether Arg-368 and Lys-369 are relevant proteolysis sites in cells endogenously expressing CDCP1, we mutated these and nearby residues by site-directed mutagenesis of the CDCP1-Flag expression construct. The following amino acid substitutions were introduced: single amino acid changes R362A, K365A, R368A, K369A, F370W; double amino acid changes R368A/K369A, K369A/F370W; and triple amino acid change R368A/K369A/F370W (Fig. 4A). When considering proteolytic processing at Arg-368, residues Arg-362, Lys-365, Arg-368, Lys-369, and Phe-370 are located in the P7, P4, P1, P1′, and P2′ positions, respectively (numbering of substrate residues from Schechter and Berger (32)). For cleavage at Lys-369, amino acids Arg-362, Lys-365, Arg-368, Lys-369, and Phe-370 are located in the P8, P5, P2, P1, and P1′ positions, respectively.
FIGURE 4.
Examination of the cellular proteolytic processing sites of CDCP1. A, schematic showing the location of single, double, and triple amino acid mutations introduced into the CDCP1-Flag sequence between CUB-like domain 1 and 2 (CUB-L1 and CUB-L2), R362A, K365A, R368A, K369A, F370W, R368A-K369A, K369A-F370W, and R368A-K369A-F370W. B, PC3 and C, DU145 cell lysates from cells either untransfected or transiently transfected with either vector or wild type or mutant CDCP1-Flag expression constructs analyzed by anti-Flag Western blot analysis. Anti-GAPDH Western blot analysis was performed to examine protein loading. Graphs were generated from densitometric analysis of at least three separate experiments. For 70 and 135 kDa CDCP1 and GAPDH, signal intensities from cells transfected with mutant constructs were normalized against the signal from cells transfected with wild-type CDCP1. These normalized values were used to calculate the ratio of 70 to 135 kDa CDCP1.
Experiments were performed by transiently transfecting wild-type and mutant CDCP1-Flag expression constructs into two cell lines that endogenously express both 70 and 135 kDa CDCP1 (PC3 and DU145). Lysates from these cells were examined by anti-Flag Western blot analysis and representative images and graphical representations of data averaged from three experiments are shown in Fig. 4, B and C. As shown in these panels (lanes 9 and 11), it was only when transfections were performed with the R368A/K369A double and R368A/K369A/F370W triple mutant constructs that proteolytic processing generating 70 kDa CDCP1 was completely blocked in both PC3 and DU145 cells. These data confirm the existence of two cleavage sites for CDCP1 in PC3 and DU145 cells at Arg-368 and Lys-369, consistent with our N-terminal sequence data showing that CDCP1 is proteolytically processed at these two sites (Fig. 3B). In addition, transfection with the single mutant K369A construct resulted in a reduction in CDCP1 processing at Arg-368 of ∼50% in both cell types (Fig. 4, B and C, lane 7). Also of note, transfection with the R369A/F370W double mutant construct resulted in reduction in CDCP1 processing at R368 in PC3 and DU145 cells of ∼90 and 75%, respectively (Fig. 4, B and C, lane 10). These reductions in processing indicate that for cleavage at Arg-368, the residues located in the P1′ (Lys-369) and P2′ (Phe-370) positions are important. Interestingly, in both cell types processing at Lys-369 was more than doubled by mutation of Arg-368 to alanine (Fig. 4, B and C, lane 6) suggesting that the P2 position is important for processing at this lysine. Altogether, these data confirm Arg-368 and Lys-369 as cellular processing sites of CDCP1 and indicate that cleavage at each site is impacted upon differently by nearby residues, suggesting the likelihood that cleavage at these sites is mediated by different serine proteases.
The CDCP1 Ectodomain Is Shed Intact from the Cell Surface
The next experiments aimed to examine the consequences of proteolytic processing on the N-terminal of CDCP1. This required an antibody capable of detecting the N-terminal fragment of CDCP1. As shown in Fig. 5A, a goat antibody generated against the CDCP1 extracellular domain detected 135 kDa but not 70 kDa CDCP1 in lysates from the same prostate cell lines examined in Fig. 1A, indicating that the epitope recognized by this antibody is indeed located within the N-terminal portion of CDCP1 that is shed after proteolysis and not in the cell-retained C-terminal fragment. Using this antibody to analyze conditioned media from these cells indicated that a CDCP1 ectodomain of 65 kDa is present at highest levels in DU145-conditioned medium, at medium levels in PC3 and at low and barely detectable levels, respectively, in LNCaP and 22Rv1-conditioned medium, with no CDCP1 ectodomain present in the conditioned medium of RWPE1 and RWPE2 cells (Fig. 5B). To confirm that the antibody actually detected CDCP1, we used it to perform an immunoprecipitation from the conditioned medium of DU145 cells and analyzed the purified 65 kDa fragment by mass spectroscopy. This analysis identified a 12-residue tryptic fragment, LEDKQPGNMAGN, that matched exactly CDCP1 amino acids 297–308. In addition, this antibody specifically detected only a protein of 65 kDa in the conditioned medium of HeLa cells transiently transfected with expression constructs encoding CDCP1 truncated after either Arg-368 or Lys-369, the identified cleavage sites (Fig. 5C). These data indicate that the CDCP1 ectodomain is shed intact from the cell surface as a fragment of 65 kDa.
FIGURE 5.
The CDCP1 ectodomain is shed intact from the cell surface. Western blot analyses using a goat antibody generated against the extracellular domain of CDCP1 (R&D Systems AF2666). A, analysis of lysates from prostate cell lines passaged with EDTA demonstrating that this antibody recognizes 135 kDa but not 70 kDa CDCP1 indicating that the cognate antigen is located in the shed portion of CDCP1. B, analysis of conditioned media from these cell lines. Cells were cultured in serum-free medium for 3 days. The medium was then centrifuged briefly to remove intact cells, and proteins were recovered by acetone precipitation. C, analysis of serum-free conditioned media from HeLa cells transiently transfected with expression constructs encoding CDCP1 truncated after either Arg-368 (designated R368*) or Lys-369 (K370*).
The Serine Protease Matriptase Is an Efficient but Not Essential Cellular Processor of CDCP1
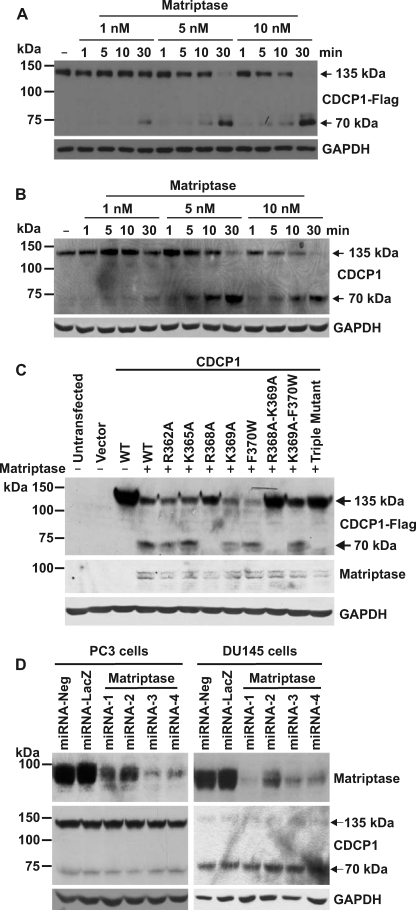
It has previously been reported that CDCP1 can be cleaved at Arg-368 by the serine protease matriptase (4). These in vitro experiments were performed using the purified recombinant extracellular domain of CDCP1 and the purified recombinant catalytic domain of matriptase, and thus provided limited insight as to whether cleavage would occur in a cellular setting. To examine whether matriptase is able to process cell expressed CDCP1, HeLa-CDCP1-Flag cells, which express only 135 kDa CDCP1, and prostate 22Rv1 cells, which predominantly express only this CDCP1 species, were incubated with increasing concentrations of recombinant matriptase protease domain over 1, 5, 10, and 30 min. Significantly, low concentrations of matriptase were able to completely process CDCP1 to 70 kDa in stable overexpressing (Fig. 6A) and endogenously expressing (Fig. 6B) cells within 30 min with an EC50 of 4.0 ± 1.8 nm in HeLa-CDCP1-Flag cells.
FIGURE 6.
The serine protease matriptase is an efficient but not essential processor of CDCP1. A, anti-Flag Western blot analysis of lysates from HeLa-CDCP1-Flag cells either untreated (−) or incubated with 1, 5, or 10 nm matriptase for 1, 5, 10 or 30 min. B, Western blot analysis of lysates from 22Rv1 cells either untreated (−) or incubated with 1, 5, or 10 nm matriptase for 1, 5, 10, or 30 min using a goat anti-CDCP1 antibody (Abcam ab1377). Anti-GAPDH Western blot analysis was performed to examine protein loading. C, HeLa cells were either untransfected, transfected with vector alone, or co-transfected with matriptase and either wild type (WT) CDCP1-Flag expression construct or constructs encoding the indicated single and double residue mutant CDCP1-Flag constructs or triple mutant CDCP1-Flag-R368A-K369A-F370W. After 24 h, lysates were collected and analyzed by anti-Flag, anti-matriptase, and anti-GAPDH Western blot analysis. D, polyclonal populations of PC3 and DU145 cells were stably transfected with one of four CDCP1 knockdown miRNA constructs. After 4 weeks of selection in blasticidin, cell lysates were analyzed by Western blot analysis using rabbit anti-matriptase, goat anti-CDCP1 (Abcam ab1377), and anti-GAPDH antibodies.
To identify the site of matriptase cleavage of cell-expressed CDCP1, HeLa cells were transiently co-transfected with a matriptase encoding expression construct and either a wild-type CDCP1 construct or a construct encoding one of the single, double, or triple CDCP1 mutants described above. As matriptase is a type II transmembrane serine protease that can also be post-translationally shed from the cell surface (33), transfections were performed with an expression construct encoding the full-length protein. Anti-Flag Western blot analysis showed that of the single mutations only R368A abolished processing, indicating that matriptase cleaves CDCP1 at Arg-368 but not at nearby arginine or lysine residues including Lys-369 shown by us to be a site at which CDCP1 is endogenously proteolytically cleaved in PC3 and DU145 cells (Fig. 6C).
To directly examine whether matriptase is a cellular processor of CDCP1, we generated PC3 and DU145 cells in which matriptase expression had been stably reduced using 4 different silencing constructs. Anti-matriptase Western blot analysis of stable polyclonal PC3 cell populations showed that two of the constructs (miRNA-3 and -4) reduced matriptase expression by ∼90% while the two remaining constructs (miRNA-1 and -2) reduced matriptase expression by ∼50% (Fig. 6D, left panel). However, anti-CDCP1 Western blot analysis failed to detect a consistent change in the levels of 70 kDa CDCP1 (Fig. 6D, left panel). Similarly with DU145 cells, reduction of matriptase expression had no impact on CDCP1 proteolysis (Fig. 6D, right panel). These data indicate either that matriptase does not proteolytically process CDCP1 in PC3 and DU145 cells or that this protease is one of at least two that cleaves CDCP1 at Arg-368 or Lys-369.
Proteolysis Mediates Tyrosine Phosphorylation of 70 kDa CDCP1 and Binding of Src and PKCδ
It has previously been reported that CDCP1 is a substrate of Src family kinases (SFKs) (2, 3, 15) and that trypsin-mediated cell de-adhesion results in phosphorylation of CDCP1-Y734 and phosphorylation-dependent binding of the kinases Src and PKCδ (3). We therefore examined the effect of proteolysis, independent of cell de-adhesion, on these processes. First, interactions between CDCP1, Src, and PKCδ were examined in adherent DU145 cells (a cell line that expresses high levels of 70 kDa CDCP1) by performing immunoprecipitations with anti-CDCP1 and anti-Src antibodies from lysates prepared in the presence and absence of the phosphatase inhibitors sodium vanadate and sodium fluoride.
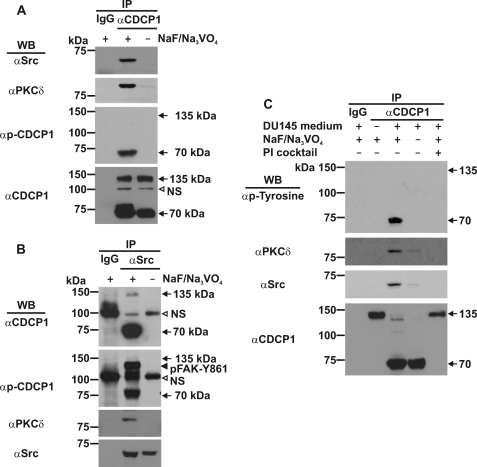
As shown in Fig. 7A, Western blot analysis of immunoprecipitations performed with an anti-CDCP1 antibody, demonstrated that CDCP1 binds Src and PKCδ in adherent DU145 cells. In the absence of phosphatase inhibitors binding was abolished confirming that formation of the CDCP1·Src·PKCδ complex is phosphorylation-dependent (10, 15). Interestingly, Western blot analysis with an antibody that detects both p-CDCP1-Y734 and p-FAK-Y861 (3), indicated that 70 kDa but not 135 kDa CDCP1-Y734 is phosphorylated in these cells (Fig. 7A, second bottom panel). Importantly, analysis of these immunoprecipitates with an anti-CDCP1 antibody indicated that both 70 and 135 kDa CDCP1 were efficiently immunoprecipitated in these experiments (Fig. 7A, lower panel).
FIGURE 7.
Examination of tyrosine phosphorylation of CDCP1 and binding of Src and PKCδ induced by DU145 media. A, anti-Src, -PKCδ, -p-CDCP1-Y734 (performed with an antibody that detects both p-CDCP1-Y734 and p-FAK-Y861), and -CDCP1 (CST 4115) Western blot analysis of anti-CDCP1 immunoprecipitates (CST 4115) obtained from DU145 cells. B, anti-CDCP1 (CST 4115), -p-CDCP1-Y734 (performed with an antibody that detects both p-CDCP1-Y734 and p-FAK-Y861), -PKCδ, and -Src Western blot analysis of anti-Src immunoprecipitates obtained from DU145 cells. C, anti-phosphotyrosine, anti-PKCδ, anti-Src, and anti-CDCP1 (CST 4115) Western blot analysis of anti-CDCP1 immunoprecipitates obtained from 22Rv1 cells either untreated (−) or treated (+) with 3 day serum-free conditioned medium from DU145 cells for 36 h. The media was either untreated (−) or treated (+) with protease inhibitor (PI) mixture before incubation with 22Rv1 cells. Lysates from all experiments were collected in the presence (+) or absence (−) of sodium vanadate and sodium fluoride. All experiments included control immunoprecipitations performed with species matched IgG. NS, nonspecific.
Several interesting observations were made from Western blot analyses of immunoprecipitations performed with an anti-Src antibody. In addition to supporting the existence of a phosphorylation-dependent CDCP1·Src·PKCδ complex in adherent DU145 cells (Fig. 7B), these analyses showed that Src binds almost exclusively to 70 kDa CDCP1 (Fig. 7B, upper panel). In addition, analysis with the antibody that detects both p-CDCP1-Y734 and p-FAK-Y861 (3), demonstrated that Src-bound 70 kDa CDCP1 is phosphorylated on Tyr-734 (Fig. 7B, second panel from top). Consistent with a previous report (34), p-FAK-Y861 co-immunoprecipitated with Src (Fig. 7B, second panel from top). However, based on the data shown in Fig. 7A (second bottom panel), p-FAK-Y861 is not present in the 70 kDa CDCP1·Src·PKCδ complex.
Together the data in Fig. 7, A and B suggest that phosphorylated 70 kDa CDCP1-Y734 forms a complex with Src and PKCδ in DU145 cells. However, it is not yet known whether proteolysis is a driver of phosphorylation of 70 kDa CDCP1 and formation of the 70 kDa p-CDCP1·Src·PKCδ complex. To examine this question, we performed anti-CDCP1 antibody immunoprecipitations from 22Rv1 cells treated with DU145-conditioned medium (which cleaves after both Arg-368 and Lys-369 converting CDCP1 to 70 kDa). To ensure that any effects were due to proteolytic activity, we included a control experiment in which DU145 medium had been pretreated with protease inhibitor mixture before incubation with 22Rv1 cells. As CDCP1 contains five intracellular tyrosine residues (Tyr-707, Tyr-734, Tyr-743, Tyr-762, Tyr-806), to examine the total tyrosine phosphorylation state of CDCP1, rather than only Tyr-734, Western blot analyses were performed with anti-p-tyrosine antibody PY20, as well as with anti-Src, -PKCδ, and -CDCP1 antibodies.
Significantly, as shown in Fig. 7C, Western blot analysis indicated that DU145 medium induced complete conversion of 135 kDa CDCP1 to 70 kDa (bottom panel), which was accompanied by tyrosine phosphorylation of 70 kDa CDCP1 (upper panel), and binding of PKCδ (second panel from top) and Src (second panel from bottom). Importantly, the 70 kDa p-CDCP1· Src·PKCδ complex was not detected in the absence of phosphatase inhibitors (Fig. 7C, lane 4), indicating that proteolysis-induced formation of this complex is dependent on phosphorylation. Similarly, data obtained in the presence of protease inhibitors (Fig. 7C, lane 5) indicated that formation of this complex is dependent on proteolytic activity and not some other factor present in DU145 media. In all experiments, DU145 medium treatment had no effect on cell adhesion (data not shown).
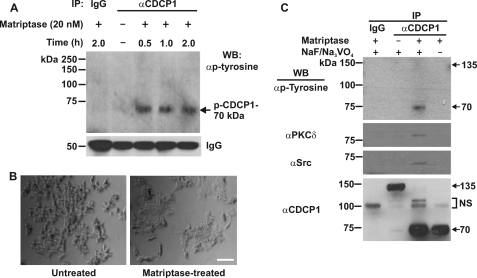
As DU145 medium cleaves CDCP1 at both Arg-368 and Lys-369, we were interested to examine the effect of proteolysis at Arg-368 only. For this purpose we performed a time course treatment of 22Rv1 cells with recombinant matriptase, which we have shown cleaves CDCP1 exclusively at this site. As shown in Fig. 8A, matriptase (20 nm) induced tyrosine phosphorylation of 70 kDa CDCP1 in 22Rv1 cells within 30 min. However, in contrast to an equal concentration of trypsin, which induced complete detachment of cells in 10 min (data not shown), matriptase had no impact on 22Rv1 cell attachment after 2 h of treatment (Fig. 8B). As shown in Fig. 8C, after 30 min matriptase completely converted CDCP1 to 70 kDa and, like DU145 medium, this was accompanied by tyrosine phosphorylation of 70 kDa CDCP1 and binding of Src and PKCδ. These data indicate that cleavage at Arg-368 promotes formation of the 70 kDa p-CDCP1·Src·PKCδ complex.
FIGURE 8.
Matriptase proteolysis mediates tyrosine phosphorylation and binding of Src and PKCδ to 70 kDa CDCP1. A, anti-phosphotyrosine Western blot analysis of proteins immunoprecipitated from 22Rv1 cell lysates using a rabbit anti-CDCP1 antibody (CST 4115). Lysates were collected from 22Rv1 cells either untreated (−) or treated (+) with 20 nm matriptase for the indicated times. The blot was reprobed with an anti-rabbit secondary antibody to detect rabbit IgG to assess consistency in the amount of antibody used for immunoprecipitations. B, photographic images of 22Rv1 cells either untreated (left panel) or treated for 2 h with 20 nm matriptase (right panel). Bar, 50 μm. C, anti-phosphotyrosine, -PKCδ, -Src, and -CDCP1 (CST 4115) Western blot analysis of anti-CDCP1 (CST 4115) immunoprecipitates obtained from 22Rv1 cells either untreated (−) or treated (+) with matriptase (20 nm; 0.5 h). Lysates were collected in the presence (+) or absence (−) of sodium vanadate and sodium fluoride. All experiments included control immunoprecipitations performed with species matched IgG. NS, nonspecific.
DISCUSSION
CDCP1 is an integral membrane glycoprotein with potential as a marker (1, 2, 5–9) and therapeutic target (10–13) for a number of cancers. Currently its function and the mechanisms regulating CDCP1 in cancer and normal physiology are not known (14). Here we show that full-length, 135 kDa CDCP1 is proteolytically processed in cultured cell lines by endogenous serine protease activity to a C-terminal 70 kDa fragment. We also show for the first time that, independent of cell detachment, proteolysis is accompanied by tyrosine phosphorylation of 70 kDa CDCP1 and recruitment of Src and PKCδ to this cell-retained fragment. Importantly, examination of conditioned medium indicates that proteolytic processing also generates a shed 65 kDa N-terminal CDCP1 ectodomain. Our detection of this CDCP1 fragment in conditioned medium by Western blot analysis is consistent with our previous immunohistochemical detection of soluble CDCP1 in vivo within the lumen of normal colonic crypts and colon adenocarcinoma (2). These data provide support for the proposal that soluble/shed CDCP1 may be useful as a marker for diseases in which CDCP1 processing is dysregulated (2).
Our data raise the possibility that proteolytic processing of CDCP1 establishes two cellular signaling pathways. One involving the direct relay of signals across the plasma membrane via the CDCP1 70 kDa cell-retained C-terminal region. As shown in Fig. 9, in this pathway proteolysis induces tyrosine phosphorylation of 70 kDa CDCP1 and recruitment of Src and PKCδ to this fragment forming a 70 kDa p-CDCP1·Src·PKCδ complex. As previous reports suggest that phosphorylation of CDCP1 and formation of CDCP1 signaling complexes are important in a number of settings, it is likely that proteolytically induced formation of the 70 kDa p-CDCP1·Src·PKCδ complex seen by us will also be functionally important. For example, Uekita et al. (10) demonstrated that PKCδ, phosphorylated by a CDCP1·SFK complex, is essential for CDCP1-mediated A549 lung cancer cell resistance to anoikis. Furthermore, 135 kDa p-CDCP1-Y734 levels were increased in gastric cancer 44As3 tumor nodules in nude mice compared with these cells grown in culture and p-CDCP1-Y734 levels were elevated in ∼30% of 10 analyzed scirrhous-type gastric cancer tissues compared with normal tissue (11). In addition, phosphorylation of CDCP1 at Tyr-743, while rare in normal epithelium, is a common event in many cancers of epithelial origin and has been proposed to be of importance in invasion and metastasis of these malignancies (16). In addition, phosphorylation of 70 kDa CDCP1 has been noted in trypsin induced cell rounding and detachment (3, 17) and is a consequence of loss of cell anchorage common to events such as mitosis and physiological cell shedding occurring at the apices of colonic villi (17).
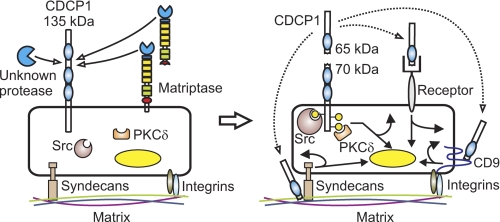
FIGURE 9.
Protease-mediated processing of CDCP1. Serine proteolysis generates a 65 kDa shed ectodomain and a tyrosine-phosphorylated cell retained 70 kDa CDCP1 fragment. Proteolysis occurs at Arg-368 and Lys-369 between CUB-like domain 1 and 2 of CDCP1 (the 3 CDCP1 CUB-like domains are shown as blue ovals). Proteolysis results in tyrosine phosphorylation of 70 kDa CDCP1 (yellow-filled circles) and recruitment of Src and PKCδ in a phosphorylation-dependent manner. We propose that the CDCP1 ectodomain may function as a ligand or competitive inhibitor for a cell surface receptor (autocrine receptor binding is shown, but paracrine and endocrine signaling may also be relevant) or as a matrix-interacting protein potentially modulating cell:matrix interactions occurring via known CDCP1 interacting proteins such as syndecan 1 and 4 and the tetraspannin CD9 (dotted arrows). Potential signaling downstream of the 70 kDa CDCP1 cell-retained fragment and ectodomain resulting in direct cellular responses and changes in gene expression are represented by curved and straight arrows, respectively. Currently the endogenous serine protease or proteases mediating cleavage of CDCP1 at Arg-368 and Lys-369 are not known. However, we have shown that the cell surface and shed serine protease matriptase efficiently cleaves CDCP1 exclusively at Arg-368.
As shown in Fig. 9, we are also proposing that proteolytic processing of CDCP1 may initiate a second signaling pathway via the shed CDCP1 65 kDa ectodomain. It is possible that this domain may act as a ligand either inducing cell signaling by binding to a plasma membrane receptor or modulating signaling by functioning as a competitive inhibitor for a receptor. Alternatively, it may function as a matrix interacting protein. In this capacity modulating cell:matrix interactions occurring via known CDCP1 interacting proteins such as syndecan 1 and 4 (4) and the integrin binding tetraspannin CD9 (35) (Fig. 9).
The work reported here clarifies data from a number of recent reports on CDCP1 species of different apparent molecular weights. Specifically, it has previously been demonstrated that CDCP1 is expressed with a reported molecular mass of 135 or 140 kDa (2–4, 10, 15) and as shorter species variously described as 70, 80, or 85 kDa that are expressed endogenously or induced through the action of exogenous serine proteases such as trypsin and plasmin (3, 10, 11, 16, 17). By analyzing cell lines exclusively passaged non-enzymatically, we show that the short endogenously expressed CDCP1 protein has a molecular mass of 70 kDa. We have also shown that trypsin and another serine protease, matriptase, generate CDCP1 of the same molecular mass (70 kDa). The difference in molecular mass reported by us for low molecular mass CDCP1 (70 kDa) and previously reported assignments of 80 and 85 kDa is likely due to the use of different molecular weight standards for protein sizing or possibly due to differential glycosylation or other post-translational modifications.
A significant finding from this study, obtained through immunopurification of 70 kDa CDCP1, N-terminal sequencing, and mutagenesis, is that proteolytic processing of this integral membrane protein occurs at adjacent sites, Arg-368 and Lys-369. These sites are located between two regions of unknown function that have low homology to CUB domains and span residues 221–348 and 417–544, respectively (14). Based upon the high degree of variation in sequence either side of Arg-368 and Lys-369 (KQSR↓KFVP and QSRK↓FVPG; arrow indicates cleavage site), we propose that processing at these sites is mediated by different serine proteases. This proposal is supported by our mutagenesis studies showing that for cleavage at Arg-368, the residues located in the P1′ (Lys-369) and P2′ (Phe-370) positions are important, while for cleavage at Lys-369 the P2 (Arg-368) position is important. Although we have shown that the proteolytic activity present in DU145, which cleaves at both Arg-368 and Lys-369, and the serine protease matriptase, which cleaves exclusively at Arg-368, both induce phosphorylation of 70 kDa CDCP1 and recruitment of Src and PKCδ, an interesting possibility is that cleavage at these two sites will induce different cellular responses via activation of different signaling pathways and release of ectodomains that differ by one residue at the C terminus.
Another interesting observation was our finding that the membrane-anchored and cell surface-shed serine protease matriptase is an efficient but not essential processor of CDCP1. Our finding that cleavage occurs at Arg-368 is supported by Bhatt et al. (4) who previously showed that the recombinant serine protease domain of matriptase, is able to cleave the purified recombinant extracellular domain of CDCP1 at this site in vitro. However, it is clear from our experiments using four different silencing constructs in two prostate cancer cell lines that reduction of matriptase expression does not reduce CDCP1 processing. What is not yet clear is whether endogenous matriptase cleaves CDCP1 in cultured cells or in vivo.
A possibility is that even though we reduced matriptase protein levels by up to ∼90%, this was not sufficient to reduce CDCP1 processing in PC3 and DU145 cells at Arg-368. Of relevance, it has previously been shown that, whereas deletion of the matriptase gene in mice results in neonatal death due to loss of epidermal and oral barrier function (36), hypomorphic mice, expressing matriptase mRNA at ∼1% of the levels seen in wild-type mice, survive normally with only a moderately impaired skin barrier function (37). Although this suggests that matriptase-mediated proteolysis must be completely reduced to block its normal role in maintenance of skin function, it is important to note that this study from List et al. (37) was not able to quantitatively measure residual matriptase proteolytic activity. Therefore it is possible that matriptase proteolytic activity in these hypomorphic mice was relatively high despite low mRNA levels.
We also note that if our proposal is correct that cleavage at Arg-368 and Lys-369 requires the action of two different serine proteases, silencing of one of these enzymes would not be expected to prevent CDCP1 processing, as cleavage at the second site would still occur. Accordingly, it remains feasible that matriptase is involved in CDCP1 processing in cells and in vivo, and it will not be practical to address this question further until it is possible to silence both matriptase and proteases capable of cleaving CDCP1 at Lys-369. Further, we note that there is additional published data supporting the proposal that matriptase is a relevant processor of CDCP1 at Arg-369. For example, Bhatt et al. (4) demonstrated that matriptase and CDCP1 co-immunoprecipitate from MDA-MB-468 breast cancer cells inducibly overexpressing CDCP1. This observation is supported by our immunoprecipitations showing that endogenous 135 kDa (but not 70 kDa) CDCP1 and matriptase co-immunoprecipitate from PC3 cells suggesting that these proteins interact via the shed CDCP1 fragment (residues 30–368) (supplemental Fig. S1). The existence of interactions between CDCP1 and matriptase is also supported by a recent report showing that these proteins co-immunoprecipitate from SW480 and SW620 colon cancer cell lysates using an antibody against the tetraspanin CD9 (35). In addition, it has been reported that the mRNA expression patterns of this protease:substrate pair are significantly correlated in many tissue types (38). It has also been shown in separate reports that both matriptase (39) and CDCP1 (12) are expressed during prostate cancer progression. In particular, as it is known that matriptase expression increases with increasing prostate cancer grade, with concomitant reduction in expression of its cognate inhibitor, hepatocyte growth factor activator inhibitor-1 (HAI-1) (39), it is possible that increased matriptase activity will result in increased CDCP1 processing during prostate cancer progression. Furthermore, it has been recognized that the residues N terminus to the CDCP1 R368 cleavage site (KQSR) match exactly with the peptide substrate specificity of matriptase determined using a positional scanning-synthetic combinatorial library approach (40). Finally, it has been shown that the trypsin-fold serine protease inhibitor ecotin, at 7 μm, markedly reduced processing in MDA-MB-468 cells stably overexpressing CDCP1 (4). These findings suggest that although matriptase silencing does not reduce proteolytic processing of CDCP1 in PC3 and DU145 cells, it is possible that matriptase cleavage of CDCP1 at Arg-368 may be relevant in pathological and physiological settings.
Based on our data it will be important to identify the endogenous serine proteases that cleave CDCP1 in normal and pathological settings. For example, it appears likely that loss of regulation of these proteases will lead to increased processing of CDCP1 and inappropriate amplification of downstream signaling (including recruitment of Src and PKCδ), potentially leading to cellular responses critical for disease progression. In addition to matriptase, plasmin, and trypsin, candidate proteases include secreted and shed members of the proprotein convertase family as these proteases cleave after arginine or lysine, preceded by a basic residue at the P2, P4, P6, and/or P8-position (41). Other candidates include the sheddases ADAM10 and ADAM17, which process a number of other cell surface proteins after basic residues (42) and the TTSP hepsin, which is structurally related to matriptase (43), has preference for cleavage after arginine/lysine residues (44), and is up-regulated in (45, 46) and proposed to promote prostate cancer progression (46–48).
Of relevance, it is clear from our inhibitor profile, that serine proteases mediating cleavage of CDCP1 at both Arg-368 and Lys-369 are sensitive to the serine protease inhibitor aprotinin (> 5 μm), with lower sensitivity to another serine protease inhibitor PMSF (> 2 mm) and the mixed serine/cysteine protease inhibitor leupeptin (> 50 μm). Although not completely analogous to inhibition of cleavage of macromolecular substrates such as CDCP1, the inhibitor profiles of matriptase and hepsin against a fluorogenic tripeptide substrate (QAR) overlap our observations of inhibition of CDCP1 processing suggesting that these proteases may be cellular processors of this cell surface protein-0.3 μm aprotinin and 1 μm leupeptin inhibit the activity of both proteases against this tripeptide substrate (49). In addition, significantly hepsin cleavage of the macromolecular substrate factor VII is inhibited by 8 μm aprotinin (50).
In summary, we have identified two residues, Arg-368 and Lys-369, as sites at which the integral membrane protein CDCP1 is proteolytically processed in cells. We have also shown that proteolysis induces recruitment of Src and PKCδ to tyrosine-phosphorylated 70 kDa CDCP1. These data provide a platform for further studies examining the biological function, molecular regulators, and downstream signaling events coupled to proteolytic processing of CDCP1 and whether these contribute to disease progression. Also, the observation that low and high molecular weight CDCP1 species are expressed in neonatal mouse epidermis (3), indicates that it will be important to understand mechanisms regulating CDCP1 processing in normal physiological settings.
Supplementary Material
Acknowledgments
We thank Dr. Georgina Giannikopoulos and Dr. Bernie McInerney for performing N-terminal sequencing, which was facilitated by access to the Australian Proteome Analysis Facility which is supported under the Australian Government National Collaborative Research Infrastructure Strategy (NCRIS). We also thank Carina Walpole and Dr. Sally-Anne Stephenson (Queensland University of Technology) for providing breast cancer cell lines, and James Broadbent for assistance with mass spectroscopy analysis.
This work was supported by the National Health and Medical Research Council of Australia (Fellowship 339732 to J. D. H.), by a Canadian Institutes of Health Research (CIHR) grant (to R. L.), and by funding supplied by the Cancer & Bowel Research Trust (scholarship to A. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- CDCP1
- CUB domain-containing protein 1
- HAI-1
- hepatocyte growth factor activator inhibitor-1
- TTSP
- type II transmembrane serine protease
- TPCK
- tosyl-l-phenylalanine chloromethyl ketone
- TLCK
- tosyl-l-lysine chloromethyl ketone.
REFERENCES
- 1.Scherl-Mostageer M., Sommergruber W., Abseher R., Hauptmann R., Ambros P., Schweifer N. (2001) Oncogene 20, 4402–4408 [DOI] [PubMed] [Google Scholar]
- 2.Hooper J. D., Zijlstra A., Aimes R. T., Liang H., Claassen G. F., Tarin D., Testa J. E., Quigley J. P. (2003) Oncogene 22, 1783–1794 [DOI] [PubMed] [Google Scholar]
- 3.Brown T. A., Yang T. M., Zaitsevskaia T., Xia Y., Dunn C. A., Sigle R. O., Knudsen B., Carter W. G. (2004) J. Biol. Chem. 279, 14772–14783 [DOI] [PubMed] [Google Scholar]
- 4.Bhatt A. S., Erdjument-Bromage H., Tempst P., Craik C. S., Moasser M. M. (2005) Oncogene 24, 5333–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bühring H. J., Kuçi S., Conze T., Rathke G., Bartolović K., Grünebach F., Scherl-Mostageer M., Brümmendorf T. H., Schweifer N., Lammers R. (2004) Stem. Cells 22, 334–343 [DOI] [PubMed] [Google Scholar]
- 6.Ikeda J. I., Morii E., Kimura H., Tomita Y., Takakuwa T., Hasegawa J. I., Kim Y. K., Miyoshi Y., Noguchi S., Nishida T., Aozasa K. (2006) J. Pathol. 210, 75–84 [DOI] [PubMed] [Google Scholar]
- 7.Perry S. E., Robinson P., Melcher A., Quirke P., Bühring H. J., Cook G. P., Blair G. E. (2007) FEBS Lett. 581, 1137–1142 [DOI] [PubMed] [Google Scholar]
- 8.Awakura Y., Nakamura E., Takahashi T., Kotani H., Mikami Y., Kadowaki T., Myoumoto A., Akiyama H., Ito N., Kamoto T., Manabe T., Nobumasa H., Tsujimoto G., Ogawa O. (2008) J. Cancer Res. Clin. Oncol. 134, 1363–1369 [DOI] [PubMed] [Google Scholar]
- 9.Ikeda J., Oda T., Inoue M., Uekita T., Sakai R., Okumura M., Aozasa K., Morii E. (2009) Cancer Sci. 100, 429–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uekita T., Jia L., Narisawa-Saito M., Yokota J., Kiyono T., Sakai R. (2007) Mol. Cell Biol. 27, 7649–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uekita T., Tanaka M., Takigahira M., Miyazawa Y., Nakanishi Y., Kanai Y., Yanagihara K., Sakai R. (2008) Am. J. Pathol. 172, 1729–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siva A. C., Wild M. A., Kirkland R. E., Nolan M. J., Lin B., Maruyama T., Yantiri-Wernimont F., Frederickson S., Bowdish K. S., Xin H. (2008) Cancer Res. 68, 3759–3766 [DOI] [PubMed] [Google Scholar]
- 13.Deryugina E. I., Conn E. M., Wortmann A., Partridge J. J., Kupriyanova T. A., Ardi V. C., Hooper J. D., Quigley J. P. (2009) Mol. Cancer Res. 7, 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wortmann A., He Y., Deryugina E. I., Quigley J. P., Hooper J. D. (2009) IUBMB Life 61, 723–730 [DOI] [PubMed] [Google Scholar]
- 15.Benes C. H., Wu N., Elia A. E., Dharia T., Cantley L. C., Soltoff S. P. (2005) Cell 121, 271–280 [DOI] [PubMed] [Google Scholar]
- 16.Wong C. H., Baehner F. L., Spassov D. S., Ahuja D., Wang D., Hann B., Blair J., Shokat K., Welm A. L., Moasser M. M. (2009) Clin. Cancer Res. 15, 2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spassov D. S., Baehner F. L., Wong C. H., McDonough S., Moasser M. M. (2009) Am. J. Pathol. 174, 1756–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bugge T. H., List K., Szabo R. (2007) Front Biosci. 12, 5060–5070 [DOI] [PubMed] [Google Scholar]
- 19.Désilets A., Béliveau F., Vandal G., McDuff F. O., Lavigne P., Leduc R. (2008) J. Biol. Chem. 283, 10535–10542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchhofer D., Peek M., Li W., Stamos J., Eigenbrot C., Kadkhodayan S., Elliott J. M., Corpuz R. T., Lazarus R. A., Moran P. (2003) J. Biol. Chem. 278, 36341–36349 [DOI] [PubMed] [Google Scholar]
- 21.Szabo R., Netzel-Arnett S., Hobson J. P., Antalis T. M., Bugge T. H. (2005) Biochem. J. 390, 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunitz M., Northrop J. H. (1936) J. Gen. Physiol. 19, 991–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed S., Jin X., Yagi M., Yasuda C., Sato Y., Higashi S., Lin C. Y., Dickson R. B., Miyazaki K. (2006) FEBS J. 273, 615–627 [DOI] [PubMed] [Google Scholar]
- 24.Coote K., Atherton-Watson H. C., Sugar R., Young A., MacKenzie-Beevor A., Gosling M., Bhalay G., Bloomfield G., Dunstan A., Bridges R. J., Sabater J. R., Abraham W. M., Tully D., Pacoma R., Schumacher A., Harris J., Danahay H. (2009) J. Pharmacol. Exp. Ther. 329, 764–774 [DOI] [PubMed] [Google Scholar]
- 25.Cowan F. M., Broomfield C. A., Smith W. J. (2002) Cell Biol. Toxicol. 18, 175–180 [DOI] [PubMed] [Google Scholar]
- 26.Hultsch T., Ennis M., Heidtmann H. H. (1988) Agents Actions 23, 198–200 [DOI] [PubMed] [Google Scholar]
- 27.Umezawa H. (1976) Methods Enzymol. 45, 678–695 [DOI] [PubMed] [Google Scholar]
- 28.Leppert D., Waubant E., Galardy R., Bunnett N. W., Hauser S. L. (1995) J. Immunol. 154, 4379–4389 [PubMed] [Google Scholar]
- 29.Olaso E., Ikeda K., Eng F. J., Xu L., Wang L. H., Lin H. C., Friedman S. L. (2001) J. Clin. Invest. 108, 1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi K., Chang W. J. (1976) J. Biochem. 80, 497–506 [DOI] [PubMed] [Google Scholar]
- 31.Smith R. A., Copp L. J., Donnelly S. L., Spencer R. W., Krantz A. (1988) Biochemistry 27, 6568–6573 [DOI] [PubMed] [Google Scholar]
- 32.Schechter I., Berger A. (1968) Biochem. Biophys. Res. Commun. 32, 898–902 [DOI] [PubMed] [Google Scholar]
- 33.Benaud C., Dickson R. B., Lin C. Y. (2001) Eur. J. Biochem. 268, 1439–1447 [DOI] [PubMed] [Google Scholar]
- 34.Calalb M. B., Zhang X., Polte T. R., Hanks S. K. (1996) Biochem. Biophys. Res. Commun. 228, 662–668 [DOI] [PubMed] [Google Scholar]
- 35.André M., Le Caer J. P., Greco C., Planchon S., El Nemer W., Boucheix C., Rubinstein E., Chamot-Rooke J., Le Naour F. (2006) Proteomics 6, 1437–1449 [DOI] [PubMed] [Google Scholar]
- 36.List K., Haudenschild C. C., Szabo R., Chen W., Wahl S. M., Swaim W., Engelholm L. H., Behrendt N., Bugge T. H. (2002) Oncogene 21, 3765–3779 [DOI] [PubMed] [Google Scholar]
- 37.List K., Currie B., Scharschmidt T. C., Szabo R., Shireman J., Molinolo A., Cravatt B. F., Segre J., Bugge T. H. (2007) J. Biol. Chem. 282, 36714–36723 [DOI] [PubMed] [Google Scholar]
- 38.Bhatt A. S., Welm A., Farady C. J., Vásquez M., Wilson K., Craik C. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5771–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saleem M., Adhami V. M., Zhong W., Longley B. J., Lin C. Y., Dickson R. B., Reagan-Shaw S., Jarrard D. F., Mukhtar H. (2006) Cancer Epidemiol. Biomarkers Prev. 15, 217–227 [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi T., Harris J. L., Huang W., Yan K. W., Coughlin S. R., Craik C. S. (2000) J. Biol. Chem. 275, 26333–26342 [DOI] [PubMed] [Google Scholar]
- 41.Chrétien M., Seidah N. G., Basak A., Mbikay M. (2008) Exp. Opin. Ther. Targets 12, 1289–1300 [DOI] [PubMed] [Google Scholar]
- 42.Caescu C. I., Jeschke G. R., Turk B. E. (2009) Biochem. J. 424, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooper J. D., Clements J. A., Quigley J. P., Antalis T. M. (2001) J. Biol. Chem. 276, 857–860 [DOI] [PubMed] [Google Scholar]
- 44.Zhukov A., Hellman U., Ingelman-Sundberg M. (1997) Biochim. Biophys. Acta 1337, 85–95 [DOI] [PubMed] [Google Scholar]
- 45.Dhanasekaran S. M., Barrette T. R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2001) Nature 412, 822–826 [DOI] [PubMed] [Google Scholar]
- 46.Xuan J. A., Schneider D., Toy P., Lin R., Newton A., Zhu Y., Finster S., Vogel D., Mintzer B., Dinter H., Light D., Parry R., Polokoff M., Whitlow M., Wu Q., Parry G. (2006) Cancer Res. 66, 3611–3619 [DOI] [PubMed] [Google Scholar]
- 47.Klezovitch O., Chevillet J., Mirosevich J., Roberts R. L., Matusik R. J., Vasioukhin V. (2004) Cancer Cell 6, 185–195 [DOI] [PubMed] [Google Scholar]
- 48.Li W., Wang B. E., Moran P., Lipari T., Ganesan R., Corpuz R., Ludlam M. J., Gogineni A., Koeppen H., Bunting S., Gao W. Q., Kirchhofer D. (2009) Cancer Res. 69, 8395–8402 [DOI] [PubMed] [Google Scholar]
- 49.Béliveau F., Désilets A., Leduc R. (2009) FEBS J. 276, 2213–2226 [DOI] [PubMed] [Google Scholar]
- 50.Kazama Y., Hamamoto T., Foster D. C., Kisiel W. (1995) J. Biol. Chem. 270, 66–72 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.