Abstract
Reactive molecules have diverse effects on cells and contribute to several pathological conditions. Cells have evolved complex protective systems to neutralize these molecules and restore redox homeostasis. Previously, we showed that association of nuclear factor (NF)-erythroid-derived 2 (E2)-related factor 2 (NRF2) with the nuclear matrix protein NRP/B was essential for the transcriptional activity of NRF2 target genes in tumor cells. The present study demonstrates the molecular mechanism by which NRP/B, via NRF2, modulates the transcriptional activity of antioxidant response element (ARE)-driven genes. NRP/B is localized in the nucleus of primary brain tissue and human neuroblastoma (SH-SY5Y) cells. Treatment with hydrogen peroxide (H2O2) enhances the nuclear colocalization of NRF2 and NRP/B and induces heme oxygenase 1 (HO1). Treatment of NRP/B or NRF2 knockdowns with H2O2 induced apoptosis. Co-expression of NRF2 with members of the Kelch protein family, NRP/B, MAYVEN, or MAYVEN-related protein 2 (MRP2), revealed that the NRF2-NRP/B complex is important for the transcriptional activity of ARE-driven genes HO1 and NAD(P)H:quinine oxidoreductase 1 (NQO1). NRP/B interaction with Nrf2 was mapped to NRF2 ECH homology 4 (Neh4)/Neh5 regions of NRF2. NRP/B mutations that resulted in low binding affinity to NRF2 were unable to activate NRF2-modulated transcriptional activity of the ARE-driven genes, HO1 and NQO1. Thus, the interaction of NRP/B with the Neh4/Neh5 domains of NRF2 is indispensable for activation of NRF2-mediated ARE-driven antioxidant and detoxifying genes that confer cellular defense against oxidative stress-induced damage.
Keywords: Antioxidant; Neurobiology; Oxidative Stress; Protein-Protein Interactions; Reactive Oxygen Species (ROS); BTB Domain; Nrf2; Nuclear Matrix Protein, NRP/B
Introduction
Mammalian cells are exposed to reactive molecules originating from several exogenous and endogenous sources (1, 2). These molecules include reactive oxygen species, electrophilic chemicals, and heavy metals, which induce oxidative stress that impairs cellular functions and results in diverse pathological conditions, such as cancer, cardiovascular disease, diabetes, and neurodegenerative disorders (2–5). In response to these reactive molecules, cells have evolved multiple protective mechanisms to neutralize and clear toxic molecules and restore cellular redox homeostasis. One of these mechanisms is modulated by the nuclear factor (NF)2-E2-related factor 2 (NRF2) protein (6), which binds to ARE promoter sequences, leading to the coordinated up-regulation of ARE-driven detoxifying and antioxidant genes (7–11). The NRF2 system confers a protective effect on mammalian cells from a variety of toxic insults including carcinogens, reactive oxygen species, diesel exhaust, inflammation, calcium disturbance, UV light, and cigarette smoke (6).
Kelch-related proteins contain an N-terminal BTB (Broad-complex, Tramtrack, Bric-a-brac) domain that is primarily found in zinc finger proteins and functions as a homo- or heterodimerization domain (12, 13) and a C-terminal Kelch repeat or β-propeller domain that binds actin tails and has important roles in protein folding and as a protein-protein interaction module (12, 13). Multiprotein complexes formed through contact sites in β-propeller domains (14) are believed to be important for gene expression, cytoskeletal maintenance (15, 16), and controlling cellular organization and morphology (15, 16). Importantly, alterations or mutations in Kelch-related proteins have been found in numerous tumors (14, 17) and neurodegenerative disorders (18). The cytoplasmic protein Kelch-like ECH-associated protein 1 (KEAP1) and the nuclear NRP/B are both members of the Kelch-related family (8, 19). The localization of these proteins is important for regulation of the NRF2 pathway in which KEAP1 and NRP/B/ENC1 suppress and enhance, respectively, NRF2-mediated phase II detoxifying and antioxidant enzymes in response to oxidative stress (8, 17, 20).
Six functional domains of NRF2, termed NRF2 ECH homology (Neh) 1–6, have been identified and found to be highly conserved in various species (8). Neh1 is a bZip motif that mediates DNA binding and participates in heterodimerization with small MAF proteins (21). The Kelch domain of KEAP1 interacts with Neh2 suppressing NRF2 activity. Neh4 and Neh5 interact with the CREB-binding protein (cAMP responsive element-binding protein) to synergistically induce strong NRF2 transcriptional activation (22). Under conditions of oxidative stress, NRF2 is phosphorylated and released by KEAP1 into the nucleus where it associates with NRP/B and activates phase II detoxifying and antioxidant genes (17, 20). The molecular mechanism by which NRF2 activates ARE-driven genes has remained elusive.
In the present study, we describe the mechanism of NRP/B-modulated activation of NRF2 target genes. We also identify an interaction between the NRP/B BTB domain and the NRF2 Neh4/Neh5 region that may play a crucial role in modulating NRF2-dependent ARE-driven phase II detoxifying and antioxidant genes.
EXPERIMENTAL PROCEDURES
Reagents
Anti-NRF2 (catalog number sc-13032) and anti-Myc (catalog number sc-40) antibodies, and NRF2 siRNA (catalog number sc-37030) were purchased from Santa Cruz Biotechnology. Anti-FLAG (M2) antibody was obtained from Sigma (catalog number F1804). NRP/B siRNA (UUAACUGGAUCAGCUAUGA) was purchased from Dharmacon. The Luciferase assay kit was purchased from Promega (catalog number E4030).
Cell Cultures
SH-SY5Y cells were purchased from the American Type Cell Collection (ATCC) and prepared as recommended. Cells were cultured in a 1:1 mixture of ATCC-formulated Eagle's minimum essential medium (catalog number 30-2003) and F-12 medium supplemented with 10% fetal bovine serum at 37 °C in a 5% CO2 incubator. 293T cells were grown in RPMI 1640 medium with 2 mm l-glutamine adjusted to contain 1.5 g/liter of sodium bicarbonate, 4.5 g/liter of glucose, 10 mm HEPES, 1.0 mm sodium pyruvate, and supplemented with 0.2 units/ml of bovine insulin and 10% fetal bovine serum.
DNA Constructs
For analysis of the NRF2 interaction with NRP/B, we generated several truncations of the NRF2 and NRP/B proteins. NRF2 truncations were generated by PCR using a pEGFP-NRF2 template, a gift from Dr. Laurie Zipper (23), and primer pairs as shown in Table 1. The PCR products were cloned into vector pGEX-KT at the BamHI and EcoRI restriction sites. All plasmids were verified by sequencing and designated pGEX-NRF2(1–339), pGEX-NRF2(338–606), pGEX-NRF2(1–110), pGEX-NRF2(109–210), pGEX-NRF2(209-33), pGEX-NRF2(75–125), pGEX-NRF2(100–150), and pGEX-NRF2(125–175). GST-NRF2 truncation proteins were expressed in Escherichia coli BL21(DE3) cells by induction with isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h, purified on gluthatione-Sepharose beads (GE Healthcare) according to the manufacturer's instructions, and stored at −80 °C. FLAG-tagged constructs corresponding to the BTB domain, intervening sequence (IVS), BTB-IVS, and IVS regions of each of these domains were generated by PCR using a NRP/B cDNA template (17, 20) and specific primers (Table 1). The PCR products were purified, digested with HindIII and EcoRV restriction enzymes, ligated into pFLAG-CMV4 (catalog number E1775, Sigma), and designated pCMV4-NRP/B BTB, pCMV4-NRP/B BTB-IVS, and pCMV4-NRP/B IVS (Table 1).
TABLE 1.
Primers
DNA Transfection
HEK293T cells were transfected with pCMV4-NRP/B or pMyc-NRP/B constructs (17). Protein expression was confirmed by Western blotting using anti-FLAG or anti-Myc antibody. In addition, we also transfected pCMV4-MRP2 (24), pEGFP-MAYVEN (25), DsRed-KEAP1 (26), ARE-Ti-luc promoter (11), and pGL2-HO1 (27). HEK293T and SH-SY5Y cells were grown in 6-well plates and transfected using Lipofectamine 2000 reagent (Invitrogen).
GST Pull-down Assay
HEK293T cells transfected with pCMV4-NRP/B or pMyc-NRP/B constructs were lysed in RIPA buffer and solubilized for 1 h at 4 °C. Protein extracts were pre-cleaned by incubation with gluthatione-Sepharose beads for 30 min at 4 °C. After centrifugation, the supernatants were incubated with GST or GST-NRF2 truncation proteins at room temperature for 3 min. After washing the precipitates were subjected to immunoblot analysis using anti-FLAG or anti-Myc antibody.
Immunoprecipitation
HEK293T cells were seeded in 6-well plates overnight and co-transfected with DNA constructs. 24 h after transfection, the cultures were washed twice with PBS and lysed with ice-cold lysis buffer (RIPA). Cell extracts were solubilized for 30 min at 4 °C. After pre-clearing, the extracts were immunoprecipitated with anti-NRF2 antibody. Precipitates were blotted and probed with anti-Myc antibody.
Immunoblot Analysis
Cell cultures were lysed with cold lysis buffer (100 mm KCl, 300 mm sucrose, 10 mm Pipes, pH 6.8, 3 mm MgCl2, 1.2 mm phenylmethylsulfonyl fluoride, 0.5% Triton X-100, and 1 mm EGTA). The lysates were transferred to a fresh tube and solubilized for 1 h at 4 °C and clarified by centrifugation at 12,000 × g for 20 s at 4 °C. Total cell extract protein concentration was determined using the Bradford assay. Equal amounts of proteins were electrophoresed on SDS-PAGE gels, blotted onto polyvinylidene difluoride membrane, and incubated with anti-NRP/B (VD2), anti-NRF2, anti-FLAG, anti-Myc, or anti-CSK antibodies. After washing, the blots were incubated with horseradish peroxidase-conjugated anti-IgG antibody.
Immunocytochemistry
Cell cultures were washed with PBS, fixed with 4% paraformaldehyde, and treated with 0.5% Triton X-100 in PBS for 30 min. After 3 washes with PBS, cells were treated with 10% goat serum in PBS for 2 h and incubated with anti-NRP/B (VD2) and/or anti-NRF2 antibodies for 1 h at room temperature. After washing with PBS, the cells were incubated with fluorescein isothiocyanate-conjugated IgG and/or Texas Red-conjugated IgG antibodies for 1 h. The cells were then washed, mounted on slides, and imaged by confocal microscopy (Zeiss LSM 510 Meta).
RNA Isolation
Total RNA isolation was performed using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Briefly, cells were seeded on 10-cm dishes and treated accordingly. One ml of TRIzol reagent was added, followed by addition of 0.2 ml of chloroform after 15 min. Samples were vigorously inverted by hand for 15 s, incubated at room temperature for 3 min, and centrifuged at 12,000 × g for 15 min at 4 °C. 0.5 ml of isopropyl alcohol was added to the supernatant. After a 10-min incubation at room temperature, samples were centrifuged at 12,000 × g for 10 min at 4 °C. The pellets were washed with 75% ethanol, dissolved in RNase-free water, and incubated at 60 °C for 10 min. Total RNA concentration was measured and the samples were stored at −80 °C for use in Northern blotting and RT-PCR analysis.
RT-PCR Analysis
Transcript levels were semi-quantified by one-step RT-PCR according to the manufacturer's instructions (Clontech) using primers specific for HO1 and GAPDH (Table 1). The temperature conditions were 50 °C for 1 h, 94 °C for 5 min, followed by 25 cycles of 94 °C for 30 min, 68 °C for 30 min, and 68 °C for 60 min with an extension of 68 °C for 2 min.
Northern Blot Analysis
20 μg of total RNA was electrophoresed on a 1% agarose gel, and transferred onto Hybond-N+ (Amersham Biosciences) in 20× SSC buffer overnight. Membranes were cross-linked and pre-hybridized in pre-hybridization solution (5× SSPE, 2% SDS, and 5× Denhardt's reagent containing 100 μg/ml of denatured salmon testes DNA) at 65 °C for 6 h. Purified PCR-generated NRP/B (BTB domain) serving as a probe, was labeled with α-32P using the NEblot kit (catalog number N1500L, New England Biolabs) according to the manufacturer's instructions. The membranes were incubated with hybridization solution containing the NRP/B BTB domain-labeled probe at 65 °C overnight, and then washed in washing solution (0.5× SSPE, 1% SDS) for 20 min, and twice in washing solution (0.1× SSPE, 1% SDS) for 20 min each. Membranes were exposed to x-ray film at −80 °C.
Apoptotic DNA Ladder
DNA fragmentation in apoptotic cells was detected using the Apoptotic DNA Ladder Detection Kit (Chemicon International). The procedure was performed per the manufacturer's instructions. Briefly, cells were treated with an apoptosis inducing agent appropriate to the purposes of our experiment. Treated cells were washed with PBS, collected, and centrifuged. The pellets were resuspended in TE lysis buffer and treated with RNase A and proteinase K. DNA was precipitated with isopropyl alcohol and redissolved in DNA Suspension Buffer. DNA fragmentation was visualized on a 1% agarose gel stained with ethidium bromide.
Luciferase Assay
SH-SY5Y or HEK293T cells were co-transfected with a combination of plasmids as indicated in figure legends using Lipofectamine 2000 or Lipofectatime. In addition, pCMV β-galactosidase was co-transfected for the purpose of controlling the transcriptional activity of the promoter(s). Cell lysates were prepared and luciferase and β-galactosidase activities were quantified using a Luciferase assay kit (Promega, Madison, WI) according to the manufacturer's instructions. The effects of the proteins on the promoter(s) transcriptional activity were normalized to the ratio of the luciferase versus β-galactosidase activity.
Statistical Analysis
Data are presented as the mean ± S.D. The Student's t test and one-way analysis of variance were used to assess the significance of independent experiments. p < 0.05 represents the statistical significance.
RESULTS
Expression of NRP/B in Neuronal Cells
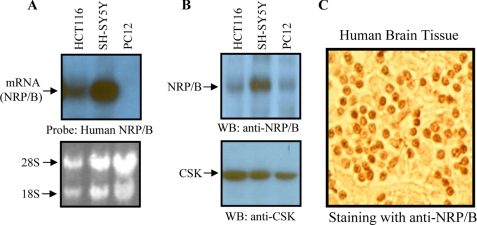
NRP/B has been shown to be expressed in various tissues and cells, particularly neuronal cells (17, 19, 20). We determined the molecular mechanism by which NRP/B mediates cellular protection against agents of oxidative stress in the brain. We examined expression of NRP/B in SH-SY5Y and PC12 cells by Northern and Western blot analyses. HCT-116 cells reported to express NRP/B (28) served as a positive control. Using a human NRP/B probe, Northern blotting showed that NRP/B mRNA was detected in HCT-116 and SH-SY5Y cells (Fig. 1A). Protein extracts from HCT-116, SH-SY5Y, and PC12 cells were subjected to Western blot analysis using a monoclonal antibody against NRP/B (DV2) (17). This analysis showed that NRP/B was expressed in all cell lines tested (Fig. 1B). NRP/B expression was detected in PC12 by Western blot, but not by Northern blot (Fig. 1, A and B) due to the high specificity of the human NRP/B probe and the high contingency of the Northern blot condition. The human NRP/B probe used did not hybridize rat NRP/B mRNA in PC12 cells (Fig. 1A), indicating that the Northern blot condition was highly specific.
FIGURE 1.
Expression and localization of NRP/B in neuronal origin-derived cells and human brain specimen. A, for Northern blot analysis, total RNA was purified from HCT-116, SH-SY5Y, and PC12 cells using TRIzol reagent. Ten μg of RNA was blotted on a Hybond-N+ membrane and hybridized with an α-32P-labeled human NRP/B probe. B, total cell lysates were prepared from several cell lines, and 100-μg samples were blotted on a polyvinylidene difluoride membrane. The blot was incubated with anti-NRP/B (VD2) or anti-c-src tyrosine kinase (CSK) antibodies. CSK was used to confirm equal protein loading. C, localization of NRP/B in sample specimens of normal brains. Normal brain specimens were obtained from the Cooperative Human Tissue Network. The paraffin-embedded samples were immunostained with anti-NRP/B (VD2) antibody. WB, Western blot.
Immunohistochemical staining with anti-NRP/B (VD2) antibody was carried out to detect expression and localization of NRP/B in human brain specimens. NRP/B was expressed in the nucleus of the brain specimen (Fig. 1C). In addition to our previous report (17), expression of NRP/B mRNA and protein was detected in human SH-SY5Y. Thus, we selected SH-SY5Y cells for further analysis of NRP/B function and mechanism in the oxidative stress response.
H2O2 Increased the Colocalization of NRF2 and NRP/B in SH-SY5Y Cells
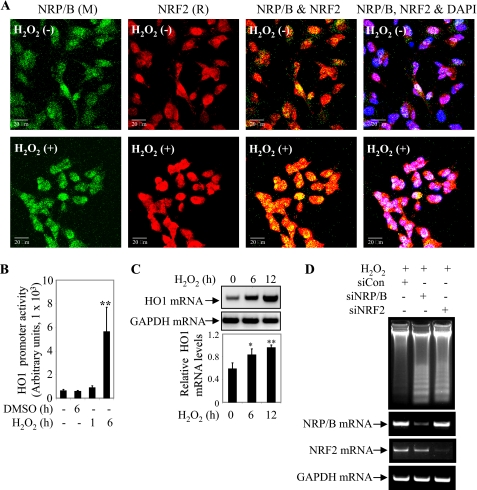
H2O2 treatment has been shown to increase the co-localization of NRP/B and Nrf2 in breast cancer cells (20). We examined the effect of H2O2 on NRF2 and NRP/B localization in SH-SY5Y cells. SH-SY5Y cells were treated with 5 μm H2O2 for 12 h, fixed, and immunostained with anti-NRF2 and anti-NRP/B (VD2) antibodies (Fig. 2A). Fixed cells stained with either mouse or rabbit IgG antibody served as control (supplemental Fig. S1). Colocalization of NRF2 and NRP/B intensified in the nucleus following H2O2 treatment (Fig. 2A).
FIGURE 2.
NRP/B-NRF2 mediates cellular defense against H2O2 in SH-SY5Y cells. A, subcellular localization of NRP/B and NRF2 upon treatment of 5 μm H2O2. SH-SY5Y cells were plated on coated glass slides and treated with H2O2 for 12 h, fixed, and immunostained with anti-NRF2, anti-NRP/B (VD2) antibodies, and 4′,6-diamidino-2-phenylindole (DAPI). The images were taken using a confocal microscope (Zeiss LSM 510 Meta). M, mouse. R, rabbit. Bar, 20 μm. B, H2O2 treatment induced HO1 transcriptional activity. SH-SY5Y cells were transfected with HO1 promoter. Cells were treated with 5 μm H2O2 for 6 h. HO1 promoter (luciferase) activity was measured, and then the relative luciferase activity versus β-galactosidase activity (derived from three individual experiments) was expressed in arbitrary units. The bars in the graphs represent the mean ± S.D. **, p < 0.01. C, H2O2 treatment up-regulated the HO1 transcription level. SH-SY5Y cells were treated with 5 μm H2O2 in time-dependent manner. Following the treatment cells were harvested at 0, 6, and 12 h, and RNA was purified and analyzed by semi-quantitative RT-PCR. HO1 mRNA was up-regulated over the course of H2O2 treatment. Relative HO1 mRNA levels versus GAPDH levels upon H2O2 treatment, as derived from 3 individual experiments, were converted into arbitrary units. The bars in the graphs represent the mean ± S.D. *, p < 0.05; **, p < 0.01. D, reduction of NRP/B or NRF2 expression rendered SH-SY5Y cells sensible to H2O2-induced apoptosis. Cells were plated on 6-well plates and treated with siNRP/B, siNRF2, or siCTL (control). After 12 h of 10 μm H2O2 treatment cells were harvested, and DNA fragmentation was detected using the Apoptotic DNA Ladder Detection Kit or RNA was purified and RT-PCR was performed to detect transcription levels of NRP/B and NRF2 in siRNA-treated cells. DMSO, dimethyl sulfoxide.
NRF2 and NRP/B Conferred Cellular Protection against H2O2 via HO1 Activation
To define the mechanism by which the association of NRF2 and NRP/B induces HO1 activation, we examined the effect of H2O2 treatment on HO1 promoter activity by luciferase assay. SH-SY5Y cells were transfected with the HO1 promoter and treated with dimethyl sulfoxide or 5 μm H2O2 in a time-dependent manner as indicated (Fig. 2B). Cell lysates were prepared and luciferase activity was measured. Six hours after treatment, relative luciferase activity was up to 7-fold higher in cells treated with H2O2 compared with those treated with dimethyl sulfoxide (Fig. 2B). Next, we measured the transcription levels of HO1 mRNA induced by H2O2 treatment. SH-SY5Y cells were harvested 0, 6, and 12 h after treatment with H2O2. Total RNA was purified and semi-quantitative RT-PCR analysis showed that HO1 mRNA was up-regulated over the course of H2O2 treatment (Fig. 2C). We then examined whether NRF2 and NRP/B expression protected cells from apoptosis induced by H2O2 treatment. SH-SY5Y cells were treated with siNRP/B, siNRF2, or siCon (control). After 24 h, cells were treated with 10 μm H2O2 for 12 h. DNA fragmentation resulting from H2O2-induced apoptosis was detected using the DNA ladder kit. Intense DNA fragmentation was observed in siNRP/B- and siNRF2-treated cells when compared with siCon-treated controls in response to H2O2 treatment (Fig. 2D). These data provide strong evidence that expression of both NRP/B and NRF2 is crucial for cellular protection against H2O2-induced apoptosis.
Specific Activation of NRF2 Target Genes by NRP/B
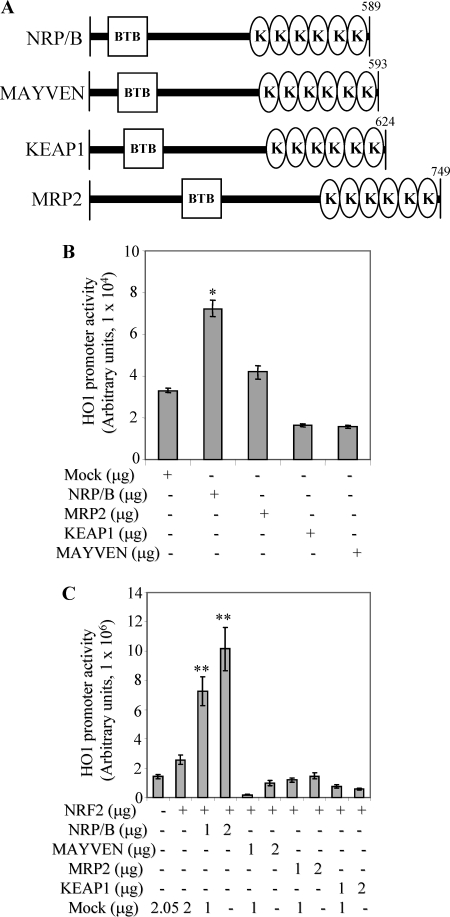
We previously cloned and characterized several Kelch-related family members including NRP/B (19), MRP2 (24), and MAYVEN (29). These proteins have similar domain organization (Fig. 3A) consisting of a N-terminal BTB domain and a C-terminal Kelch repeat domain separated by an IVS. BTB domain mediates homo- and heterodimerization (12) whereas, the C-terminal Kelch repeat region functions to bind actin tails, and is involved in protein-protein interactions (30). KEAP1, another Kelch family member, functions as transcriptional repressor for NRF2 (8), whereas NRP/B is an enhancer for NRF2 activity (17, 20). MRP2 is involved in glycogen synthase kinase 3β-mediated neuronal differentiation (24) and process elongation in oligodendrocytes (25, 31). MAYVEN is also essential for FYN-modulated process elongation in oligodendrocytes (25). Whereas KEAP1, MRP2, and MAYVEN are endogenously localized in the cytoplasm (8, 24, 25), NRP/B is endogenously localized in the nucleus (19, 32). We examined the differential effects of these proteins on NRF2-mediated HO1 transcriptional activity using a luciferase reporter assay. The HO1 promoter plasmid was co-transfected with NRP/B, MRP2, KEAP1, MAYVEN, or mock (control). Cell lysates were prepared and luciferase activity was measured after 24 h. Relative luciferase activity was up to 2-fold higher in cells transfected with NRP/B compared with cells transfected with mock or MRP2, and up to 4-fold higher when compared with cells transfected with KEAP1 or MAYVEN (Fig. 3B).
FIGURE 3.
Transcriptional induction of HO-1 mediated by NRF2-NRP/B. A, schematic diagrams of the Kelch-related protein family, as indicated. The Kelch-related protein family consists of two main domains, BTB (responsible for homo- or hetero-dimerization) and Kelch repeats (responsible for actin binding). B, differential effect of Kelch-related proteins on HO1 promoter activity. NRP/B, MRP2, KEAP1, MAYVEN, or mock were co-transfected with HO1 promoter in SH-SY5Y cells. Following 24 h, cell extracts were prepared. HO1 promoter (luciferase) activity was measured, and then the relative luciferase activity versus β-galactosidase activity (derived from three individual experiments) was expressed in arbitrary units. The bars in the graphs represent the mean ± S.D. *, p < 0.05. C, differential effect of Kelch-related proteins on NRF2-mediated HO1 promoter activity, SH-SY5Y cells. NRP/B, MRP2, KEAP1, MAYVEN, or mock were co-transfected with HO1 promoter and NRF2 in SH-SY5Y cells. Following 24 h, cell extracts were prepared. HO1 promoter activity was measured, and then the relative luciferase activity versus β-galactosidase activity (derived from three individual experiments) was expressed in arbitrary units. The bars in the graphs represent the mean ± S.D. **, p < 0.01.
A similar set of experiments performed using cells co-transfected with the HO1 promoter and NRF2 in conjunction with NRP/B, MAYVEN, MRP2, KEAP1, or mock (control) in a dose-dependent manner (Fig. 3C) showed a more than 10-fold increase in HO1 transcriptional activity in cells transfected with NRP/B as compared with those transfected with MAYVEN, MRP2, KEAP1, or mock (Fig. 3C). NRP/B was important in up-regulating HO1 transcriptional activity, whereas KEAP1 showed an inhibitory effect on the activity of HO1. MAYVEN and MRP2 did not significantly effect the transcriptional activity as compared with control counterparts. These data together with previous reports (17, 20) indicate that NRP/B specifically activates NRF2 target genes.
Mapping Interacting Domains of NRF2 and NRP/B
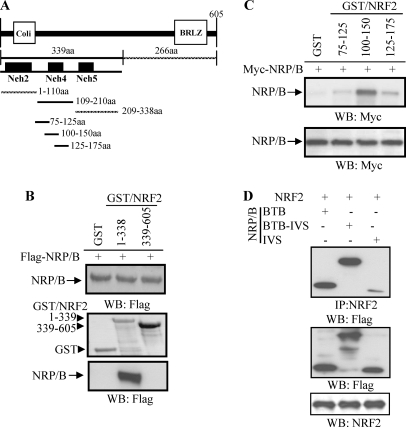
We previously showed that the NRF2-NRP/B complex was crucial for induction of the NRF2 target gene, NQO1 (17, 20). In this study, co-expression of NRF2 with KEAP1 inhibited HO1 transcriptional activity, whereas co-expression of NRF2 with NRP/B increased the transcriptional activity (Fig. 3). Thus, the interaction of NRF2 with NRP/B is an important regulator of NRF2 target genes. Here we identified the interacting domains of NRF2 and NRP/B. For this purpose, we generated several truncation mutants of NRF2 and NRP/B and studied their interaction by GST pull-down assay and immunoprecipitation. First, GST fusion proteins encompassing the N-terminal region of NRF2 from amino acid 1 to 339 and the C-terminal region containing amino acids 338 to 605 were generated and designated GST-NRF2(1–339) and GST-NRF2(338–605), respectively (Fig. 4A). HEK293T cells were transfected with FLAG-NRP/B (17, 20). After 24 h, cell extracts were incubated with purified GST-NRF2(1–339), GST-NRF2(338–605), or GST alone. The complexes were subjected to Western blot analysis using anti-FLAG antibody. Pull-down assays showed that NRP/B interacted with the NRF2 N-terminal region (1–339) but not with the NRF2 C-terminal region (338–605) (Fig. 4B). Next, we generated several truncation mutants within the N-terminal region of NRF2 to further delineate the interacting region with NRP/B. Pull-down assays revealed that NRP/B interacted with a GST fusion protein containing NRF2 spanning amino acids 1 to 110 (GST-NRF2(1–110)) and 109 to 210 (GST-NRF2(109–210)), but not amino acids from 209 to 338 (GST-NRF2(209–338)) (data not shown). We further mapped the interaction within the N-terminal 175 amino acids of NRF2. The GST fusion proteins NRF2(75–125), NRF2(100–150), and NRF2(125–175) were expressed, purified, and used to perform pull-down assays with NRP/B and Western blotted with anti-Myc antibody. Although a weak interaction with NRF2(75–125) and NRF2(125–175) was seen, NRP/B interacted strongly with NRF2(100–150) (Fig. 4C). The N-terminal 339 amino acids of NRF2 encompasses the Neh2, Neh4, and Neh5 domains (Fig. 4A). This deletion mapping approach demonstrated that the Neh4 domain of NRF2 interacted specifically with NRP/B.
FIGURE 4.
Interaction of NRP/B with NRF2. A, generation of NRF2 truncations. The truncations were generated with PCR using primers as indicated in Table 1. PCR products were constructed into pGEX-KT and designated GST-NRF2(1–339), GST-NRF2(1–110), GST-NRF2(109–210), GST-NRF2(209–338), GST-NRF2(75–125), GST-NRF2(100–150), GST-NRF2(125–175), and GST-NRF2(338–605). Sequences were confirmed by sequencing from both orientations and expressions were verified by isopropyl 1-thio-β-d-galactopyranoside treatment. These purified GST-NRF2 truncations were used for pull-down assay. B, interaction of NRP/B with N-terminal NRF2 (1–399 amino acids). Protein extracts from 293T cells transfected with FLAG-NRP/B were incubated with GST-NRF2(1–339), GST-NRF2(338–605), or GST. The complexes were subjected to Western blotting (WB) and probed with anti-FLAG antibody. Upper panel indicates NRP/B expression in the cell extract. Middle panel is GST, GST-NRF2(1–339), and GST-NRF2(338–605) expressions induced by isopropyl 1-thio-β-d-galactopyranoside. Lower panel is the interaction of NRP/B with NRF2. C, interaction of NRP/B within N-terminal NRF2. Protein extracts from 293T cells transfected with Myc-NRP/B were incubated with GST-NRF2(75–125), GST-NRF2(100–150), GST-NRF2(125–175), or GST. The complexes were subjected to Western blotting and probed with anti-Myc antibody. Upper panel indicates the interaction of NRP/B with NRF2 truncations. Lower panel indicates NRP/B expression in the cell extract. D, association of NRF2 with the BTB domain of NRP/B. NRF2 was cotransfected with NRP/B BTB, NRP/B BTB/IVS, or NRP/B IVS in HEK293T cells, and protein extracts were prepared and immunoprecipitated with anti-NRF2 antibody. The complexes were subjected to Western blotting with anti-FLAG antibody (upper panel). Cell extracts were Western blotted with anti-FLAG antibody (middle panel) and anti-NRF2 antibody (lower panel).
Next we examined the interaction of the NRP/B BTB domain with NRF2. For this purpose, HEK293T cells were transfected with NRP/B constructs containing the BTB domain (NRP/B BTB), the BTB domain and central linker region (NRP/B BTB-IVS), and the central linker region alone (NRP/B IVS). Cell extracts were prepared, immunoprecipitated with anti-NRF2 antibody, and subjected to Western blot analysis using anti-FLAG antibody. Immunoprecipitation assays showed that the NRP/B BTB and NRP/B BTB-IVS regions strongly interacted with NRF2, whereas an NRP/B IVS was less obvious (Fig. 4D). Thus, NRF2 may interact with the BTB domain of NRP/B. Taken together our data indicate that interaction of NRP/B with the region encompassing the Neh4 domain of NRF2 may interact to induce NRF2-mediated HO1 activity.
Homology Modeling of the NRP/B BTB Domain
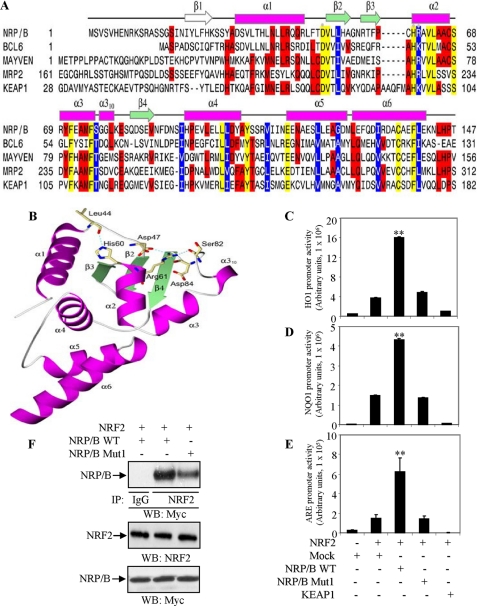
A homology model of the NRP/B BTB domain containing residues 1 to 147 was generated using the program MODELLER (33). The crystal structure of BCL6 BTB domain (Protein Data Bank code 1R29), which shares 33% sequence identity with the NRP/B BTB domain, was used as a template structure (Fig. 5A). The homology model was selected from three computed models based upon the lowest values of the MODELLER objective function. The secondary structure of the model contains the BTB core elements of three β-strands (β2–β4) forming a mixed β-sheet, and five α-helices (α2–α6) (Fig. 5A). A short α310 helix interrupts the α3-β4 loop. The region from Asp-30 to Arg-43 of NRP/B forms an N-terminal α-helical extension of the core BTB domain. The β1 strand observed in the crystal structure of the BCL6 BTB domain is not present in the NRP/B BTB homology model.
FIGURE 5.
Mutation in the BTB domain of NRP/B impairs ARE-driven transcriptional activity. A, sequence alignment of the NRP/B, BCL6, MAYVEN, MRP2, and KEAP1 BTB domains. The protein sequences were aligned using the program CLUSTAL W and manual intervention. Hyphens represent gaps inserted for optimum alignment. The secondary structure elements of the NRP/B BTB domain were assigned using the program STRIDE. Helices are depicted as magenta rectangles and β-strands as green arrows. Conserved amino acids are shaded yellow, identical amino acids in three of more sequences are shaded red, and similar amino acids are white letters on a blue background. The secondary structure elements are labeled according to the BCL6 assignments. The β-strand seen in the structure of the BCL6 BTB domain, but not the NRP/B BTB model is shown as a white arrow. The positions of the three mutants, D47A, H60A, and R61D are highlighted with gray dots. B, ribbon representation of the NRP/B BTB domain. The loop regions are gray coils. Amino acids Asp-47, Ser-57, His-60, Arg-61, Ser-82, and Asp-84, involved in hydrogen bonding interactions are shown as khaki sticks. Nitrogen and oxygen atoms are colored blue and red, respectively. Hydrogen bonds are shown as dashed cyan lines. The figure was made using POVSCRIPT and POV-Ray. C–E, BTB mutation of NRP/B reduces the ARE-driven transcriptional activity. HO1 (C), NQO1 (D), or ARE (E) promoter and NRF2 were cotransfected with NRP/B, NRP/B Mut1, KEAP1, or mock (control) in HEK293T cells. After 24 h, total cell lysates were prepared, and luciferase activity was measured. Relative luciferase versus β-galactosidase activity (derived from three individual experiments) was expressed in arbitrary units. The bars in the graphs represent the mean ± S.D. **, p < 0.01. F, effect of D47A, H60A, and R61D of NRP/B BTB domain on its association with NRF2. NRF2 was cotransfected with either NRP/B WT or NRP/B Mut1 in HEK293T cells, and protein extracts were prepared and immunoprecipitated with anti-NRF2 antibody. The complexes were subjected to Western blotting with anti-Myc antibody (upper panel). For expression control, cell extracts were Western blotted with anti-NRF2 antibody (middle panel) and anti-Myc antibody (lower panel).
As shown in Fig. 5B, Asp-47 is a conserved amino acid and sits in the loop region connecting the α1 and β2 secondary structure elements. Its side chain makes a hydrogen bond to the backbone of Arg-61. Amino acid position 61 is invariably an arginine or a lysine. Arg-61 in the NRP/B BTB domain is part of the α2 helix and makes up to three hydrogen bonds, one to the carbonyl oxygen of Asp-47 in the α1-β2 loop, one to Ser-82 and one to Asp-84 in the α3-β4 loop. The side chain of His-60 at the N-terminal end of α2 hydrogen bonds to the backbone oxygen of Leu-44 in the α1-β2 loop. These interactions serve to link some of the core secondary structure elements and stabilize the overall BTB domain fold. The mutations D47A, H60A, and R61D would break this hydrogen bonding network, disturb the fold, and disrupt the BTB domains ability to associate with interacting proteins such as NRF2.
Involvement of NRP/B BTB Domain in NRF2-mediated ARE-driven Transcriptional Activity
Homology modeling showed that Asp-47, His-60, and Arg-61 of the NRP/B BTB domain have important roles in maintaining the overall fold (Fig. 5B). Here, we examined whether mutations D47A, H69A, and R61D altered ARE-driven transcriptional activity. The triple mutant D47A,H69A,R61D, designated NRP/B Mut1, was generated, and its expression was verified by Western blotting using anti-Myc antibody. The HO1 promoter in conjunction with NRF2 was co-transfected with wild-type NRP/B (NRP/B WT), NRP/B Mut1, KEAP1, or Mock (control) in HEK293T cells. Cell lysates were prepared and luciferase activity was measured 24 h after transfection. Relative luciferase activity was ∼5-fold higher in cells transfected with NRP/B (wild-type) (17) as compared with cells receiving NRP/B Mut1 or mock (Fig. 5C). KEAP1 inhibited HO1 promoter activity as expected (Fig. 5C).
We also examined the effect of these mutations on transcriptional activity of the NQO1 promoter and ARE-driven luciferase activity. NQO1 promoter (34) or ARE-driven luciferase (27) in conjunction with NRF2 was co-transfected with NRP/B WT, NRP/B Mut1, KEAP1, or mock (control) in HEK293T cells. NRP/B Mut1 was unable to induce NRF2-mediated transcriptional activity of NQO1 (Fig. 5D) and ARE-driven gene expression (Fig. 5E).
To further examine the effect of the mutations in the BTB domain of NRP/B on its association with NRF2, NRF2 was cotransfected with either NRP/B WT or NRP/B Mut1. Protein extracts were prepared, immunoprecipitated with anti-NRF2 antibody, and subjected to Western blot analysis using anti-Myc antibody. For expression verification, cell extracts were blotted and probed with anti-NRF2 or anti-Myc antibody. As shown in Fig. 5F, NRP/B Mut1 had lower binding affinity with NRF2 as compared with the NRP/B WT counterpart (Fig. 5F). Taken together, these results indicate that the NRP/B BTB domain played an important role in modulating ARE-driven antioxidant gene expression in an NRF2-dependent fashion.
DISCUSSION
In the present study we elucidated the molecular mechanism by which NRP/B activates transcriptional activity of ARE-driven gene(s) through the NRF2 pathway. We observed that the NRP/B BTB domain interacts with the activation domains, Neh4/Neh5, of NRF2. Mutations that are predicted to disturb the secondary structure of the NRP/B BTB domain fail to induce NRF2 target genes (NQO1 and HO1) and ARE-driven transcriptional activity. This study illustrates that the association of the NRP/B with the Nhe4/Neh5 domains of NRF2 is indispensable for activation of NRF2-mediated ARE-driven phase II enzymes that confer cellular protection against oxidative stress-induced damage.
More than 60 Kelch-related proteins have been identified in organisms from viruses to mammals. Forming multiprotein complexes through contact sites within their BTB and Kelch domains, this family of proteins regulates cell morphology and organization and gene expression (14, 35). Previously, we cloned and characterized several members of the Kelch-related protein family, namely, NRP/B, MRP2, and MAYVEN. These proteins and another Kelch-related protein, KEAP1 have similar domain organization (Fig. 3A). We investigated the functions of these proteins in the context of oxidative stress responses. Despite similar domain organization their cellular localizations were found to be different. While MAYVEN, MRP2, and KEAP1 are localized in the cytoplasm, NRP/B is confined to the nucleus (17, 20, 24). In the presence of NRF2, NRP/B actively induced the transcriptional activity of HO1, whereas KEAP1 inhibited this activity (Fig. 3, B and C). Our previous report showed that GFP-NRF2 shuttled from the nucleus to the cytoplasm when it was cotransfected with KEAP1 and inhibited transcriptional activity of the NRF2 target gene, NQO1 (17). However, the phenomenon did not occur when NRF2 was contransfected with MAYVEN or MRP2 (data not shown). These findings illustrated a role for NRP/B and KEAP1 in modulating the NRF2 pathway in response to reactive oxygen species.
The interaction of NRP/B and NRF2 was essential for the activation of NRF2 target genes (Fig. 3C). We examined which structural domain of NRF2 was responsible for induction of NRF2 target genes. Several truncation mutants of NRP/B were analyzed for their ability to interact with NRF2. The NRP/B BTB and BTB-IVS regions strongly interacted with NRF2, whereas the IVS region displayed a much weaker association (Fig. 4D). The mutations D47A, H60A, and R61D in the BTB domain (NRP/B Mut1), which we predicted from homology modeling to destabilize the BTB fold, reduced the level of NRP/B interaction with NRF2 (Fig. 5F). Furthermore, wild-type NRP/B, but not NRP/B Mut1, induced ARE-driven transcriptional activity of the phase II detoxifying and antioxidant enzymes, HO1 and NQO1 (Fig. 5, C and D). The NRB/B BTB domain may, therefore, significantly contribute to the activation of NRF2 target genes.
NRF2 contains 6 domains, Neh2, Neh4, and Neh5 at the N-terminal, and Neh6, Neh1, and Neh3 at the C-terminal (22). KEAP1 interacts with Neh2 of NRF2 to inhibit transcriptional activity (8). Neh4 and Neh5 are conserved in several species and interact with the co-activator CREB-binding protein to synergistically activate the transcription of NRF2 target genes (22). To determine which domain(s) of NRF2 is associated with NRP/B, we adopted a deletion mapping approach, which revealed that the N-terminal region of NRF2 spanning amino acids 1–339 containing Neh2, Neh4, and Neh5 interacted specifically with NRP/B (Fig. 4). Furthermore, the NRF2 region spanning amino acids 100 to 150 containing Neh4 strongly associated with NRP/B (Fig. 4C) (22). It was observed that NRP/B also interacted with the Neh5 activation domain of NRF2, although the affinity of the association was not as strong as the Neh4 domain. These data reveal the core domains of NRP/B and NRF2 that are critical for activation of NRF2 target genes HO1 and NQO1.
In conclusion, we elucidated the molecular mechanism by which NRP/B activates NRF2-mediated ARE-driven transcription of phase II detoxifying and antioxidant enzymes NQO1 and HO1. The association of the NRP/B BTB domain with the NRF2 Neh4/Neh5 domains is central to NRF2-mediated cellular protection against damage caused by oxidative stress. NQO1 is an activating enzyme for some anti-cancer drugs and has been implicated in cancer prevention (36–38). HO1 exerts antioxidant effects, including inhibition of the proliferation of vascular and airway smooth muscle cells (39, 40), and has been implicated in the endogenous defense against oxidative stress (41). Understanding the molecular mechanism by which the NRF2-NRP/B interaction mediates enzyme activation may have potential therapeutic consequences for the treatment of cancer and neurodegenerative disorders.
Supplementary Material
Acknowledgment
We thank Makara Men for assistance in editing this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL80699 (to S. A.), CA096805 (to H. K. A.), CA135226 (to H. K. A.), a Department of Defense Cancer Concept Award (to S. A.), and Flight Attendant Medical Research Institute Young Clinical Scientist Faculty Award (to S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- NF
- nuclear factor
- NRF2
- nuclear factor erythroid-derived 2-related factor 2
- ARE
- antioxidant response element
- BTB
- Broad-complex, Tramtrack, Bric-a-brac
- CSK
- c-src tyrosine kinase
- HO1
- heme oxygenase 1
- KEAP1
- Kelch-like ECH-associated protein
- MRP2
- MAYVEN-related protein 2
- NRP/B
- nuclear-restricted protein in brain
- NQO1
- NAD(P)H:quinine oxidoreductase 1 (nicotinamide quinine oxidoreductase)
- CREB
- cAMP responsive element-binding protein
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- Neh
- NRF2 ECH homology
- GST
- glutathione S-transferase
- IVS
- intervening sequence
- PBS
- phosphate-buffered saline
- Pipes
- 1,4-piperazinediethanesulfonic acid
- RT
- reverse transcription
- siRNA
- small interfering RNA
- WT
- wild-type.
REFERENCES
- 1.Finkel T., Holbrook N. J. (2000) Nature 408, 239–247 [DOI] [PubMed] [Google Scholar]
- 2.Jackson A. L., Loeb L. A. (2001) Mutat. Res. 477, 7–21 [DOI] [PubMed] [Google Scholar]
- 3.Ames B. N., Shigenaga M. K., Hagen T. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7915–7922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghanbari H. A., Ghanbari K., Harris P. L., Jones P. K., Kubat Z., Castellani R. J., Wolozin B. L., Smith M. A., Perry G. (2004) Aging Cell 3, 41–44 [DOI] [PubMed] [Google Scholar]
- 5.Mhatre M., Floyd R. A., Hensley K. (2004) J. Alzheimers Dis. 6, 147–157 [DOI] [PubMed] [Google Scholar]
- 6.Lee J. M., Li J., Johnson D. A., Stein T. D., Kraft A. D., Calkins M. J., Jakel R. J., Johnson J. A. (2005) FASEB J. 19, 1061–1066 [DOI] [PubMed] [Google Scholar]
- 7.Dalton T. P., Shertzer H. G., Puga A. (1999) Annu. Rev. Pharmacol. Toxicol. 39, 67–101 [DOI] [PubMed] [Google Scholar]
- 8.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999) Genes Dev. 13, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen T., Sherratt P. J., Huang H. C., Yang C. S., Pickett C. B. (2003) J. Biol. Chem. 278, 4536–4541 [DOI] [PubMed] [Google Scholar]
- 10.Venugopal R., Jaiswal A. K. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasserman W. W., Fahl W. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5361–5366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deweindt C., Albagli O., Bernardin E., Dhordain P., Quief S., Lantoine D., Kerckaert J. P., Leprince D. (1995) Cell Growth Differ. 12, 1495–1503 [PubMed] [Google Scholar]
- 13.Bardwell V. J., Treisman R. (1994) Genes Dev. 8, 1664–1677 [DOI] [PubMed] [Google Scholar]
- 14.Liang X. Q., Avraham H. K., Jiang S., Avraham S. (2004) Oncogene 23, 5890–5900 [DOI] [PubMed] [Google Scholar]
- 15.Collins T., Stone J. R., Williams A. J. (2001) Mol. Cell. Biol. 21, 3609–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson D. N., Cant K., Cooley L. (1994) Development 120, 2015–2025 [DOI] [PubMed] [Google Scholar]
- 17.Seng S., Avraham H. K., Birrane G., Jiang S., Li H., Katz G., Bass C. E., Zagozdzon R., Avraham S. (2009) Oncogene 28, 378–789 [DOI] [PubMed] [Google Scholar]
- 18.Bomont P., Cavalier L., Blondeau F., Ben Hamida C., Belal S., Tazir M., Demir E., Topaloglu H., Korinthenberg R., Tüysüz B., Landrieu P., Hentati F., Koenig M. (2000) Nat. Genet. 26, 370–374 [DOI] [PubMed] [Google Scholar]
- 19.Kim T. A., Lim J., Ota S., Raja S., Rogers R., Rivnay B., Avraham H., Avraham S. (1998) J. Cell Biol. 141, 553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seng S., Avraham H. K., Jiang S., Yang S., Sekine M., Kimelman N., Li H., Avraham S. (2007) Cancer Res. 67, 8596–8604 [DOI] [PubMed] [Google Scholar]
- 21.Motohashi H., Yamamoto M. (2004) Trends Mol. Med. 10, 549–557 [DOI] [PubMed] [Google Scholar]
- 22.Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M. (2001) Genes Cells 6, 857–868 [DOI] [PubMed] [Google Scholar]
- 23.Zipper L. M., Mulcahy R. T. (2002) J. Biol. Chem. 277, 36544–36552 [DOI] [PubMed] [Google Scholar]
- 24.Seng S., Avraham H. K., Jiang S., Venkatesh S., Avraham S. (2006) Mol. Cell. Biol. 26, 8371–8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S., Avraham H. K., Park S. Y., Kim T. A., Bu X., Seng S., Avraham S. (2005) J. Neurochem. 92, 1191–2003 [DOI] [PubMed] [Google Scholar]
- 26.Furukawa M., Xiong Y. (2005) Mol. Cell. Biol. 25, 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung S. W., Chen Y. H., Perrella M. A. (2005) J. Biol. Chem. 280, 4578–4584 [DOI] [PubMed] [Google Scholar]
- 28.Polyak K., Xia Y., Zweier J. L., Kinzler K. W., Vogelstein B. (1997) Nature 389, 300–305 [DOI] [PubMed] [Google Scholar]
- 29.Soltysik-Espanola M., Rogers R. A., Jiang S., Kim T. A., Gaedigk R., White R. A., Avraham H., Avraham S. (1999) Mol. Biol. Cell 10, 2361–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams J., Kelso R., Cooley L. (2000) Trends Cell Biol. 10, 17–24 [DOI] [PubMed] [Google Scholar]
- 31.Jiang S., Seng S., Avraham H. K., Fu Y., Avraham S. (2007) J. Biol. Chem. 282, 12319–12329 [DOI] [PubMed] [Google Scholar]
- 32.Kim T. A., Jiang S., Seng S., Cha K., Avraham H. K., Avraham S. (2005) J. Cell Sci. 118, 5537–5548 [DOI] [PubMed] [Google Scholar]
- 33.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 34.Nioi P., McMahon M., Itoh K., Yamamoto M., Hayes J. D. (2003) Biochem. J. 374, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bork P., Doolittle R. F. (1994) J. Mol. Biol. 236, 1277–1282 [DOI] [PubMed] [Google Scholar]
- 36.Begleiter A., Leith M. K., Curphey T. J., Doherty G. P. (1997) Oncol. Res. 9, 371–382 [PubMed] [Google Scholar]
- 37.Riley R. J., Workman P. (1992) Biochem. Pharmacol. 43, 1657–1669 [DOI] [PubMed] [Google Scholar]
- 38.Ross D., Siegel D., Beall H., Prakash A. S., Mulcahy R. T., Gibson N. W. (1993) Cancer Metastasis Rev. 12, 83–101 [DOI] [PubMed] [Google Scholar]
- 39.Zhang M., Zhang B. H., Chen L., An W. (2002) Cell Res. 12, 123–132 [DOI] [PubMed] [Google Scholar]
- 40.Taillé C., Almolki A., Benhamed M., Zedda C., Mégret J., Berger P., Lesèche G., Fadel E., Yamaguchi T., Marthan R., Aubier M., Boczkowski J. (2003) J. Biol. Chem. 278, 27160–27168 [DOI] [PubMed] [Google Scholar]
- 41.Poss K. D., Tonegawa S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10925–10930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.