Abstract
Wnt signaling pathways play important roles in various stages of developmental events and several aspects of adult homeostasis. Aberrant activation of Wnt signaling has also been associated with several types of cancer. We have recently identified Traf2- and Nck-interacting kinase (TNIK) as a novel activator of Wnt signaling through a comprehensive proteomic approach in human colorectal cancer cell lines. TNIK is an activating kinase for T-cell factor-4 (TCF4) and essential for the β-catenin-TCF4 transactivation and colorectal cancer growth. Here, we report the essential role of TNIK in Wnt signaling during Xenopus development. We found that Xenopus TNIK (XTNIK) was expressed maternally and that the functional knockdown of XTNIK by catalytically inactive XTNIK (K54R) or antisense morpholino oligonucleotides resulted in significant malformations with a complete loss of head and axis structures. XTNIK enhanced β-catenin-induced axis duplication and the expression of β-catenin-TCF target genes, whereas knockdown of XTNIK inhibited it. XTNIK was recruited to the promoter region of β-catenin-TCF target genes in a β-catenin-dependent manner. These results demonstrate that XTNIK is an essential factor for the transcriptional activity of the β-catenin-TCF complex and dorsal axis determination in Xenopus embryos.
Keywords: Beta-Catenin, T-cell Factor (TCF), Tumor, Wnt Pathway, Xenopus, TNIK, XTNIK, Kinase
Introduction
Wnt signaling pathways are known to control both embryonic development and adult homeostasis. The canonical Wnt pathway is the best characterized among the Wnt signaling pathways and consists of a large number of factors. In short, secreted Wnt molecules are known to evoke downstream intracellular signaling events by binding to cell surface receptors, including the Frizzled family of proteins. These signals inhibit phosphorylation of β-catenin by glycogen synthase kinase 3β and subsequent degradation of β-catenin through the ubiquitin-proteasome system, resulting in the accumulation of cytoplasmic β-catenin protein. The accumulated β-catenin protein is then translocated into the nucleus, where it acts as a transcriptional co-activator by forming complexes with the T-cell factor (TCF)2/lymphoid enhancer factor (LEF) family of nuclear proteins. The β-catenin and TCF/LEF complexes have been shown to contain a variety of protein components that modulate transcriptional activity (1–5).
During Xenopus embryogenesis, the canonical Wnt pathway plays numerous important roles. It is well established that accumulation of β-catenin at the dorsal side of the embryo (referred to as “early” canonical Wnt signaling) induces organizer formation (6). In fact, down-regulation of β-catenin by antisense oligodeoxynucleotide or antisense morpholino oligonucleotides inhibits dorsal induction (7, 8). β-Catenin accumulation is dependent on dorsal enrichment of intracellular molecules including Dishevelled (Dsh) caused by cortical rotation that is triggered by sperm entry. Moreover, it has been shown that an extracellular ligand, maternal Wnt11, also asymmetrically activates the early canonical Wnt pathways (9). Early Xenopus embryos ubiquitously express three XTcf family members, XTcf1, XTcf3, and XTcf4, which are inherited by fertilized eggs as maternal transcripts. Maternal depletion of the XTcf genes has revealed their molecular functions. XTcf1 and XTcf3 have been shown to act cooperatively as repressors of dorsal genes including Siamois and Xnr-3 in the ventral and lateral regions of early embryos in the absence of nuclear β-catenin accumulation, whereas XTcf1 and XTcf4 dorsally transactivate dorsal genes in cooperation with nuclear β-catenin (10, 11). The functional diversity of the XTcf genes may be due to differences in their DNA binding properties and binding co-factors. However, the mechanisms underlying these functions are yet to be precisely defined.
We and other researchers have recently identified that the germinal center kinase family protein, Traf2- and Nck-interacting kinase (TNIK) (12), comprised one component of the β-catenin-TCF4 complex (13, 14). TCF4 is a member of the TCF/LEF family implicated in intestinal epithelial cell renewal and colorectal carcinogenesis. We have revealed that TNIK phosphorylates TCF4 and activates the transcriptional activity of the β-catenin-TCF4 complex. Colorectal cancer cells were also shown to be highly dependent on the expression levels and kinase activity of TNIK for proliferation (15).
In the present study, we investigated the role of XTNIK during Xenopus embryogenesis. We show that XTNIK plays an essential role in β-catenin-mediated dorsal axis determination and transactivation.
EXPERIMENTAL PROCEDURES
Preparation of Xenopus Embryos
Preparation of Xenopus laevis embryos and microinjection were performed as described previously (16). Eggs were fertilized in vitro and dejellied with 1% sodium thioglycolate solution. Embryos were staged according to Nieuwkoop and Faber (17). Capped mRNA was synthesized by in vitro transcription (mMESSAGE mMACHINE kit; Ambion, Austin, TX).
Experiments were carried out according to the guidelines of the National Cancer Center Research Institute (Tokyo, Japan), which meet all the ethical requirements stipulated by Japanese law. The experimental protocols were reviewed and approved by the institutional ethics and recombination safety committees.
Real-time RT-PCR
Total RNA was isolated with TRIzol (Invitrogen). DNase-I-treated RNA was random-primed and reverse-transcribed using SuperScript II reverse transcriptase (Invitrogen). The TaqMan universal PCR master mix and custom TaqMan gene expression probe and primer sets were purchased from Applied Biosystems (Foster City, CA). The sequences of probe and primer sets were listed in supplemental Table S1. Amplification data measured as an increase in reporter fluorescence were collected using the PRISM 7000 sequence detection system (Applied Biosystems). mRNA expression levels relative to the internal control (ornithine decarboxylase (odc)) was calculated using the comparative threshold cycle (CT) method (18).
Whole-mount in Situ Hybridization
Whole-mount in situ hybridization was performed according to Harland (19). Antisense and sense digoxigenin-labeled RNA probes were generated by in vitro transcription (MEGASCRIPT kit; Ambion) from linearized plasmids encoding XTNIK, Xnr3, Siamois, chordin, and Xvent-1. Injected embryos were traced by β-galactosidase staining with Red-Gal solution (Research Organics, Cleveland, OH) as described previously (15). The percentage of embryos showing unreduced expression at the Red-Gal staining region and the total number of examined embryos are indicated in supplemental Table S2.
Immunoblot Analysis
Protein samples were fractionated and immunoblotted as described previously (20). Anti-β-catenin (sc-7199), anti-Myc (9E10), anti-HA (12CA5), and anti-β-actin (AC-15) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), Zymed Laboratories Inc. (South San Francisco, CA), Abgent (San Diego, CA), and Abcam (Cambridge, MA), respectively.
Plasmids
pCS2-FLAG-Xenopus β-catenin (Xβ-catenin) (21) and -833pSia-Luc (22) were kindly provided by Dr. S. Sokol (Mount Sinai School of Medicine, New York, NY), and pCS2-nuclear β-gal (n-β-gal) (23, 24) was kindly provided by Drs. D. Turner, R. Rupp, and J. Lee (Fred Hutchinson Cancer Research Center, Seattle, WA). pCS2+-Myc and pCS2+-HA were kindly provided by Dr. M. Taira (University of Tokyo, Tokyo, Japan). pCS2+-Xnr5 (25) and pCS2+-BMP4-HA (26) were kindly provided by Dr. M. Asashima (University of Tokyo, Tokyo, Japan).
For the whole-mount in situ hybridization probes, partial sequences of XTNIK, Xnr3, Siamois, chordin, and Xvent-1 were PCR-amplified using the following primer sets: XTNIK, 5′-aaagaaccctcctggaatgg-3′ and 5′-actgatctgttgtgggtctt-3′; Xnr3, 5′-ctatccaacctggaacatcctact-3′ and 5′-gaacagcttctggccaagac-3′; Siamois, 5′-tgccacgctgaaattattggg-3′ and 5′-gtggaaagtggttgctcttgg-3′; chordin, 5′-gttgttaaagggcttctatggg-3′ and 5′-gtctgccagttccataggac-3′; Xvent-1, 5′-ccaaccaaatatgccaaggaga-3′ and 5′-agccaccagggtaataaggg-3′. The amplified fragments were then subcloned into the EcoRV site of pcDNA3.1 (Invitrogen).
The 5′-UTR and ORF of LOC443633 (XTNIK), which are recognized by antisense morpholino oligonucleotide (MO) XTNIK-MO1 or -MO2, were PCR-amplified from the X. laevis IMAGE cDNA clone MXL1736-98358477 (Open Biosystems, Huntsville, AL) and subcloned into pCS2+-Myc (pCS-XTNIK-WT-Myc). The mutant form of pCS-XTNIK-WT-Myc (pCS-XTNIK-K54R-Myc) was constructed by mutagenesis with oligonucleotides 5′-AGGGTCATGGATGTCACAGGGGATG-3′ and 5′-AATAGCTGCAAGCTGTCCGGTTTTAAC-3′ to alter the lysine 54 residue to arginine.
The ORF sequence of XTNIK, which is not recognized by XTNIK-MO1 or -MO2, was constructed by mutagenesis with oligonucleotides 5′-ATGGCcAGtGAtTCtCCGGCTCGTAGCCTGGATGA-3′ (lowercase letters indicate modifications) and 5′-ATCGATGGGATCCTGCAAAAAGAACAA-3′ (pCS2-XTNIKORF-HA). XTCF4 was PCR-amplified and subcloned into the EcoRV site of pCS2+-HA plasmid.
Antisense and Control MOs
The antisense MOs for Xenopus TNIK (XTNIK-MO1 and -MO2) and the corresponding control MOs (carrying five nucleotide substitutions (underlined nucleotides) within the XTNIK-MO1 and -MO2 sequences (5mis-Control-1 and -2, where “5mis” designates five mismatched nucleotides)) were obtained from Gene Tools (Philomath, OR). A database search confirmed the absence of a significant homologous sequence to the complements of XTNIK-MO1 and -MO2 in X. laevis. The sequences of the MOs used in this study were: XTNIK-MO1 (5′-GGGAGTCGCTCGCCATGTTTCCTTT-3′), XTNIK-MO2 (5′-CCCCGTTCTTTCCACCTTGCGGCTG-3′), 5mis-Control-1 (5′-GGCAGTGGCTCCCCATCTTTCGTTT-3′), and 5mis-Control-2 (5′-CCGCGTTGTTTCGACCTTCCGCCTG-3′).
Luciferase Assay
Embryos were injected with -833pSia-Luc and pRL-TK plasmids (Promega, Madison, WI) with mRNA in the animal pole at the four-cell stage. Animal caps were dissected at stage 9, and luciferase activity was measured with the Dual-Luciferase reporter assay system (Promega) and normalized using Renilla reniformis luciferase activity as an internal control.
Chromatin Immunoprecipitation
Injected or uninjected embryos were fixed with 1% formaldehyde for 15 min at room temperature, washed twice with PBS, and lysed with radioimmune precipitation buffer lysis (Upstate Biotech Millipore, Lake Placid, NY) containing protease inhibitor mixture (Roche Diagnostics, Mannheim, Germany) and phosphatase inhibitor cocktails (Sigma-Aldrich) as reported previously (27). The solution was then sheared using a BioRuptor sonicator (Cosmo Bio Co., Tokyo, Japan) with 30-s pulses at 1-min intervals for 8 min at the maximum setting and then centrifuged with 14,000 × g at 4 °C for 10 min. The supernatant was diluted 10 times with ChIP dilution buffer (ChIP assay kit, Millipore, Billerica, MA), and 50 μl of the dilute was separated as the input sample. The remainder was prewashed with protein A beads and incubated with 4 μg of anti-Myc antibody (9E10, Sigma) overnight at 4 °C. Immunoprecipitates were washed, and cross-links were removed according to the manufacturer's instructions (Millipore). Input and immunoprecipitated DNA were subjected to real-time PCR with probe/primers overlapping the promoter region of the Xnr3 or Siamois gene. Immunoprecipitated DNA values relative to the input values was calculated using the comparative threshold cycle (CT) method as mentioned previously (27).
Immunoprecipitation
Injected embryos were harvested at stage 10 and lysed with lysis buffer (0.5% Triton X-100, 250 mm NaCl, 50 mm Tris-HCl, pH 7.0) containing protease inhibitor mixture (Roche Diagnostics) and phosphatase inhibitor mixture (Sigma). After preclearing the lysates with mouse IgG-protein G complex, they were incubated with 4 μg of anti-Myc antibody (9E10), anti-HA antibody (H3663, Sigma), or normal mouse IgG overnight at 4 °C and precipitated with protein G-Sepharose (GE Healthcare, Buckinghamshire, UK).
Statistical Analysis
Results were obtained from at least three experiments, and statistical significance of the difference between groups was determined using Student's t test. Differences were considered significant for p values < 0.05.
RESULTS
Identification of Xenopus TNIK
A previously described registry has demonstrated the significant homology between human TNIK and Xenopus XTNIK in the UniGene Xenopus laevis database (termed hypothetical protein LOC443633). Although XTNIK lacks the portion equivalent to the C-terminal half of the human TNIK, the kinase domain spanning amino acids 25–289 was found to be highly conserved (98.9%) between human and Xenopus (supplemental Fig. S1).
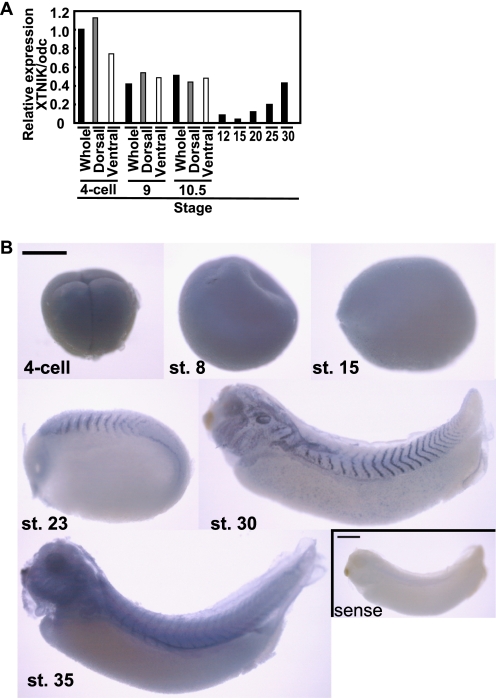
We first examined the expression of XTNIK mRNA during Xenopus embryogenesis. We found that the expression of XTNIK was detected maternally. At the gastrula stages, zygotic XTNIK expression was observed and maintained until the tadpole stage (Fig. 1A). Whole-mount in situ hybridization demonstrated that XTNIK was ubiquitously expressed until the gastrula stage. At the neurula stage, localized expression was observed in the somatic mesoderm. In the tadpole stage, expression was localized to the somatic mesoderm, pronephros, branchial arches, eyes and otic vesicles (Fig. 1B).
FIGURE 1.
Expression of XTNIK in Xenopus embryo. A, expression of XTNIK was quantified using real-time PCR and expressed as a ratio relative to the expression of the ornithine decarboxylase (odc) gene. Embryos at the four-cell stage, stage 9, and stage 10.5 were divided into dorsal (gray) and ventral (white) sides. B, whole-mount in situ hybridization for the expression of XTNIK. Lateral views are presented. As a negative control, sense probe staining is presented in the right bottom panel. Scale bars = 0.5 mm. st., stage.
Involvement of XTNIK in Dorsal Axis Determination
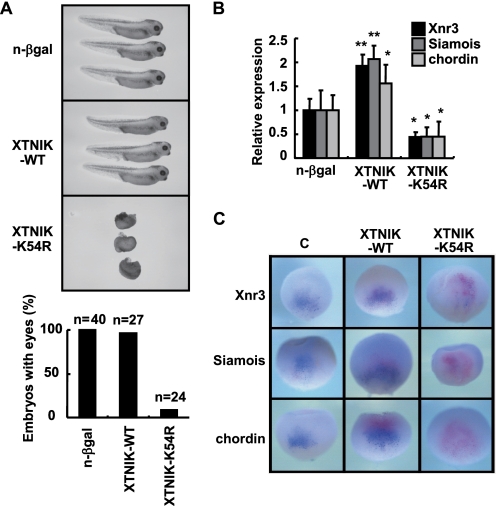
To examine the effects of TNIK and the catalytically inactive form of TNIK (K54R), we injected XTNIK or XTNIK (K54R) mRNA into Xenopus embryos. Injection of XTNIK (K54R) mRNA into dorsal blastomeres of eight-cell stage embryos inhibited the initiation of gastrulation at stage 10 (supplemental Fig. S2) and resulted in significant axis defects with a complete loss of head and axis structures (Fig. 2A). Injection of n-β-gal or XTNIK (WT) mRNA failed to affect axis formation (Fig. 2 and supplemental Fig. S2). Embryos injected with XTNIK or XTNIK (K54R) mRNA into the ventral blastomeres were also found to develop normally (data not shown). Consistent with the morphological defects, dorsal injection of TNIK (K54R) mRNA suppressed the expression of Siamois, Xnr3, and chordin, whereas dorsal injection of XTNIK (WT) increased the expression (Fig. 2, B and C, and supplemental Table S2). These results suggest involvement of XTNIK in dorsal axis determination in the Xenopus embryo.
FIGURE 2.
XTNIK in dorsal axis determination. mRNA for n-β-gal (500 pg), XTNIK-WT-Myc (500 pg), or XTNIK-K54R-Myc (500 pg) was injected into the dorsal marginal zone of eight-cell stage embryos. A, representative appearance of tadpoles and the ratio of tadpoles with eyes at stage 35. B, embryos were harvested at stage 9, and the relative mRNA expression of Siamois, Xnr3, and chordin was determined by real-time PCR. The statistical significance with respect to n-β-gal-injected control (*, p < 0.05; **, p < 0.01) was determined using Student's t test. Bars represent mean ± S.D. C, embryos were injected with XTNIK-WT-Myc (500 pg) or XTNIK-K54R-Myc (500 pg) together with n-β-gal (300 pg) mRNA at the four-cell stage and harvested at stage 9.5. The mRNA expression of Xnr3, Siamois, and chordin was examined by whole-mount in situ hybridization and stained blue. Red-Gal staining was used as a tracer. Dorsal views are presented.
XTNIK Enhanced β-Catenin-induced Secondary Axis Formation
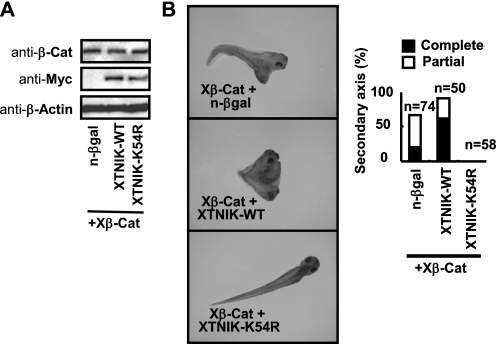
We next examined whether XTNIK was involved in Wnt signaling. Co-injection of XTNIK (WT) mRNA with a low dose of Xβ-catenin (25 pg) into the ventral marginal zone of eight-cell stage embryos enhanced the formation of secondary axis with complete head structures. Catalytically inactive XTNIK (K54R) completely inhibited secondary axis formation (Fig. 3, A and B). The expression of Xnr3, Siamois, and chordin induced by ventral injection of Xβ-catenin was inhibited by XTNIK (K54R) (supplemental Fig. S3). Embryos injected with XTNIK (WT) or XTNIK (K54R) only (without Xβ-catenin) were found to develop normally (data not shown). These observations indicate that XTNIK enhances Wnt signaling in a β-catenin-dependent manner.
FIGURE 3.
Regulation of Wnt signaling by XTNIK in Xenopus embryos. mRNA for Xβ-catenin (Xβ-Cat, 25 pg), n-β-gal (500 pg), XTNIK-WT-Myc (500 pg), or XTNIK-K54R-Myc (500 pg) was injected in the indicated combinations into the ventral marginal zone of eight-cell stage embryos. A, expression of Xβ-catenin, Myc-tagged XTNIK (WT or K54R), and β-actin (loading control) proteins was determined by immunoblotting. B, representative appearance of tadpoles and the ratio of tadpoles with secondary axis formation. Complete indicates axis formation with head-to-trunk structures. Partial indicates axis formation without heads. Secondary axes with or without cement glands were counted as complete or partial, respectively.
Translational Blockage of XTNIK Inhibits β-Catenin-induced Axis Formation
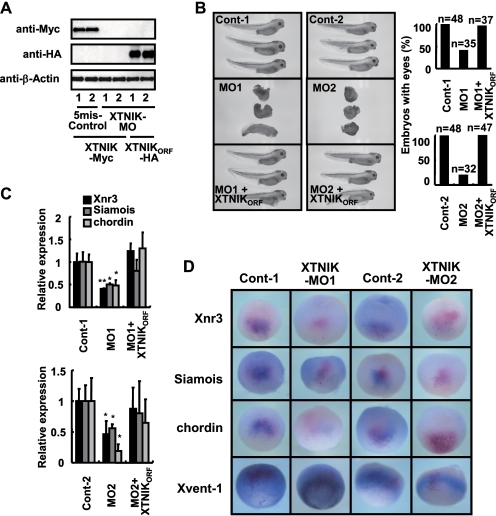
To block the translation of XTNIK, we designed two antisense MOs for XTNIK (namely MO1 and MO2), as well as their corresponding control MOs (5mis-Control-1 and -Control-2) in which five mismatched nucleotides were inserted into the XTNIK-MO1 and -MO2 sequences, respectively. We found that XTNIK-MO1 and -MO2 blocked the translation of XTNIK-(WT)-Myc that contained the 5′-UTR and ORF sequences recognized by the XTNIK-MO1 and -MO2 mRNA (Fig. 4A). Neither 5mis-Control-1 nor 5mis-Control-2 blocked this translation. Translation of HA-tagged XTNIKORF (XTNIKORF-HA) mRNA (ORF of LOC443633 (XTNIK) lacking the 5′-UTR targeted by XTNIK-MO1 and -MO2) was not blocked by either XTNIK-MO1 or XTNIK-MO2. Therefore, we used XTNIKORF-HA for the rescue of XTNIK protein level reduced by XTNIK-MO1 and -MO2 in the following experiments.
FIGURE 4.
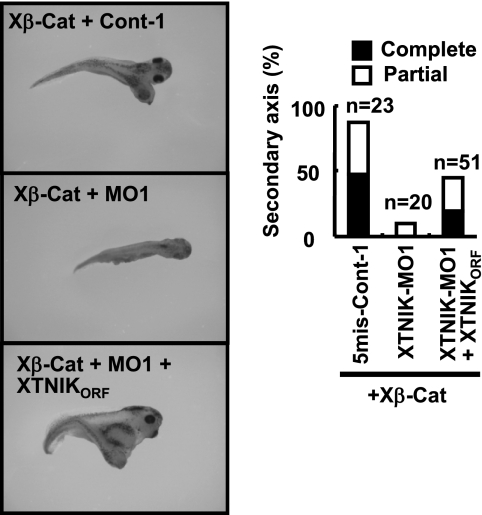
Translational blockage of XTNIK inhibits dorsal axis formation. 5mis-Control-1 (40 ng), 5mis-Control-2 (40 ng), XTNIK-MO1 (40 ng), or XTNIK-MO2 (40 ng) and XTNIKORF-HA (500 pg) mRNA were co-injected in the indicated combinations into the dorsal marginal zone of eight-cell stage embryos. A, expression of Myc-tagged XTNIK (WT), HA-tagged XTNIKORF, and β-actin (loading control) proteins was determined by immunoblotting. B, representative appearance of tadpoles and the ratio of tadpoles with eyes. C, relative mRNA expression of Siamois, Xnr3, and chordin at stage 9 was determined by real-time PCR. The statistical significance with respect to the control MO-injected control (*, p < 0.05; **, p < 0.01) was determined using Student's t test. Bars represent mean ± S.D. D, embryos injected with 5mis-Control-1 (40 ng), 5mis-Control-2 (40 ng), XTNIK-MO1 (40 ng), or XTNIK-MO2 (40 ng) together with n-β-gal (300 pg) mRNA at the four-cell stage and harvested at stage 9.5 (for Xnr3, Siamois, and chordin) or 10.5 (for Xvent-1). The mRNA expression of Xnr3, Siamois, chordin, and Xvent-1 was examined by whole-mount in situ hybridization and stained blue. Red-Gal staining was used as a tracer. Dorsal views (Xnr3, Siamois, and chordin) and ventral-vegetal views (Xvent-1) are presented.
We found that embryos injected dorsally with XTNIK-MOs (MO1 and MO2) failed to initiate gastrulation at stage 10 (supplemental Fig. S4) and developed into abnormal tadpoles with significantly reduced head and axis structures (Fig. 4B). Embryos injected with control MOs developed normally. The defects caused by XTNIK-MO were rescued by co-injection with XTNIKORF-HA (Fig. 4B and supplemental Fig. S4). The expression of Siamois, Xnr3, and chordin was also reduced by XTNIK-MO (Fig. 4, C and D, and supplemental Table S2), and the expression was reversed by co-injection with XTNIKORF-HA (Fig. 4C). Ventral injection of XTNIK-MO did not affect Xvent-1 expression (Fig. 4D) and dorsal marker expression (data not shown).
XTNIK-MO1 and -MO2, but not their corresponding control MOs, blocked secondary axis formation induced by Xβ-catenin when co-injected into the ventral marginal zone of eight-cell stage embryos. The blockage was abrogated by co-injection of XTNIKORF-HA mRNA (Fig. 5 and supplemental Fig. S5). These results suggest that endogenous XTNIK is essential for β-catenin-dependent axis formation.
FIGURE 5.
Translational blockage of XTNIK inhibits β-catenin-induced secondary axis formation. 5mis-Control-1 (40 ng), XTNIK-MO1 (40 ng), or XTNIKORF-HA (500 pg) mRNA and Xβ-catenin (Xβ-Cat) mRNA (50 pg) were co-injected in the indicated combinations into the ventral marginal zone of eight-cell stage embryos. The representative appearance of tadpoles and the ratio of tadpoles with secondary axis formation are shown. Complete indicates axis formation with head-to-trunk structures. Partial indicates axis formation without heads. Secondary axes with or without cement glands were counted as complete or partial, respectively.
XTNIK Is an Essential, Conserved Enhancer of Canonical Wnt Signaling
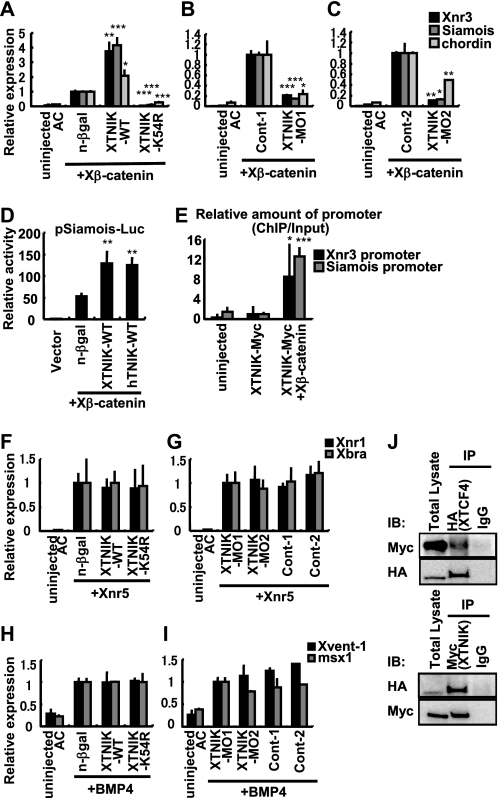
In the animal cap assay, injection of Xβ-catenin induced expression of known Wnt signaling target genes, including Siamois and Xnr3, and dorsal gene, chordin. Co-injection with XTNIK (WT) enhanced the expression of these genes, whereas XTNIK (K54R) or XTNIK-MO down-regulated their expression (Fig. 6, A–C). Consistent with this, XTNIK (WT) enhanced the luciferase activity promoted by the Siamois promoter (22), as well as by human TNIK (Fig. 6D), suggesting functional conservation. Furthermore, ChIP assay revealed that XTNIK was recruited to the promoter region of Wnt signaling target genes (Xnr3 or Siamois) in a β-catenin-dependent manner (Fig, 6E), which has been suggested in mouse crypt (14). Neither overexpression nor knockdown of XTNIK affected the target gene expression induced by Xnr5 or BMP4 (Fig. 6, F–I), suggesting a specific role of XTNIK in Wnt signaling. Finally, we also confirmed the interaction between XTNIK and XTCF4 in Xenopus embryos (Fig. 6J). These results suggest an essential, conserved role of XTNIK in Wnt signaling.
FIGURE 6.
XTNIK is an essential and specific enhancer for β-catenin-Tcf-transactivation. A–C, mRNA for Xβ-catenin (25 pg), nuclear β-galactosidase (500 pg), XTNIK-WT-Myc (500 pg), XTNIK-K54R-Myc (500 pg), 5mis-Control-MO1 (Cont-1) (40 ng), XTNIK-MO1 (40 ng), 5mis-Control-MO2 (Cont-2) (40 ng), or XTNIK-MO2 (40 ng) was injected in the indicated combinations into the animal poles of four-cell stage embryos. Twenty animal caps (AC) per group were dissected at stage 9, cultured for 30 min, and harvested for RNA isolation. Relative mRNA expression of Siamois, Xnr3, and chordin was determined by real-time PCR. The statistical significance with respect to n-β-gal mRNA or control MO-injected controls (*, p < 0.05; **, p < 0.01; ***, p < 0.001) was determined using Student's t test. D, -833pSia-Luc plasmid (50 pg) and pRL-TK plasmid (10 pg) were co-injected with Xβ-catenin (25 pg), n-β-gal (500 pg), XTNIK (WT) (500 pg), or human TNIK (WT) (hTNIK-WT, 500 pg) mRNA in the indicated combinations into the animal poles of four-cell stage embryos. Ten animal caps per group were dissected at stage 9, cultured for 30 min, and harvested for the luciferase assay. The statistical significance with respect to the n-β-gal mRNA-injected control (**, p < 0.01) was determined using Student's t test. E, XTNIK-WT-Myc (500 pg) was injected with or without Xβ-catenin (100 pg) mRNA in the ventral marginal zone of embryos. Embryos were harvested at stage 10, and a ChIP assay was performed. F–I, mRNA for Xnr5 (5 pg), BMP4 (500 pg), nuclear β-galactosidase (500 pg), XTNIK-WT-Myc (500 pg), or XTNIK-K54R-Myc (500 pg) and for 5mis-Control-MO1 (Cont-1) (40 ng), XTNIK-MO1 (40 ng), 5mis-Control-MO2 (Cont-2) (40 ng), or XTNIK-MO2 (40 ng) was injected in the indicated combinations into the animal poles of four-cell stage embryos. Twenty animal caps per group were dissected at stage 9, cultured for 1.5 h, and harvested for RNA isolation. Relative mRNA expression of Xnr1, Xbra, Xvent-1, and msx1 was determined by real-time PCR. Bars represent mean ± S.D. J, mRNA for XTNIK-WT-Myc (500 pg) and HA-XTCF4 (500 pg) was injected into the dorsal marginal zone of four-cell stage embryos and harvested at stage 9, and immunoprecipitation was performed.
DISCUSSION
Our previous study identified TNIK as one of the protein components of TCF4-containing complexes in the colorectal cancer cell lines DLD1 and HCT-116, which each have a respective truncating mutation in the APC gene and a missense mutation in the CTNNB1 gene (12, 13). Recently, other investigators have also identified the presence of TNIK in TCF4-containing complexes in proliferative crypts of the murine small intestine (14). We and others have also demonstrated that TNIK interacts with and phosphorylates TCF4, in addition to activating transcription by the β-catenin-TCF4 complex. TNIK interacts with TCF4 through amino acids 1–289 and phosphorylates TCF4, whereas amino acids 100–215 of TCF4 are necessary for interaction with TNIK (15). The TCF4-interacting domain (amino acids 1–289) of human TNIK is highly conserved with XTNIK, sharing 99% identity. The TNIK-interacting domain (amino acids 100–215) of human TCF4 is also conserved in XTcf4 and XTcf3, which share 68 and 50% identity, respectively. These data indicate that the function of TNIK is most likely conserved between human and Xenopus in relation to TCF phosphorylation. In the current study, we showed that XTNIK interacted with XTCF4 and was essential for β-catenin-TCF-mediated transactivation and dorsal axis formation.
In early Xenopus embryo stages, three XTcf family members, XTcf1, XTcf3, and XTcf4, cooperatively regulate dorsal-ventral specification. XTcf1 and XTcf3 cooperatively act as repressors of β-catenin-TCF target genes by binding at the promoter region in the ventral region, whereas XTcf1 and XTcf4 transactivate β-catenin-TCF target genes in cooperation with nuclear β-catenin on the dorsal side (11). Considering the similarities between the TNIK-interacting domain of human TCF4 and the relevant domains of the XTcf genes, XTcf4 and XTcf3 are hypothesized to interact with, and be phosphorylated by, XTNIK. We demonstrated that dorsal inhibition of XTNIK by XTNIK (K54R) or XTNIK-MO suppresses dorsal axis formation and expression of β-catenin-TCF target genes. In contrast, ventral inhibition of XTNIK resulted in no obvious effects. When XTNIK (K54R) or XTNIK-MO was injected with β-catenin in the ventral marginal zone, β-catenin-induced secondary axis formation was blocked. These results suggest that XTNIK is not activated in the absence of nuclear β-catenin but is essential for β-catenin-mediated transactivation of its target genes. It has been reported that TNIK is recruited to the promoter region of its target genes in cooperation with TCF4 and β-catenin in a β-catenin-dependent manner (14). We also showed that XTNIK is recruited to the promoter region of the Xnr3 and Siamois genes in a β-catenin-dependent manner. Taken together, our results suggest that the XTNIK promotes dorsal axis formation in cooperation with XTcf4 and β-catenin. These results demonstrate the conserved functional involvement of TNIK during canonical Wnt signaling in vivo and the essential function of TNIK for transactivation of β-catenin and TCF target genes. Aberrant activation of the Wnt pathway has been associated with several types of human cancers and disease. Thus, TNIK may prove useful as a therapeutic target molecule for specific inhibition of the Wnt signaling pathway for cancer therapeutics.
Supplementary Material
Acknowledgments
We thank Dr. S. Sokol (Mount Sinai School of Medicine, New York, NY) for providing pCS2+Xβ-catenin and -833pSia-Luc; Drs. D. Turner, R. Rupp, and J. Lee (Fred Hutchinson Cancer Research Center, Seattle, WA) for providing pCS2+n-β-gal; Dr. M. Taira (University of Tokyo, Tokyo, Japan) for providing pCS2+ vectors and for technical advice; and Dr. M. Asashima (University of Tokyo, Tokyo, Japan) for providing pCS2+-Xnr5 and pCS2+-BMP4-HA.
This work was supported by grants from the “Program for Promotion of Fundamental Studies in Health Sciences” conducted by the National Institute of Biomedical Innovation of Japan and the “Third-Term Comprehensive Control Research for Cancer” and “Research on Biological Markers for New Drug Development” conducted by the Ministry of Health, Labor, and Welfare of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
- TCF
- T-cell factor
- LEF
- lymphoid enhancer factor
- MO
- morpholino oligonucleotide
- n-β-gal
- nuclear β-gal
- TNIK
- Traf2- and Nck-interacting kinase
- XTNIK
- Xenopus TNIK
- Xβ-catenin
- Xenopus β-catenin.
REFERENCES
- 1.Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., Peifer M., Bejsovec A. (1998) Nature 395, 604–608 [DOI] [PubMed] [Google Scholar]
- 2.Sun Y., Kolligs F. T., Hottiger M. O., Mosavin R., Fearon E. R., Nabel G. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12613–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato S., Idogawa M., Honda K., Fujii G., Kawashima H., Takekuma K., Hoshika A., Hirohashi S., Yamada T. (2005) Gastroenterology 129, 1225–1236 [DOI] [PubMed] [Google Scholar]
- 4.Idogawa M., Masutani M., Shitashige M., Honda K., Tokino T., Shinomura Y., Imai K., Hirohashi S., Yamada T. (2007) Cancer Res. 67, 911–918 [DOI] [PubMed] [Google Scholar]
- 5.Shitashige M., Hirohashi S., Yamada T. (2008) Cancer Sci. 99, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moon R. T., Kimelman D. (1998) Bioessays 20, 536–545 [DOI] [PubMed] [Google Scholar]
- 7.Heasman J., Crawford A., Goldstone K., Garner-Hamrick P., Gumbiner B., McCrea P., Kintner C., Noro C. Y., Wylie C. (1994) Cell 79, 791–803 [DOI] [PubMed] [Google Scholar]
- 8.Heasman J., Kofron M., Wylie C. (2000) Dev. Biol. 222, 124–134 [DOI] [PubMed] [Google Scholar]
- 9.Tao Q., Yokota C., Puck H., Kofron M., Birsoy B., Yan D., Asashima M., Wylie C. C., Lin X., Heasman J. (2005) Cell 120, 857–871 [DOI] [PubMed] [Google Scholar]
- 10.Houston D. W., Kofron M., Resnik E., Langland R., Destree O., Wylie C., Heasman J. (2002) Development 129, 4015–4025 [DOI] [PubMed] [Google Scholar]
- 11.Standley H. J., Destrée O., Kofron M., Wylie C., Heasman J. (2006) Dev. Biol. 289, 318–328 [DOI] [PubMed] [Google Scholar]
- 12.Fu C. A., Shen M., Huang B. C., Lasaga J., Payan D. G., Luo Y. (1999) J. Biol. Chem. 274, 30729–30737 [DOI] [PubMed] [Google Scholar]
- 13.Shitashige M., Satow R., Honda K., Ono M., Hirohashi S., Yamada T. (2008) Gastroenterology 134, 1961–1971 [DOI] [PubMed] [Google Scholar]
- 14.Mahmoudi T., Li V. S., Ng S. S., Taouatas N., Vries R. G., Mohammed S., Heck A. J., Clevers H. (2009) EMBO J. 28, 3329–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shitashige M., Satow R., Jigami T., Aoki K., Honda K., Shibata T., Ono M., Hirohashi S., Yamada T. (2010) Cancer Res. 70, 5024–5033 [DOI] [PubMed] [Google Scholar]
- 16.Satow R., Chan T. C., Asashima M. (2002) Biochem. Biophys. Res. Commun. 295, 85–91 [DOI] [PubMed] [Google Scholar]
- 17.Nieuwkoop P. D., Faber J. (eds) (1956) Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of the Metamorphosis, North Holland Publishing Co., Amsterdam [Google Scholar]
- 18.Huang L., Shitashige M., Satow R., Honda K., Ono M., Yun J., Tomida A., Tsuruo T., Hirohashi S., Yamada T. (2007) Gastroenterology 133, 1569–1578 [DOI] [PubMed] [Google Scholar]
- 19.Harland R. M. (1991) Methods Cell Biol. 36, 685–695 [DOI] [PubMed] [Google Scholar]
- 20.Honda K., Yamada T., Hayashida Y., Idogawa M., Sato S., Hasegawa F., Ino Y., Ono M., Hirohashi S. (2005) Gastroenterology 128, 51–62 [DOI] [PubMed] [Google Scholar]
- 21.Sokol S. Y. (1996) Curr. Biol. 6, 1456–1467 [DOI] [PubMed] [Google Scholar]
- 22.Fan M. J., Grüning W., Walz G., Sokol S. Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5626–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner D. L., Weintraub H. (1994) Genes Dev. 8, 1434–1447 [DOI] [PubMed] [Google Scholar]
- 24.Rupp R. A., Snider L., Weintraub H. (1994) Genes Dev. 8, 1311–1323 [DOI] [PubMed] [Google Scholar]
- 25.Takahashi S., Yokota C., Takano K., Tanegashima K., Onuma Y., Goto J., Asashima M. (2000) Development 127, 5319–5329 [DOI] [PubMed] [Google Scholar]
- 26.Haramoto Y., Tanegashima K., Onuma Y., Takahashi S., Sekizaki H., Asashima M. (2004) Dev. Biol. 265, 155–168 [DOI] [PubMed] [Google Scholar]
- 27.Blythe S. A., Reid C. D., Kessler D. S., Klein P. S. (2009) Dev. Dyn. 238, 1422–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.