Abstract
During tumor progression, malignant cells must repeatedly survive microenvironmental stress. Hypoxia-inducible factor-1 (HIF-1) signaling has emerged as one major pathway allowing cellular adaptation to stress. Recent findings led to the hypothesis that HIF-1α may enhance the metastatic potential of tumor cells by a survival-independent mechanism. So far it has not been shown that HIF-1α also directly regulates invasive processes during metastasis in addition to conferring a survival advantage to metastasizing tumor cells. In a hypoxia-tolerant tumor cell line (L-CI.5s), which did not rely on HIF-1 signaling for viability in vitro and in vivo, knockdown of Hif-1α reduced invasiveness of the tumor cells in vitro as well as extravasation and secondary infiltration in vivo. Liver metastases associated induction of proinvasive receptor tyrosine kinase Met phosphorylation as well as gelatinolytic activity were Hif-1α-dependent. Indeed, promoter activity of the matrix metalloproteinase-9 (mmp-9) was shown to be Hif-1α-dependent. This study uncovers a new survival-independent biological function of HIF-1α contributing to the efficacy of metastases formation.
Keywords: Matrix Metalloproteinase, Metalloprotease, Protease, Tumor Metastases, Tumor Promoter, HIF-1α, Hypoxia Tolerance, Invasion, MET, MMP-9
Introduction
During the metastatic cascade, tumor cells encounter several kinds of microenvironmental stress, namely lack of oxygen and nutrients (1). Hypoxia-inducible factor-1 (HIF-1)2 is a transcription factor known to mediate the adaptation to microenvironmental stress in general (2) as well as during tumor progression in particular (1). HIF-1 consists of a constitutively expressed β-subunit and a highly regulated α-subunit that is, under physiological conditions, degraded by the proteasome (3, 4). During stress situations such as hypoxia, HIF-1α is stabilized and translocates to the nucleus where it forms together with HIF-1β the heterodimeric transcription factor HIF-1 (5). Via its interaction with hypoxia-responsive elements, HIF-1 regulates the expression of molecules such as the major pH-regulating enzyme carbonic anhydrase IX (CAIX) (6), which allows the metabolic adaptation on the cellular level (7, 8). Furthermore, HIF-1 signaling is also a major determinant of adaptation on the tissue level by induction of the “angiogenic switch,” thereby overcoming the limited supply of oxygen and nutrients in expanding neoplasias (1).
Recent findings led us to the hypothesis that HIF-1α not only impacts on metastasis formation by securing survival but also directly impacts on metastasis in a survival-independent manner. This hypothesis was based on the following findings. Reduced levels of HIF-1α correlate with decreased invasive potential of tumor cells in vitro (9–11). HIF-1α knock-out reduced primary tumor onset and growth in a transgenic model of cancer initiation (12). This correlated with a later onset of pulmonary metastasis (12). Furthermore, constitutive expression of HIF-1α has been shown to enhance bone metastasis in a breast cancer model (13). Although these data have suggested that HIF-1α increases the metastatic potential of tumor cells, recent reviews by Bertout et al. (14) and Ruan et al. (15) have pointed out that it has so far remained unclear whether HIF-1α regulates tumor cell invasiveness directly in vivo and, if so, which molecular mechanisms determine this metastasis-promoting function (14, 15).
The difficulty in clarifying this topic has been caused mainly by the fact that under microenvironmental stress, the survival of tumor cells usually depends on HIF-1 signaling (16). Consequently, so far, it could not be excluded that reduced metastasis upon HIF-1α depletion was simply a reflection of decreased survival. To elucidate whether HIF-1α only promotes metastasis by securing survival (8, 16, 17) or whether HIF-1α really promotes the invasive potential of tumor cells as one specific hallmark of cancer (17), it is advantageous to use a hypoxia-tolerant tumor cell line, which does not rely on HIF-1 signaling for survival under hypoxic conditions.
Several studies have proposed a link between HIF-1 signaling and the expression of proteolytic enzymes such as matrix metalloproteinases (MMPs) (10, 18, 19). This phenomenon is of great relevance as the invasive steps of tumor progression are characterized by imbalances in the “protease web,” comprising the complex network of proteases, their inhibitors, and effector molecules (20–22). Specifically, up-regulation of MMP-9 in the tumor microenvironment (23, 24) accounts for the degradation of the basal membrane and the remodeling of the extracellular matrix (ECM) to clear the path for tumor cells during the invasive steps of metastasis (25). Another important mediator of tumor cell scattering from already established metastases is the hepatocyte growth factor (HGF) signaling pathway (26), initiated by binding of HGF, originally also described as the “scatter factor” (27), to its receptor MET. MET expression was shown to be up-regulated by elevated levels of HIF-1α (28). Taken together, HIF-1α may be able to act as a promoter of tumor cell invasion via both ways, the induction of proteolytic activity as well as triggering HGF signaling.
The L-CI.5s murine T-lymphoma cell line (29) was shown here to be independent from HIF-1 signaling for survival under hypoxic conditions, therefore representing the necessary model to study the effects of HIF-1α on invasiveness of tumor cells without interference with survival. We here were able to show for the first time that Hif-1α knockdown in tumor cells indeed drastically reduced their invasiveness in vitro and in vivo. HIF-1α knockdown reduced the efficacy of extravasation, formation of metastatic colonies, and secondary infiltration from established metastases. Decreased proteolytic activity in primary tumors and metastases upon Hif-1α knockdown in tumor cells correlated with diminished expression of Mmp-9 in vivo and reduced mmp-9 promoter activity in vitro. Furthermore, Hif-1α deficiency impaired HGF signaling. Similar antimetastatic effects of Hif-1α knockdown were shown in a lung metastasis model of CT-26L murine colon carcinoma cells. Taking into consideration that HIF-1 signaling is detected in a large variety of cancers (30) and often correlates with poor prognosis (31), our findings support the usefulness of HIF-1α-directed therapies in advanced tumor disease.
EXPERIMENTAL PROCEDURES
Cell Lines
293T (32) as well as lacZ-tagged L-CI.5s (29) and CT26L cells (33) were cultured as described previously. Stable knockdown of HIF-1α was achieved by lentiviral gene transfer of two shRNAs (shHIF-1α, TRCN0000054450; shHIF-1α_2, TRCN0000054451). A control cell line was generated by lentiviral gene transfer of a non-targeting shRNA sequence (shNT) that does not target any murine gene. Plasmids were purchased from Sigma-Aldrich Chemie, Deisenhofen, Germany. Lentiviruses were generated using respective plasmids and the ViraPowerTM lentiviral expression system (Invitrogen, Karlsruhe, Germany). Transduction was done as described previously (23).
Experimental Metastasis Assay
5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells were inoculated into the tail vein of pathogen-free, immune-competent, syngeneic female DBA/2 mice. To analyze extravasation events during liver metastasis, 2 × 106 tumor cells were inoculated. In a second assay, pathogen-free, immune-competent, syngeneic female BALB/C mice were injected i.v. with 1 × 106 CT-26L cells. Mice were sacrificed by cervical dislocation, and livers and lungs were removed. The right lung lobe as well as three samples of each liver were snap-frozen in liquid nitrogen and stored at −80 °C for biochemical analysis. For histological analyses, two samples of each liver were analogously conserved or fixed in alcohol and then paraffin-embedded. Remaining liver and lung tissue was used for X-gal (5-bromo-4-chloro-3-indolyl-β-d-galatopyranoside) staining (see below).
Spontaneous Metastasis Assay
1 × 105 L-CI.5s-shNT or L-CI.5s-shHIF-1α were injected intradermally in the flank of DBA/2 mice. Primary tumor growth was documented using a caliper. Mice were sacrificed at primary tumor diameters of 7 mm, and primary tumors, livers, and lungs were removed. Half of the primary tumor was snap-frozen in liquid nitrogen and stored at −80 °C for biochemical analysis. The rest was fixed in alcohol and embedded in paraffin for immunohistochemistry. Livers and lungs were removed and treated as described above. All animal experiments were done in compliance with the guidelines of the Tierschutzgesetz des Landes Bayern.
Histological Analyses
Paraffin-embedded liver sections were dewaxed and rehydrated as described previously (34). For antigen retrieval, liver slices were cooked in citrate buffer (pH = 6.0) for 15 min. Immunostaining was performed according to the staining kit (R&D Systems, Wiesbaden, Germany) using rabbit polyclonal antibodies against HIF-1α (Novus Biologicals, Littleton, CO) or proliferating nuclear antigen (Abcam, Cambridge, MA), respectively. Apoptosis assay on liver and primary tumor sections was performed with the TACSTM terminal deoxynucleotidyl transferase kit (R&D Systems). In situ zymography was done as described previously (35).
X-Gal Staining
Complete liver and lung lobes were stained with X-gal (Roche Diagnostics, Penzberg, Germany) as described previously (29). Indigo blue multicellular foci on the liver surface >0.2 mm were qualified as metastases and counted. For X-gal staining of frozen liver slices (8 μm), tissue was fixed in PBS containing 2% (v/v) formaldehyde and 0.2% (v/v) glutaraldehyde, washed in PBS containing 2 mm MgCl2, and incubated in PBS containing 2 mm MgCl2, 0.01% (w/v) sodium deoxycholate, and 0.02% (v/v) Nonidet-P40 for 10 min each. Then, the slices were incubated for 12 h with X-gal as described earlier (29). After counterstaining with eosin, slices were mounted with glycerin-gelatin. To quantify the metastatic burden in the liver 3 and 6 h after tumor cell inoculation, single indigo blue-stained tumor cells were counted, and their number was normalized to the area of the section. To analyze tumor cell density in metastases, tumor cells were counted inside metastatic foci, and their number was normalized to the area of metastasis.
RNA Isolation, Reverse Transcription, and qRT-PCR
RNA was isolated using PureYieldTM RNA Midiprep system (Promega, Mannheim, Germany). Reverse transcription was done with a High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). qRT-PCR was performed as described previously (36) using primers and probes from Applied Biosystems.
In Vitro Assays
For the alamarBlue® cell viability assay (Invitrogen), L-CI.5s and CT-26L cells were seeded and cultured under normoxic (21% (v/v) O2) or hypoxic (1% (v/v) O2) conditions and counted 6, 24, and 48 h after seeding. According to standard cell culture conditions used for these assays, we cultivated cells under normoxic conditions at 21% O2. Although these conditions are rather hyperoxic and do not reflect normoxia on the tissue level, the conditions of 21% O2 and 1% O2 are suitable to activate HIF-1 signaling (4). For the Boyden chamber invasion assay, 2 × 106 tumor cells were seeded in serum-free medium in 6.5 mm Transwell® inserts with a pore size of 3 μm (Corning, Corning, NY), which had been coated with 78 μg/cm2 Matrigel (BD Biosciences, Heidelberg, Germany). Medium with FCS were given in the lower chamber of the assay. 48 h later, L-CI.5s cells in the lower chamber were counted. mmp-9 promoter activity was analyzed using a chimeric luciferase-MMP-9 promoter construct (37), cloned into the pLenti vector (Invitrogen) according to standard protocols. Transduction was performed as described above. 2 × 106 L-CI.5s cells were cultured for 20 h with or without supplementation of 100 μm desferrioxamine mesylate (DFO) (Sigma-Aldrich Chemie), RNA was isolated, and luciferase mRNA levels were assessed (see above).
Statistical Analysis
Statistical analysis was done using Student's t test when data were normally distributed. Otherwise, Mann-Whitney U rank sum test was used. p < 0.05 was considered significant.
RESULTS
Hif-1α Knockdown Decreased Invasiveness of L-CI.5s Cells in Vitro and in Vivo
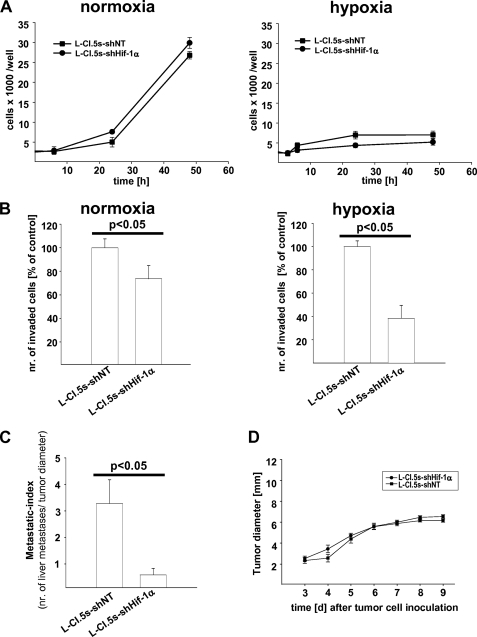
First, we assessed whether Hif-1α knockdown in L-CI.5s cells (knockdown efficacy: −90% on mRNA level as compared with the shNT control) influenced their survival in vitro. We quantified tumor cells cultured under normoxic or hypoxic conditions at several time points. The number of tumor cells was drastically diminished under hypoxia (Fig. 1A). However, Hif-1α knockdown reduced the number of tumor cells neither under hypoxia nor under normoxia (Fig. 1A), indicating that L-CI.5s cells did not require Hif-1α for survival. Next, we tested whether lack of Hif-1α reduced the invasiveness of L-CI.5s cells in vitro under both normoxic and hypoxic conditions. Hif-1α knockdown resulted in a significant reduction of invasiveness under hypoxic conditions (Fig. 1B). Importantly, even under normoxia, L-CI.5s cells were less invasive upon Hif-1α knockdown (Fig. 1B). Taken together, Hif-1α regulated the invasiveness of L-CI.5s cells in vitro in a survival-independent manner under both normoxia and hypoxia. In vivo, the metastatic index, which normalized the number of liver metastases to the diameter of primary tumors, was significantly reduced when Hif-1α-deficient tumor cells had been inoculated (Fig. 1C). In line with our in vitro results, we also observed no changes in primary tumor growth (Fig. 1D), indicating that survival of L-CI.5s cells was not affected by lack of Hif-1α.
FIGURE 1.
A, mean cell number in thousands per well ± S.E. (squares and bars), determined by alamarBlue proliferation assays of L-CI.5s-shNT and L-CI.5s-shHIF-1α cells under normoxic or hypoxic conditions (1% (v/v) O2). Proliferation of both cell lines was diminished under hypoxia, but lack of HIF-1α did not significantly diminish proliferation further. B, mean number (nr.) of invaded cells ± S.E. (columns and bars), quantified by Transwell invasion assays of L-CI.5s-shNT and L-CI.5s-shHIF-1α cells under normoxic or hypoxic conditions (1% (v/v) O2), relative to the reference group L-CI.5s-shNT; n = 4. L-CI.5s-shNT/normoxia, 100.0 ± 7.6%; L-CI.5s-shHIF-1α/normoxia, 73.6 ± 11.2%; L-CI.5s-shNT/hypoxia, 100.0 ± 2.8%; L-CI.5s-shHIF-1α/hypoxia, 38.4 ± 5.6%. C, number of spontaneous liver metastasis ± S.E. (columns and bars) normalized to diameters of the primary tumors after intradermal inoculation of 1 × 105 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells; n = 7 mice. L-CI.5s-shNT, 3.3 ± 0.9; L-CI.5s-shHIF-1α, 0.6 ± 0.3. D, mean diameter of primary tumors ± S.E. (squares and bars) after intradermal inoculation of 1 × 105 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Lack of Hif-1α did not affect primary tumor growth in vivo.
Lack of Hif-1α Reduced Experimental Liver Metastasis
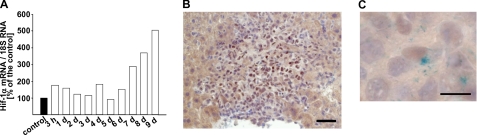
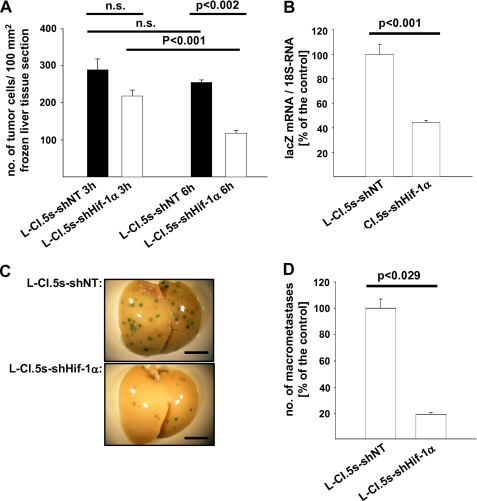
To overcome the limitation of spontaneous metastasis assays, i.e. not knowing how many tumor cells dissociate from a primary tumor at a given time point, we performed an experimental liver metastasis assay where we inoculated a defined number of tumor cells in the tail vein. qRT-PCR revealed that Hif-1α mRNA levels in the liver continuously ascended during colonization of this target organ of metastasis (Fig. 2A). Immunohistochemical analysis documented a strong staining for Hif-1α protein mainly in tumor cells located in the center of metastases (Fig. 2B). By combining immunohistochemistry with X-gal staining to identify L-CI.5s cells (29), we elucidated the presence of Hif-1α in the nucleus of tumor cells (Fig. 2C), suggesting activation of HIF-1 signaling. To determine the ability of the lacZ-tagged tumor cells to extravasate and to lodge in the liver parenchyma, invaded single tumor cells were counted at 3 and 6 h after their inoculation. At both time points, the number of tumor cells lacking Hif-1α in the liver was significantly lower (Fig. 3A), suggesting that Hif-1α mediated tumor cell extravasation. Importantly, Hif-1α mRNA levels (supplemental Fig. 1A) as well as the expression of the HIF-1 target gene (6) CAIX (supplemental Fig. 1B) were induced at 6 h after tumor cell inoculation. To elucidate the impact of Hif-1α deficiency on further progression of liver metastasis, we measured the tumor cell burden at 3 days after their inoculation. A significant decrease of the metastatic load was detected with tumor cells lacking Hif-1α (Fig. 3B). Also, a significantly reduced number of metastatic foci was found at day 7 after tumor cell inoculation (Fig. 3C). This result was confirmed with a second shRNA against Hif-1α (knockdown efficacy: −80% on mRNA level as compared with the shNT control) (supplemental Fig. 2A).
FIGURE 2.
A, Hif-1α mRNA levels continuously ascended during colonization of the liver after intravenous inoculation of 5 × 103 L-CI. 5s-shNT or L-CI.5s-shHIF-1α cells. RNA of five animals was pooled. Hif-1α mRNA levels were normalized to 18 S mRNA levels, and the Hif-1α mRNA level in the absence of tumor cells was set as 100%. B, representative microscopic image of an immunohistochemical Hif-1α staining 7 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Bar, 25 μm. C, representative microscopic image of a single tumor cell in the liver 7 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Tissue was stained with X-gal (indigo blue cytoplasmic signal) and immunohistochemically for Hif-1α (brown nuclear signal). Bar, 10 μm.
FIGURE 3.
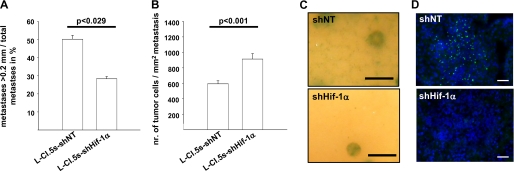
A, number of invaded tumor cells per mm2 of liver parenchyma ± S. E. (columns and bars) 3 or 6 h after intravenous inoculation of 2 × 106 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells, respectively; n = 3 mice. L-CI.5s-shNT/3h, 289.1 ± 29.3; L-CI.5s-shHIF-1α/3h, 218.4 ± 16.0; L-CI.5s-shNT/6h, 255.3 ± 6.4; L-CI.5s-shHIF-1α/6h, 117.9 ± 8.0. n.s., not significant. B, mean lacZ mRNA expression in the liver ± S.E. (columns and bars) 3 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHif-1α cells. lacZ mRNA levels were normalized to 18 S mRNA levels, and the reference group L-CI.5s-shNT was set as 100%; n = 3 mice. L-CI.5s-shNT, 100 ± 7.99%; L-CI.5s-shHIF-1α, 44.38 ± 1.51%. C and D, X-gal-stained (indigo blue signal) metastasis-bearing liver lobes 7 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. C, representative overview screens. Bars, 1 cm. D, mean number of metastatic foci > 0.2 mm ± S.E. (columns and bars) on the surfaces of the liver lobes, relative to the reference group L-CI.5s-shNT; n = 4 mice. L-CI.5s-shNT, 100.0 ± 6.9%; L-CI.5s-shHIF-1α, 19.5 ± 1.6%.
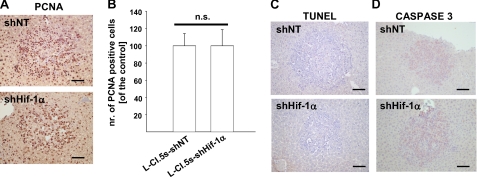
Hif-1α Knockdown Did Not Affect Tumor Cell Proliferation and Apoptosis in Vivo
To finally rule out the that the diminished number of metastases was caused by reduced survival of L-CI.5s cells upon Hif-1α knockdown in vivo, we examined proliferation and apoptosis in metastases in the liver tissue. Staining for the proliferating nuclear antigen revealed that proliferation of L-CI.5s cells within metastases was not affected by lack of Hif-1α (Fig. 4, A and B). Furthermore, TUNEL and caspase-3 analysis showed no difference in apoptosis between tumor cells with and without Hif-1α expression (Fig. 4, C and D). Thus, in analogy to the in vitro data (Fig. 1A) and the primary tumor growth (Fig. 1D), neither proliferation nor apoptosis of L-CI.5s cells in the liver was affected by Hif-1α knockdown.
FIGURE 4.
A and B, immunohistochemical proliferating nuclear antigen (PCNA) staining (brown signal) 7 days after intravenous inoculation of 5 × 103 L-CI. 5s-shNT or L-CI.5s-shHIF-1α cells. A, representative microscopic images. Bars, 25 μm. B, mean number (nr.) of proliferating nuclear antigen-positive tumor cells inside liver metastases ± S.E. (columns and bars), relative to the reference group L-CI.5s-shNT; n = 3 4mice. L-CI.5s-shNT, 100.0 ± 14.9%; L-CI.5s-shHIF-1α, 101.4 ± 18.6%. n.s., not significant. C and D, representative microscopic images of TUNEL staining (brown signal) (C) and immunohistochemical staining for caspase-3 (brown signal) (D) of liver metastases 7 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Bars, 25 μm. Lack of HIF-1α did not affect apoptosis within metastases.
Lack of Hif-1α Led to Decreased Tumor Cell Scattering
Measuring the mean diameter of established metastases revealed a significantly reduced size of metastatic foci upon Hif-1α knockdown (Fig. 5A). To clarify this finding, we counted the number of tumor cells within metastatic foci, revealing increased tumor cell density in metastases when Hif-1α was knocked down (Fig. 5B). X-Gal staining of liver lobes suggested that the increased compactness of metastases upon Hif-1α knockdown was due to decreased secondary invasion emanating from established metastases (Fig. 5C). As HGF signaling is known to be an important mediator of tumor cell scattering, we analyzed metastatic foci for Met phosphorylation. Indeed, lack of Hif-1α, as achieved by both shRNAs used, drastically reduced activation of this pathway within liver metastases (Fig. 5D and supplemental Fig. 2B), providing one explanation for decreased tumor cell scattering upon Hif-1α ablation.
FIGURE 5.
A, ratio of metastases > 0. 2 mm per total number of metastases ± S.E. (columns and bars) 7 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells; n = 4 mice. L-CI.5s-shNT, 50.3 ± 2.1%; L-CI.5s-shHIF-1α, 28.3 ± 1.2%. B, mean number of tumor cells per mm2 of metastasis ± S.E. (columns and bars) 7 days after inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells; n = 3. L-CI.5s-shNT, 592.8 ± 39.2; L-CI.5s-shHIF-1α, 913.2 ± 69.3. C, representative close-up pictures of X-gal-stained (indigo blue signal) liver lobes 7 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Bars, 1 mm. Lack of Hif-1α led to a strong reduction of tumor cell scattering from established metastases. D, immunofluorescence analysis for phosphorylated Met (green signal) in liver metastases 7 days after intravenous inoculation of 5 × 103 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Counterstaining was done with DAPI (blue signal). Bars, 50 μm. Lack of Hif-1α led to a strong reduction of Met phosphorylation within metastases.
Hif-1α Knockdown Reduced MMP-9 Expression and Gelatinolytic Activity
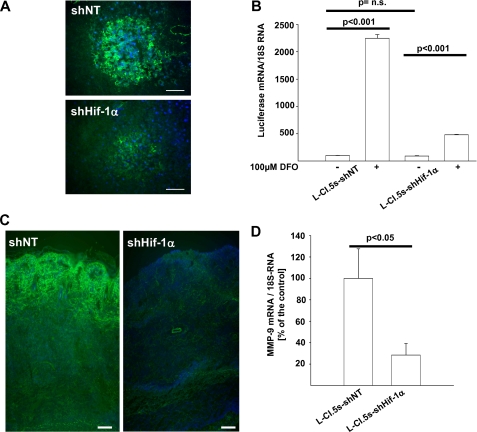
As secondary invasion of L-CI.5s cells depends on MMP-9 expression (23), we tested whether lack of Hif-1α interfered with gelatinolytic activity. In situ zymography indicated that Hif-1α deficiency, as achieved by both shRNAs used, indeed reduced gelatinolytic activity in metastases (Fig. 6A and supplemental Fig. 3A). To reveal whether Hif-1α-dependent gelatinolytic activity relied on mmp-9 expression in tumor cells, we employed an MMP-9 luciferase promoter construct. Induction of HIF-1 signaling in vitro by the HIF-1 signaling inducer DFO (38) drastically increased mmp-9 promoter activity in the L-CI.5s-shNT cells (Fig. 6B and supplemental Fig. 3B). Hif-1α knockdown drastically diminished induction of mmp-9 promoter activity in vitro, which was stimulated by DFO (Fig. 6B and supplemental Fig. 3B). This indicated that Hif-1α took part in the regulation of mmp-9 expression. qRT-PCR analysis showed that mmp-9 expression (supplemental Fig. 1C) as well as gelatinolytic activity in metastases correlated with Hif-1α mRNA levels and signaling also in vivo (supplemental Fig. 1, A and B), suggesting that Hif-1α regulated invasion of tumor cells into the liver at least in part by inducing mmp-9 expression. Next, we hypothesized that the observed reduction of spontaneous liver metastasis (Fig. 1C) also relied on decreased gelatinolytic activity and mmp-9 expression at the primary site. Indeed, in situ zymography and qRT-PCR revealed a decreased gelatinolytic activity (Fig. 6C) and reduced mmp-9 mRNA levels (Fig. 6D) in the primary tumor, again documenting an important role of MMP-9 in mediating HIF-1α-dependent invasion.
FIGURE 6.
A, in situ zymography of liver metastases 7 days after intravenous inoculation of 5 × 103 L-CI. 5s-shNT or L-CI.5s-shHIF-1α cells. Counterstaining was done with DAPI (blue signal). Bars, 25 μm. Lack of HIF-1α led to a strong reduction of gelatinolytic activity (green signal) within metastases. B, mean luciferase mRNA expression ± S.E. (columns and bars) in L-CI.5s-shNT and L-CI.5s-shHIF-1α cells, cultured with or without 100 μm DFO. Luciferase mRNA levels were normalized to 18 S mRNA levels, and the reference group L-CI.5s-shNT was set as 100%; n = 3. L-CI.5s-shNT/without DFO, 100.0 ± 4.7%; L-CI.5s-shNT/with DFO, 2246.2 ± 69.2%; L-CI.5s-shHIF-1α/without DFO, 90.4 ± 8.4%; L-CI.5s-shHIF-1α/with DFO, 479.4 ± 8.9%. n.s., not significant. C, in situ zymography of primary tumors after intradermal inoculation of 1 × 105 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Counterstaining was done with DAPI (blue signal). Bars, 100 μm. Lack of Hif-1α led to a strong reduction of gelatinolytic activity (green signal) within primary tumors. D, mean Mmp-9 mRNA expression ± S.E. (columns and bars) in primary tumors after intradermal inoculation of 1 × 105 L-CI.5s-shNT or L-CI.5s-shHIF-1α cells. Mmp-9 mRNA levels were normalized to 18 S mRNA levels, and the reference group L-CI.5s-shNT was set as 100%; n = 4 mice. L-CI.5s-shNT, 100.0 ± 7.6%; L-CI.5s-shHIF-1α, 28.8 ± 10.7%.
Hif-1α Deficiency Diminished Experimental Lung Metastasis of CT-26L Cells
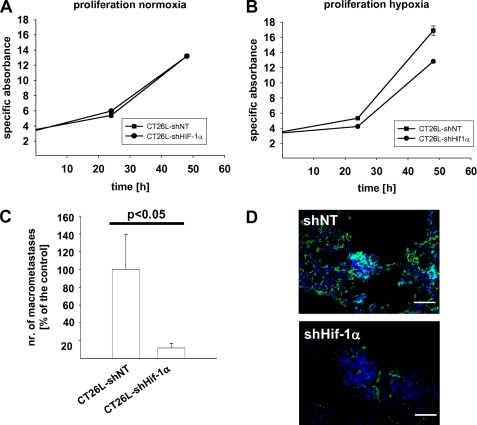
We then tested whether the observed impact of Hif-1α on tumor cell invasiveness by regulating the activity of gelatinolytic enzymes can be extended to conventional tumor models where survival is dependent on HIF-1 signaling. We employed the murine colon carcinoma cell line CT-26L, which can be tested in experimental metastasis assays in syngeneic BALB/C mice. In vitro, Hif-1α knockdown drastically reduced survival of these tumor cells under hypoxia (Fig. 7B), whereas their survival was not affected under normoxia (Fig. 7A). Consequently, the observed drastic reduction of lung metastases formation (Fig. 7C) upon Hif-1α knockdown could in part be attributed to reduced tumor cell survival in vivo. However, despite this interference with tumor cell survival, lack of Hif-1α also led to a strong reduction of gelatinolytic activity within lung metastases (Fig. 7D).
FIGURE 7.
A and B, mean cell number in thousands per well ± S. E. (dots and bars), determined by alamarBlue proliferation assay of CT-26L-shNT and CT-26L-shHIF-1α cells under normoxic (A) or hypoxic conditions (1% (v/v) O2) (B). Hif-1α knockdown drastically affected tumor cell survival only under hypoxia. C, mean number (nr.) of lung metastases >0.2 mm ± S.E. (columns and bars) 20 days after intravenous inoculation of 1 × 106 CT-26L-shNT or CT-26L-shHIF-1α cells, relative to the reference group CT-26L-shNT. CT26L-shNT, 100.0 ± 39.6%, n = 5 mice; CT-26L-shHIF-1α, 11.9 ± 5.2%, n = 7 mice. D, in situ zymography of lung metastases 20 days after intravenous inoculation of 1 × 106 CT-26L-shNT or CT-26L-shHIF-1α cells. Counterstaining was done with DAPI (blue signal). Bars, 25 μm. Lack of Hif-1α led to a strong reduction of gelatinolytic activity (green signal) within metastases.
DISCUSSION
Recent findings (9–13) led us to the hypothesis that HIF-1α increases the metastatic potential of tumor cells in a survival-independent manner. Based on these findings, Bertout et al. (14) and Ruan et al. (15) have also pointed out that it has so far remained unclear whether HIF-1α regulates tumor cell invasiveness directly in vivo (14) and, if so, which molecular mechanisms determine this metastasis-promoting function (15).
The reason for the difficulty in clarifying this topic until now has been the dependence of most tumor cell lines on HIF-1 signaling for survival (16, 39). By using a hypoxia-tolerant tumor cell line, here we were able to identify a survival-independent role of Hif-1α in enhancing the metastatic potential of tumor cells, namely its ability to regulate invasive processes in vivo via induction of Met signaling and Mmp-9.
It has been known for a long time that HIF-1α takes part in the cellular adaptation to stress situations such as lack of oxygen and nutrients (40). However, various studies suggest that HIF-1α not only regulates cellular metabolism, proliferation, and angiogenesis (40) but also induces tumor cell invasiveness under hypoxic conditions in vitro (9–11), thereby meeting one important hallmark of cancer (17). Liao et al. (12) showed that lack of HIF-1α resulted in delayed tumor onset, reduced tumor growth, and a diminished number of blood vessels, leading to a later onset of lung metastasis of the spontaneous PymT-model. Other studies elucidated that HIF-1α depletion reduced the size of experimental bone metastases of a human breast carcinoma cell line in nude mice (13) and that inhibition of HIF-1α by a synthetic inhibitor blocked outgrowth of pulmonary metastases of a human lung carcinoma in an experimental xenograft metastasis assay (41). However, these studies could not resolve whether the antitumorigenic and antimetastatic effect of HIF-1α deficiency can only be attributed to decreased survival of tumor cells within unfavorable microenvironments or whether HIF-1α directly impacts on tumor cell invasiveness in vivo, e.g. via induced expression of prometastatic genes and subsequent prometastatic modulation of the tumor microenvironment (15).
The impact of HIF-1α on the functional capabilities of tumor cells, survival and invasiveness, which are essential for metastasis, can therefore only be dissected by using a hypoxia-tolerant tumor cell line that does not rely on HIF-1 signaling for its survival. In contrast to the tumor cell lines, which all depend on HIF-1 signaling for survival under hypoxic conditions (42–44), L-CI.5s cells did not rely on this pathway as lack of HIF-1α in these cells did not alter survival in vitro neither under normoxic nor under hypoxic conditions. Regarding tumor cell invasiveness in vitro, L-CI.5s were absolutely comparable with the sensitivity of the other tumor cell lines to lack of Hif-1α (9, 11). L-CI.5s tumor cells also showed, upon Hif-1α knockdown, reduced spontaneous metastasis, an effect that is also observed in other tumor models (12). By using an experimental metastasis assay in which the inoculated L-CI.5s turned out to be independent from HIF-1 signaling for survival even in vivo, we here uncovered for the first time that HIF-1α is directly involved in the invasive phases of metastasis without interference with survival. Increased expression of Hif-1α and CAIX, which is a well known downstream target (6), as well as nuclear staining of Hif-1α in tumor cells documented the induction of HIF-1 signaling during these early invasive phases of metastasis. Consequently, knockdown of Hif-1α reduced the ability of L-CI.5s cells to lodge in the liver during these early phases. Furthermore, Hif-1α expression was also up-regulated at days 7–9 after tumor cell inoculation, representing the time point of secondary invasion of scattered tumor cells throughout the liver parenchyma (23). In the present study, at these time points, knockdown of Hif-1α drastically reduced invasive events emanating from established metastases into the surrounding liver parenchyma.
Furthermore, we could identify two mechanisms by which HIF-1α could influence the metastatic potential of tumor cells. First, reduced occurrence of metastases upon Hif-1α knockdown correlated with drastically decreased gelatinolytic activity in primary tumors and metastases. As the gelatinase MMP-9 is a known mediator of tumor cell invasion (25) and has recently been shown to be the crucial gelatinase for the invasive steps of metastasis (23, 24), the fact that its expression correlated with Hif-1α expression suggests that HIF-1α was involved in promoting MMP-9-dependent metastasis. Using a model of experimental lung metastasis of a murine colon carcinoma (CT-26L) (33), we further obtained evidence that Hif-1α-dependent peritumoral gelatinolytic activity was neither restricted to T-lymphoma metastasis nor organ-specific for the liver, although it is important to note that CT-26L cells were dependent on HIF-1 signaling for survival under hypoxic conditions. The revelation that lack of Hif-1α drastically reduced mmp-9 promoter activity in vitro upon induction of HIF-1 signaling by DFO supports the idea of MMP-9 as a HIF-1-regulated gene (45). Secondly we observed a reduction of Met signaling activation within metastases upon Hif-1α depletion in L-CI.5s tumor cells. This reduced activation of the proinvasive HGF-Met signaling pathway correlated with a dramatic decrease of secondary invasion during the late phase of metastasis. In principal, both impaired prometastatic mechanisms may account for such inhibition of micro-metastatic spread. Although promotion of tumor cell invasion via HIF-1α-triggered activation of the Met protooncogene and subsequent induction of HGF signaling have already been reported (28), here we were able for the first time to establish a link between MMP-9-mediated promotion of micrometastatic spread and HIF-1α-triggered MMP-9 expression.
We demonstrate here for the first time with a hypoxia-tolerant tumor cell line that HIF-1α is an important regulator of tumor cell invasion both in vitro and in vivo, thereby confirming our hypothesis that HIF-1α promotes the metastatic potential of tumor cells in a survival-independent manner. Taken together, the results from our study in addition to previous findings that HIF-1 signaling is important for survival of tumor cells explain the negative prognostic role of HIF-1α found in many malignancies (45). Consequently, interference with HIF-1 signaling may be a viable approach in the treatment not only of early but also of advanced tumor disease, encouraging further research on HIF-1α as a target for anticancer therapy.
Supplementary Material
Acknowledgments
We thank Katja Honert and Mareike Lehnhoff (Institut für Experimentelle Onkologie und Therapieforschung, Technische Universität München, Germany) for expert technical support. We also thank Dr. Yves St-Pierre for providing the murine MMP-9-luciferase promoter plasmid.
This work was supported by grants from the European Union Research Framework Programme 7, Project HEALTH-2007-201279/Microenvimet (to A. K.) and Project HEALTH-F2-2009-222741/Metoxia (to A. G.), and Deutsche Forschungsgemeinschaft grant KR2047/1-1 (to A. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- HIF-1
- hypoxia-inducible factor-1
- MMP
- matrix metalloproteinase
- HGF
- hepatocyte growth factor
- DFO
- desferrioxamine mesylate
- shNT
- non-targeting shRNA sequence
- qRT
- quantitative RT.
REFERENCES
- 1.Pouysségur J., Dayan F., Mazure N. M. (2006) Nature 441, 437–443 [DOI] [PubMed] [Google Scholar]
- 2.Semenza G. L., Wang G. L. (1992) Mol. Cell Biol. 12, 5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salceda S., Caro J. (1997) J. Biol. Chem. 272, 22642–22647 [DOI] [PubMed] [Google Scholar]
- 4.Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wykoff C. C., Beasley N. J., Watson P. H., Turner K. J., Pastorek J., Sibtain A., Wilson G. D., Turley H., Talks K. L., Maxwell P. H., Pugh C. W., Ratcliffe P. J., Harris A. L. (2000) Cancer Res. 60, 7075–7083 [PubMed] [Google Scholar]
- 7.Weidemann A., Johnson R. S. (2008) Cell Death Differ. 15, 621–627 [DOI] [PubMed] [Google Scholar]
- 8.Semenza G. L. (2010) Curr. Opin. Genet. Dev. 20, 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Victor N., Ivy A., Jiang B. H., Agani F. H. (2006) Clin. Exp. Metastasis 23, 87–96 [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara S., Nakagawa K., Harada H., Nagato S., Furukawa K., Teraoka M., Seno T., Oka K., Iwata S., Ohnishi T. (2007) Int. J. Oncol. 30, 793–802 [PubMed] [Google Scholar]
- 11.Krishnamachary B., Berg-Dixon S., Kelly B., Agani F., Feldser D., Ferreira G., Iyer N., LaRusch J., Pak B., Taghavi P., Semenza G. L. (2003) Cancer Res. 63, 1138–1143 [PubMed] [Google Scholar]
- 12.Liao D., Corle C., Seagroves T. N., Johnson R. S. (2007) Cancer Res. 67, 563–572 [DOI] [PubMed] [Google Scholar]
- 13.Hiraga T., Kizaka-Kondoh S., Hirota K., Hiraoka M., Yoneda T. (2007) Cancer Res. 67, 4157–4163 [DOI] [PubMed] [Google Scholar]
- 14.Bertout J. A., Patel S. A., Simon M. C. (2008) Nat. Rev. Cancer 8, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan K., Song G., Ouyang G. (2009) J. Cell Biochem. 107, 1053–1062 [DOI] [PubMed] [Google Scholar]
- 16.Semenza G. L. (2010) Oncogene 29, 625–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 18.Shyu K. G., Hsu F. L., Wang M. J., Wang B. W., Lin S. (2007) Exp. Cell Res. 313, 1181–1191 [DOI] [PubMed] [Google Scholar]
- 19.Furlan D., Sahnane N., Carnevali I., Cerutti R., Uccella S., Bertolini V., Chiaravalli A. M., Capella C. (2007) Surg. Oncol. 16, Suppl. 1, S25–S27 [DOI] [PubMed] [Google Scholar]
- 20.Krüger A., Kates R. E., Edwards D. R. (2010) Biochim. Biophys. Acta 1803, 95–102 [DOI] [PubMed] [Google Scholar]
- 21.Overall C. M., Kleifeld O. (2006) Nat. Rev. Cancer 6, 227–239 [DOI] [PubMed] [Google Scholar]
- 22.Krüger A. (2009) Biol. Chem. 390, 91–97 [DOI] [PubMed] [Google Scholar]
- 23.Gerg M., Kopitz C., Schaten S., Tschukes A., Kahlert C., Stangl M., von Weyhern C. W., Brücher B. L., Edwards D. R., Brand K., Krüger A. (2008) Mol. Cancer Res. 6, 341–351 [DOI] [PubMed] [Google Scholar]
- 24.Gorden D. L., Fingleton B., Crawford H. C., Jansen D. E., Lepage M., Matrisian L. M. (2007) Int. J. Cancer 121, 495–500 [DOI] [PubMed] [Google Scholar]
- 25.Björklund M., Koivunen E. (2005) Biochim. Biophys. Acta 1755, 37–69 [DOI] [PubMed] [Google Scholar]
- 26.Benvenuti S., Comoglio P. M. (2007) J. Cell Physiol. 213, 316–325 [DOI] [PubMed] [Google Scholar]
- 27.Naldini L., Weidner K. M., Vigna E., Gaudino G., Bardelli A., Ponzetto C., Narsimhan R. P., Hartmann G., Zarnegar R., Michalopoulos G. K., et al. (1991) EMBO J. 10, 2867–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennacchietti S., Michieli P., Galluzzo M., Mazzone M., Giordano S., Comoglio P. M. (2003) Cancer Cell 3, 347–361 [DOI] [PubMed] [Google Scholar]
- 29.Krüger A., Schirrmacher V., von Hoegen P. (1994) Int. J. Cancer 58, 275–284 [DOI] [PubMed] [Google Scholar]
- 30.Zhong H., De Marzo A. M., Laughner E., Lim M., Hilton D. A., Zagzag D., Buechler P., Isaacs W. B., Semenza G. L., Simons J. W. (1999) Cancer Res. 59, 5830–5835 [PubMed] [Google Scholar]
- 31.Yamamoto Y., Ibusuki M., Okumura Y., Kawasoe T., Kai K., Iyama K., Iwase H. (2008) Breast Cancer Res. Treat. 110, 465–475 [DOI] [PubMed] [Google Scholar]
- 32.DuBridge R. B., Tang P., Hsia H. C., Leong P. M., Miller J. H., Calos M. P. (1987) Mol. Cell Biol. 7, 379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerner W. A., Pearlstein E., Ambrogio C., Karpatkin S. (1983) Int. J. Cancer 31, 463–469 [DOI] [PubMed] [Google Scholar]
- 34.Kopitz C., Gerg M., Bandapalli O. R., Ister D., Pennington C. J., Hauser S., Flechsig C., Krell H. W., Antolovic D., Brew K., Nagase H., Stangl M., von Weyhern C. W., Brücher B. L., Brand K., Coussens L. M., Edwards D. R., Krüger A. (2007) Cancer Res. 67, 8615–8623 [DOI] [PubMed] [Google Scholar]
- 35.Krüger A., Arlt M. J., Gerg M., Kopitz C., Bernardo M. M., Chang M., Mobashery S., Fridman R. (2005) Cancer Res. 65, 3523–3526 [DOI] [PubMed] [Google Scholar]
- 36.Arlt M., Kopitz C., Pennington C., Watson K. L., Krell H. W., Bode W., Gansbacher B., Khokha R., Edwards D. R., Krüger A. (2002) Cancer Res. 62, 5543–5550 [PubMed] [Google Scholar]
- 37.Estève P. O., Robledo O., Potworowski E. F., St-Pierre Y. (2002) Biochem. Biophys. Res. Commun. 296, 864–869 [DOI] [PubMed] [Google Scholar]
- 38.Nguyen M. V., Pouvreau S., El Hajjaji F. Z., Denavit-Saubie M., Pequignot J. M. (2007) J. Neurosci. Res. 85, 1119–1125 [DOI] [PubMed] [Google Scholar]
- 39.Kung A. L., Wang S., Klco J. M., Kaelin W. G., Livingston D. M. (2000) Nat. Med. 6, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 40.Gatenby R. A., Gillies R. J. (2004) Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 41.Dikmen Z. G., Gellert G. C., Dogan P., Yoon H., Lee Y. B., Ahn C. H., Shay J. W. (2008) J. Cell Biochem. 104, 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akakura N. (2001) Hokkaido Igaku Zasshi 76, 375–384 [PubMed] [Google Scholar]
- 43.Carmeliet P., Dor Y., Herbert J. M., Fukumura D., Brusselmans K., Dewerchin M., Neeman M., Bono F., Abramovitch R., Maxwell P., Koch C. J., Ratcliffe P., Moons L., Jain R. K., Collen D., Keshert E., Keshet E. (1998) Nature 394, 485–490 [DOI] [PubMed] [Google Scholar]
- 44.Chen C., Yu Z. (2009) Anticancer Res. 29, 1367–1372 [PubMed] [Google Scholar]
- 45.Rankin E. B., Giaccia A. J. (2008) Cell Death Differ. 15, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.