Abstract
The identification of pathologic TDP-43 aggregates in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration, followed by the discovery of dominantly inherited point mutations in TDP-43 in familial ALS, have been critical insights into the mechanism of these untreatable neurodegenerative diseases. However, the biochemical basis of TDP-43 aggregation and the mechanism of how mutations in TDP-43 lead to disease remain enigmatic. In efforts to understand how TDP-43 alters its cellular localization in response to proteotoxic stress, we found that TDP-43 is sequestered into polyglutamine aggregates. Furthermore, we found that binding to polyglutamine aggregates requires a previously uncharacterized glutamine/asparagine (Q/N)-rich region in the C-terminal domain of TDP-43. Sequestration into polyglutamine aggregates causes TDP-43 to be cleared from the nucleus and become detergent-insoluble. Finally, we observed that sequestration into polyglutamine aggregates led to loss of TDP-43-mediated splicing in the nucleus and that polyglutamine toxicity could be partially rescued by increasing expression of TDP-43. These data indicate pathologic sequestration into polyglutamine aggregates, and loss of nuclear TDP-43 function may play an unexpected role in polyglutamine disease pathogenesis. Furthermore, as Q/N domains have a strong tendency to self-aggregate and in some cases can function as prions, the identification of a Q/N domain in TDP-43 has important implications for the mechanism of pathologic aggregation of TDP-43 in ALS and other neurodegenerative diseases.
Keywords: Amyotropic Lateral Sclerosis (Lou Gehrig Disease), Huntington Disease, Neurodegeneration, Polyglutamine Disease, Protein Degradation, RNA Splicing
Introduction
Abnormal protein aggregation is a hallmark of most inherited and acquired neurodegenerative diseases. Recently, TDP-43 was identified as a component of ubiquitinated aggregates in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD)4 (1). The subsequent finding of mutations in TDP-43 in cases of inherited ALS indicates that TDP-43 can be directly involved in the pathogenesis of at least the familial forms of this disease (2–6). It is notable that although aggregates of a particular protein are initially associated with a specific clinical and pathologic syndrome, they are often observed in multiple other neurodegenerative disorders. For example, although cytoplasmic inclusions of TDP-43 were initially described in ALS and FTLD, they have also been observed in Alzheimer disease, diffuse Lewy body disease, dementia pugilistica, Huntington disease, and even inclusion body myopathies (7–11). Whether TDP-43 translocation to the cytosol and aggregation plays a direct role in the pathogenesis of these disorders, or instead is part of a more general cellular stress response, remains to be elucidated (12).
Polyglutamine diseases are a family of neurodegenerative disorders caused by expansion of a CAG trinucleotide repeat in the coding regions of certain genes. Examples include Huntington disease, X-linked spinal-bulbar muscular atrophy, and several of the spinocerebellar ataxias (13–15). Expanded polyglutamine proteins have a strong tendency to aggregate, and although the formation of large macromolecular inclusions is likely protective to cells (16, 17), much evidence suggests that polyglutamine protein misfolding and aggregation (possibly of oligomeric or other soluble species) play a key role in pathogenesis (18). Several mechanisms of polyglutamine aggregate toxicity are proposed, including (i) altered metabolism and mitochondrial dysfunction; (ii) altered transcriptional regulation; (iii) induction of proteotoxic stress; (iv) alterations in axonal transport, and (v) disruption of normal polyglutamine protein-protein interactions (14, 18, 19). One mechanism by which polyglutamine aggregates mediate toxicity is through recruitment of heterologous proteins, including transcription factors and RNA-binding proteins, and components of the ubiquitin proteosome system (13–15, 18).
Many polyglutamine aggregate interacting proteins contain an unexpanded polyglutamine stretch or a glutamine/asparagine (Q/N)-rich region (20, 21). Q/N-rich regions are also known as “prion-related domains” because they themselves have a strong tendency to aggregate and can propagate the aggregated state from mother to daughter cells in yeast. For example, the Q/N domain mediates misfolding and aggregation of the protein Sup35, leading to inactivation of the protein, and inheritance of nonfunctional Sup35 as a non-Mendelian trait (22). Interestingly FUS/TLS, another RNA-binding protein found to be mutated in familial ALS (23, 24), was recently identified in a screen for polyglutamine-interacting proteins, and this interaction required the Q/N-rich domain in the protein (25).
In an effort to identify conditions that influence TDP-43 translocation to the cytosol, and its aggregation, we observed that TDP-43 is sequestered into polyglutamine aggregates. This causes TDP-43 to be depleted from its normal nuclear location, which in turn disrupts TDP-43-mediated splicing in the nucleus. Importantly, co-aggregation with polyglutamine proteins requires a previously uncharacterized Q/N-rich domain in the C-terminal region of TDP-43, which provides novel insight into the mechanism of self-aggregation of TDP-43.
EXPERIMENTAL PROCEDURES
Plasmids, Constructs, and Live Cell Imaging
PolyQ19-CFP and polyQ80-CFP, FLAG-tagged human TDP-43 fused to pCherry, and Htt-Q72 and Htt-Q25 YFP/CFP constructs were previously described (26–28). GFP-fused wild-type and G59S dynactin constructs were generously provided by Erika Holzbauer. The human caveolin-3 cDNA was purchased from Invitrogen and was subcloned into pcDNA6.2 YFP DEST Gateway vector to make a caveolin3-YFP fusion construct. The P104L point mutation was then generated in the pcDNA6.2-caveolin3-YFP using QuikChange mutagenesis (Stratagene), and incorporation of the point mutation was confirmed via nucleotide sequencing. TDP-43 deletion constructs were generated using PCR mutagenesis, cloned into the pCherry-C1 vector, and sequenced in their entirety.
Cell Culture, Antibodies, and Immunocytochemistry
Imaging of live cells was performed in a climate-controlled chamber (In Vivo Scientific) at 37 °C and 5% CO2, and images were acquired with a Cool Snap HQ2 CCD camera (Photometrics) mounted on a Nikon Eclipse Ti-U microscope, controlled using MetaMorph software (Molecular Devices). For proteosome inhibition experiments, COS7 cells were plated in 24-well plates, transfected with Cherry-TDP-43 constructs using TransitLT1 (Mirus) according to the manufacturer's instructions, and 24 h after transfection treated with 10 μm MG132 (Sigma) for 16 h, and immunocytochemistry for ubiquitin was performed. For immunocytochemistry, HeLa or COS7 cells were transfected the day after plating using TransitLT1 transfection reagent. Cells were rinsed in cold PBS, fixed in 4% paraformaldehyde for 15 min at room temperature, washed again in PBS, permeabilized in PBS + 0.1% Triton X-100 for 5 min, blocked in PBS with 5% normal goat serum for 60 min, and incubated in primary antibody overnight in blocking solution. After three washes with PBS, cells were incubated with the appropriate secondary antibody in blocking solution for 1 h at room temperature, washed again in PBS, and imaged. Antibodies used were as follows: rabbit anti-TDP-43 (Proteintech Group, 1:500); mouse anti-hnRNPA1 (GeneTex, 1:500); mouse anti-SAFB (Abcam, 1:500); mouse anti-ubiquitin FK2 (Millipore, 1:1000); anti-GFP (Sigma, 1:2500); goat anti-rabbit Alexa-488 or Alexa-594 (Invitrogen, 1:100).
Filter Trap Assay for Detergent-insoluble Protein Aggregates
15,000 HeLa cells/cm2 were seeded in 6-well plates. Cells were transfected 24 h later with FuGENE 6 reagent in a 1:3 (μg/μl) ratio using 500 ng of Cherry fusion constructs and 500 ng of polyglutamine constructs. 48 h later cells were washed twice with PBS with 1 mm CaCl2, 0.5 mm MgCl2 (PBSc), scraped in PBS, and pelleted at 700 × g for 10 min at 4 °C. Cell pellets were resuspended in 300 μl of PBS with 1 mm PMSF (PBS/PMSF) and sonicated with 10 pulses in an ultrasonic homogenizer model Omni-Ruptor 250 (Omni, Kennesaw, GA) with 25% power and 10% pulser settings. After centrifugation for 10 min at 700 × g at 4 °C, cell lysates were normalized to 0.2 mg/ml protein in PBS/PMSF and diluted in PBS, 2% SDS buffer. 20 or 5 μg was applied to a pre-wetted cellulose acetate 0.2-μm filter using a dot blot device (Bio-Rad). After two washes with 500 μl of PBS, 2% SDS buffer, the membrane was incubated for 1 h in blocking buffer (5% milk in PBS containing 0.05% Tween 20) with gentle rocking at room temperature. The membrane was then incubated with anti-GFP antibodies in blocking buffer for 2 h at room temperature, washed four times for 10 min with washing buffer (PBS with 0.05% Tween 20), and incubated with secondary antibodies in blocking buffer (1:5000) for 2 h at room temperature. The membrane was washed seven times, and proteins trapped in the filter were visualized using ECL reagent (GE Healthcare).
Fluorescence Resonance Energy Transfer Assays
For FRET experiments, 150,000 cells/cm2 (HEK293) or 50,000 cells/cm2 (HeLa) were seeded in 24-multiwell plates and grown for 24 h in growth media containing no antibiotics. Cells were transfected with FuGENE 6 reagent (Roche Applied Science) in a 1:3 (μg/μl) ratio according to the manufacturer's recommendations using the following amounts of plasmids per well: 50 ng of CFP, 150 ng of YFP, and 160 ng of test plasmid for FRET determinations; 100 ng CFP alone, for CFP bleed through determination from the sample FRET; 100 ng of YFP alone, for YFP crossover activation determinations; and no DNA, for background determination. After 36 h, the cells were trypsinized in 300 μl for 2 min, and the trypsin reaction was stopped by adding 700 μl of growing media. Cells were dispersed by trituration and plated in quadruplicate by transferring 1/10 of the cells per each well of a black transparent bottom 96-well plate (Costar 3603). After 36 h, cells were fixed for 20 min in PBS/paraformaldehyde 4%, washed twice with PBS, and read in an Infinite M1000 plate reader (Tecan Group Ltd., Männedorf, Switzerland). For HeLa cells, all PBS-based solutions were supplemented with 1 mm CaCl2, 0.5 mm MgCl2 to prevent detachment from the plate. For dose-response experiments, cells were similarly transfected, and the amount of total test plasmid was set to 320 ng. The specific doses utilized per well were 320, 240, 160, and 80 ng of the modifier plasmid, and the total amount of DNA was kept constant by using pcDNA3 plasmid. A control with 320 ng of pcDNA3-only was also included.
For FRET studies, the data were analyzed essentially as described before (26, 29). The background CFP, YFP, and FRET signals were first subtracted from the raw data. Corrected FRET/donor values were determined for each sample (SMPL) according to the following formula: FRET/donor = {SMPL435/527 − X·(SMPL485/527)}/SMPL435/485, where X = YFP435/527/YFP485/527. Data were represented as a percentage of FRET/donor from Cherry-transfected cells. For dose-response experiments, FRET data were represented as percentage to the FRET/donor value from transfected cells at the higher Cherry plasmid dose.
Assessment of Polyglutamine Aggregation by Fluorescence Microscopy
2,000 cells/cm2 were seeded in glass coverslips in 12-multiwell plates and incubated for 24 h in growth media containing no antibiotics. Cells were transfected with FuGENE 6 reagent in a 1:3 (μg/μl) ratio using 250 ng of Cherry-bearing construct plasmid and 250 ng of Q80-CFP plasmid, and the cells were grown for an additional 48 h. Cells were washed twice with PBS and fixed with 4% paraformaldehyde in PBS. After three washes with PBS, coverslips were dried and mounted in DAPI-containing mounting media. Pictures from 10 random fields, each one containing at least 200 transfected cells, were acquired with a Nikon Eclipse 80i microscope using an apochromat ×10 objective, and images were deconvoluted. For quantification, cells were visualized with the NIS-Elements Advanced Research 3.0 software, classified for the presence of aggregates, and counted. The data are represented as the percentage of transfected, mCherry-expressing cells showing polyglutamine aggregation.
TDP-43-mediated Alternative Splicing of CFTR Minigene Construct
The TG13T3 CFTR exon 9 reporter construct was provided by Dr. F. Baralle (30). One day after plating, the TG13T3 CFTR reporter construct was transfected into HeLa cells together with Q19-CFP, Q80-CFP, or full-length TDP-43 (1 μg each). 48 h later, total RNA was isolated from cells using TRIzol reagent (Invitrogen), and the first strand reverse-transcribed cDNA was generated using random hexamers. First strand RTs underwent PCR amplification using primers 5′-TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA-3′and 5′-CAACTTCAAGCTCCTAAGCCACTGC-3′. Amplification conditions were as follows: denaturation 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 62 °C for 60 s, and 72 °C for 90 s. Amplicons were run out on a 3% agarose gel and visualized using ethidium bromide. Images were inverted, and bands were analyzed via densitometry using ImageJ software (National Institutes of Health). For quantitation, the ratio of CFTR exon 9 exclusion/inclusion (i.e. densitometry value of the upper band/densitometry value of lower two bands = CFTR exon 9 inclusion/exclusion) was calculated from three independent experiments and averaged.
Measurement of HttQ72-induced Cell Death
COS7 were plated in 24-well plates and co-transfected with pCherry, together with the indicated constructs (HttQ72, TDP-43, TDP-43(221–414)). 24 h after transfection, three fields were imaged at ×10 magnification in duplicate wells, and the number of Cherry positive cells were counted, with >200 cells per well imaged. Using an automated XY stage (Prior) controlled with Metamorph software (Molecular Devices), the same fields were imaged at 72 h, and the percentage of Cherry-positive cells initially present at 24 h was determined for duplicate wells. Similar results were obtained from four independent experiments.
RESULTS
Polyglutamine Aggregates Sequester TDP-43 and Deplete It from the Nucleus
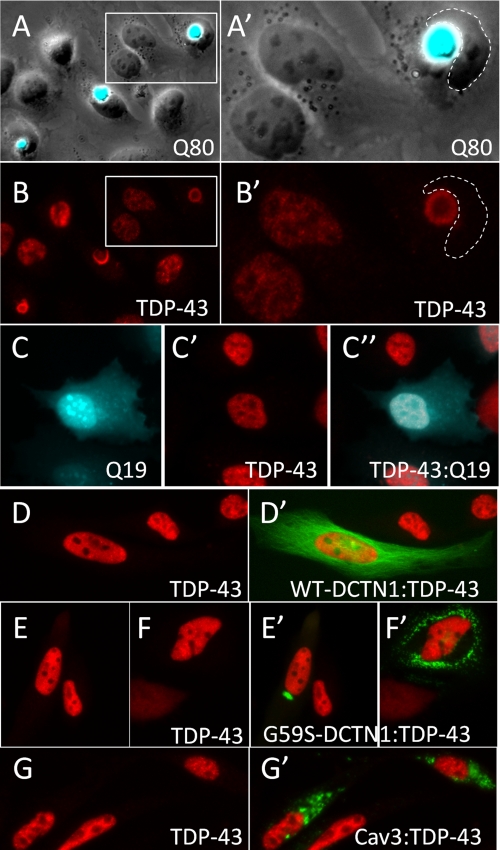
TDP-43 translocates to the cytosol under conditions of cellular stress, most notably in motor neurons after sciatic nerve axotomy (12). To test whether cellular stressors would affect translocation of TDP-43, we treated TDP-43-transfected cells with the proteasome inhibitor MG132. This led to the formation of a perinuclear ubiquitinated TDP-43 inclusion in ∼23% of cells (supplemental Fig. S1). To determine whether this represented a block in normal TDP-43 degradation in the cytosol or instead was a response to the accumulation of misfolded proteins in the cytoplasm, we expressed several proteins that form ubiquitinated cytoplasmic aggregates (polyglutamine Q80-CFP, dynactin G59S, and caveolin 3-P104L) (27, 31, 32) in HeLa cells and determined their effect on endogenous TDP-43 localization via immunostaining (Fig. 1). Cells expressing the expanded polyglutamine protein Q80-CFP formed large cytoplasmic polyglutamine inclusions (Fig. 1A). Interestingly, endogenous TDP-43 was always observed in Q80-CFP inclusions, typically with partial or complete loss of TDP-43 staining from the nucleus (Fig. 1B). Q19-CFP, which does not form ubiquitinated cytoplasmic inclusions, did not alter the localization of endogenous TDP-43 (Fig. 1C). Aggregates of an N-terminal fragment of huntingtin (Htt) containing an expanded polyglutamine tract (Htt-Q72) similarly bound and sequestered TDP-43 (data not shown). Translocation to the cytosol and sequestration of TDP-43 were specific to polyglutamine inclusions, as ubiquitinated cytoplasmic aggregates of dynactin-G59S or caveolin 3-P104L did not alter the localization of endogenous TDP-43 (Fig. 1, D–G). Importantly, co-localization of TDP-43 with polyglutamine inclusions was recently reported in cortical neurons from patients with Huntington disease, supporting that binding of TDP-43 to polyglutamine aggregates also occurs in vivo (10).
FIGURE 1.
Cytoplasmic polyglutamine aggregates bind and sequester nuclear TDP-43. A, HeLa cells expressing an expanded polyglutamine construct Q80-CFP developed large cytoplasmic polyglutamine aggregates (A, overlay of phase contrast and CFP fluorescence images). B, immunofluorescence staining to visualize TDP-43 in cells with Q80-CFP aggregates showed that endogenous TDP-43 was completely sequestered into the Q80 aggregate and was absent from the nucleus. A′ and B′ represent higher magnification images of the boxed regions shown in A and B. Cells transfected with Q19-CFP (C) showed normal nuclear localization of endogenous TDP-43 (C′, TDP-43, C″- overlay). d–G, TDP-43 immunostaining in HeLa cells transfected other aggregation prone proteins. Wild-type dynactin-1(WT-DCTN1) fused to GFP (D′) showed normal distribution along microtubules, whereas the G59S-DCTN1 mutant formed discrete focal or multifocal ubiquitinated cytoplasmic aggregates (E′ and F′). G, GFP-Caveolin-3 with the P104L point mutation (Cav3) also formed cytoplasmic ubiquitinated aggregates when transfected into HeLa cells. Unlike Q80 polyglutamine aggregates, cytoplasmic aggregates of G59S-DCTN1 (E′ and F′) and Cav3 (G′) did not induce translocation to the cytosol or sequestration of endogenous TDP-43.
Some RNA-binding proteins, including hnRNPA1, are known to exit the nucleus in the setting of osmotic or other cellular stressors (33). Thus, we examined the localization of two other nuclear RNA-binding proteins in HeLa cells expressing Q80-CFP. Q80-CFP aggregates did not alter the localization or sequester endogenous hnRNPA1 or SAFB1 (Fig. 2, A and B) (34). This is particularly notable as hnRNPA1 has a similar domain structure to TDP-43, with two RNA-binding motifs and a C-terminal glycine-rich region (35). Therefore, binding and sequestration into polyglutamine aggregates were relatively specific for TDP-43 and were not observed with other structurally and functionally related RNA-binding proteins.
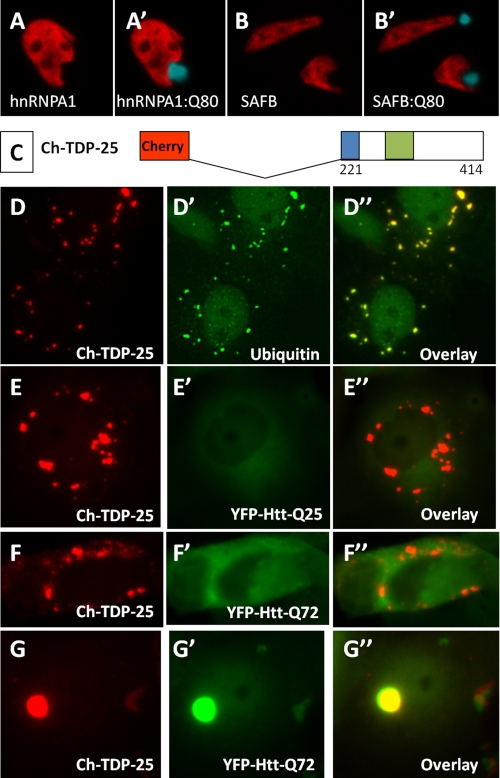
FIGURE 2.
Polyglutamine aggregates do not recruit other nuclear RNA-binding proteins, and cytoplasmic aggregates of TDP-43 C-terminal fragments do not recruit polyglutamine proteins. A and B, immunostaining of endogenous hnRNPA1 and SAFB1 in HeLa cells transfected with Q80-CFP. Unlike TDP-43, the nuclear RNA-binding proteins hnRNPA1 and SAFB did not exit the nucleus or bind to cytoplasmic aggregates of Q80-CFP. C, schematic of 25-kDa C-terminal fragment of TDP-43 fused to mCherry, Ch-TDP-25. d, expression of Ch-TDP-25 in COS7 cells showed numerous punctate cytoplasmic aggregates that stained with an antibody to ubiquitin (D′). Co-expression of Ch-TDP-25 and an N-terminal huntingtin fragment containing a normal (E, HttQ25-YFP) or expanded (F, HttQ72-YFP) polyglutamine stretch showed that cytoplasmic aggregates of the C-terminal domain of TDP-43 were common but did not seed aggregation of polyglutamine-containing proteins. By contrast, cells that spontaneously formed aggregates of HttQ72-YFP showed complete sequestration of Ch-TDP-25 into the aggregate (G).
To investigate whether TDP-43 aggregates could recruit polyglutamine-containing proteins, we expressed a C-terminal fragment of TDP-43 that forms cytoplasmic inclusions (36–38), and we determined whether co-expressed Htt-Q25 or Htt-Q72 was recruited to the TDP-43-based aggregates. A 25-kDa C-terminal fragment of TDP-43 (amino acids 221–414) formed multifocal ubiquitinated inclusions in the cytosol of transfected cells (Fig. 2, C and D). These cytoplasmic TDP-43 aggregates did not recruit Htt-Q25 or Htt-Q72 (Fig. 2, E and F). However, as with Q80-CFP, in cells with Htt-Q72 inclusions, we observed complete sequestration of TDP-43 (Fig. 2G). These data suggest that although expanded polyglutamine proteins aggregate and recruit TDP-43, the converse is not true, i.e. TDP-43-based cytoplasmic aggregates do not recruit polyglutamine proteins.
Polyglutamine Aggregate Interaction Requires a Q/N-rich Domain in the C Terminus of TDP-43
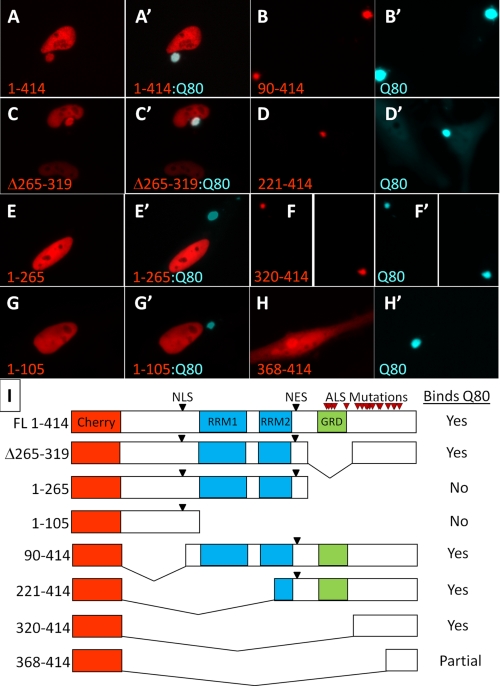
To define which regions of TDP-43 were required for interaction with polyglutamine aggregates, we generated a series of deletion constructs of TDP-43-fused red fluorescent proteins (Ch-TDP-43) and co-expressed them with Q80-CFP in HeLa cells (Fig. 3). Full-length Ch-TDP-43 co-localized with the Q80-CFP inclusions (Fig. 3A). Of note, unlike endogenous TDP-43, which was completely sequestered from the nucleus, we observed that some Ch-TDP-43 was retained in the nucleus, likely due to high Ch-TDP-43 levels from transient overexpression. Constructs containing the N terminus and RNA-binding motifs (constructs 1–265 and 1–105) maintained normal nuclear localization and were not sequestered into polyglutamine aggregates (Fig. 3, E and G). By contrast, constructs containing the C terminus of TDP-43(90–414, 221–414, 320–414) were completely sequestered into Q80-CFP aggregates (Fig. 3, B, D and F). Although the 25-kDa C-terminal fragment (221–414) normally forms multifocal cytoplasmic aggregates (see Fig. 2D), in the presence of Q80 all of the TDP-43 was sequestered into one large polyglutamine inclusion (Fig. 3D). Deletion of the glycine-rich region (Δ265–319), previously found to be required for TDP-43-mediated alternative splicing (39), did not abolish polyglutamine interaction (Fig. 3C). The core region required for binding to polyglutamine aggregates included residues 320–367, as residues 368–414 showed only minimal polyglutamine aggregate binding and remained diffusely cytoplasmic (Fig. 3H).
FIGURE 3.
Polyglutamine aggregate interaction requires amino acids 320–367 in the C-terminal domain of TDP-43. To determine the region of TDP-43 necessary for interaction with polyglutamine aggregates, a series of deletion mutants in TDP-43 fused to mCherry were individually co-transfected into HeLa cells with Q80-CFP, and imaged using fluorescence microscopy. Full-length TDP-43 (A) and deletion of the glycine-rich domain (C) both localized properly to the nucleus and were sequestered into the Q80-CFP aggregate. C-terminal deletions (1–105 and 1–265) retaining the nuclear localization signal (NLS) were localized to the nucleus but did not bind to Q80-CFP aggregates (E and G). N-terminal deletions missing the nuclear localization signal were localized to the cytoplasm (see Fig. 2D); however, in the presence of Q80-CFP aggregates all of the cytoplasmic TDP-43 fragments were incorporated into polyglutamine inclusions (B, D, and F), with the exception of 368–414, which showed only partial colocalization with Q80-CFP (H). The core domain required for binding to polyglutamine aggregates was amino acids 320–367, immediately adjacent to the GRD. I, schematic table of deletion constructs and their binding to Q80-CFP, showing location of the nuclear localization and export signals (NLS and NES), RNA-binding motifs (RRM1 and RRM2), and the location of ALS mutations in TDP-43 (red arrowheads).
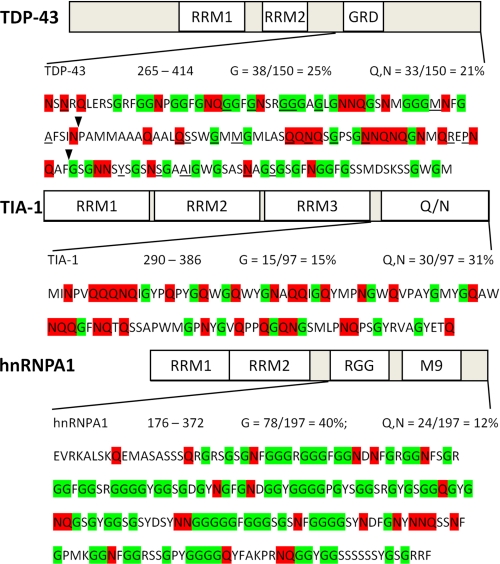
Polyglutamine inclusions are known to sequester proteins containing stretches of polyglutamine residues such as cAMP-response element-binding protein-binding protein and TATA box-binding protein (20, 40, 41) and those rich in glutamine and asparagine residues (Q/N-rich) (21). Among the polyglutamine aggregate interacting Q/N-rich proteins are TIA-1, an RNA-binding protein involved in stress granule formation (21, 42), and FUS/TLS, an RNA-binding protein recently found to be mutated in familial ALS (23–25). Examination of the C-terminal region of TDP-43(265–414) indicated that it is relatively Q/N-rich (21%), with the core region required for polyglutamine aggregate binding (320–367) having 31% Q/N content, similar to the Q/N domains in FUS/TLS (24%) and TIA-1 (31%) (Fig. 4). By contrast, the analogous C-terminal domain of hnRNPA1 is significantly richer in glycine residues than TDP-43 (40 versus 25%) but has much lower Q/N content (12%). These findings support that the sequestration of TDP-43 into polyglutamine aggregates requires a previously uncharacterized Q/N-rich region located within the TDP-43 C-terminal domain.
FIGURE 4.
Sequence of the C-terminal domains of TDP-43, TIA-1, and hnRNPA1 reveals a Q/N-rich domain in TDP-43. TDP-43 is shown at the top, followed by TIA-1, which contains a well characterized Q/N-rich domain at the C terminus, and hnRNPA1, which contains a canonical glycine-rich domain of the 2×RBD-Gly family, composed of an RGG domain and an M9 nuclear shuttling signal. Glycine (G) residues are highlighted in green and glutamine (Q) and asparagine (N) residues are in red. The overall Q/N content of the C terminus of TDP-43 is 21%. The core region required for the interaction of TDP-43 with polyglutamine aggregates identified in the deletion analysis (flanked by arrowheads; 320–367) shows 31% Q/N content, similar to the Q/N-rich prion-related domain of TIA-1. By contrast, the C-terminal region of hnRNPA1 is significantly more glycine-rich than TDP-43 but has low Q/N content and does not bind polyglutamine aggregates. All but one of the currently described mutations in TDP-43 (underlined) are located in the C-terminal domain.
TDP-43 Incorporates into Detergent-insoluble Polyglutamine Aggregates and Alters Aggregate Dynamics
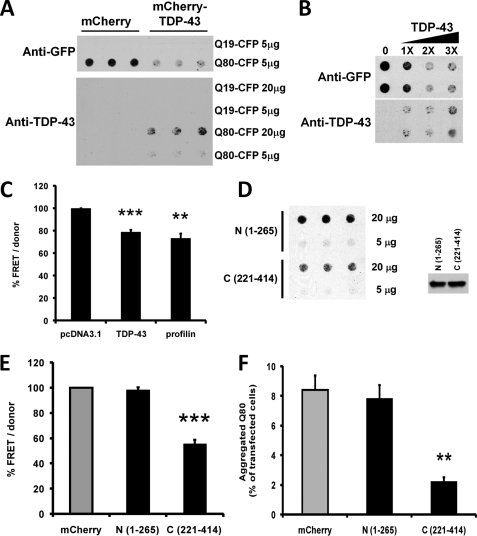
Some polyglutamine aggregate-interacting proteins, particularly polyQ and Q/N-rich proteins, become incorporated into polyglutamine aggregates and themselves become detergent-insoluble (21, 25), whereas others, including molecular chaperones, interact dynamically and are easily dissociated from polyglutamine aggregates in the presence of SDS (43, 44). To determine whether TDP-43 is incorporated into detergent-insoluble polyglutamine aggregates, we co-transfected HeLa cells with TDP-43 and either Q19-CFP or Q80-CFP and performed a filter trap assay (Fig. 5). Cells were lysed in 2% SDS buffer and vacuum filtered through a 0.2-μm cellulose acetate membrane, and detergent-insoluble aggregates unable to pass through the filter were detected using antibodies to GFP (to detect polyQ-CFP) or TDP-43. As expected, detergent-insoluble Q80-CFP aggregates were trapped on the filter, in contrast to Q19-CFP (Fig. 5A, top). TDP-43 remained soluble in Q19-CFP-transfected cells; however, cells expressing Q80-CFP strongly recruited TDP-43 into detergent-insoluble aggregates (Fig. 5A, bottom). This indicates that TDP-43 incorporates tightly into polyglutamine aggregates, similar to the Q/N-rich domain containing RNA-binding proteins TIA-1 and FUS/TLS (21, 25, 42).
FIGURE 5.
TDP-43 is sequestered into insoluble polyglutamine aggregates and TDP-43 overexpression inhibits polyglutamine aggregation. A, HeLa cells were transfected with Q19-CFP or Q80-CFP, together with either mCherry (control), or Cherry-TDP-43. Cell lysates (either 5 or 20 μg) were applied to a 0.2-μm pore cellulose acetate filter via vacuum filtration and blotted using antibodies to either GFP (upper panel) or TDP-43 (lower panel) to visualize SDS-insoluble protein aggregates. Q19-CFP, which does not aggregate, passed through the filter, whereas detergent-insoluble aggregates of Q80-CFP were trapped in the filter. Blotting with an anti-TDP-43 antibody showed that in the presence of Q80-CFP, TDP-43 also becomes trapped in the filter, consistent with sequestration into detergent-insoluble Q80-CFP aggregates. Note that TDP-43 transfection decreased the amount of insoluble Q80-CFP retained in the filter (upper panel, right). B, TDP-43 expression decreased Q80-CFP aggregation in a dose-dependent manner. HeLa cells were co-transfected with Q80-CFP and increasing amounts of FLAG-tagged TDP-43 and analyzed using the filter trap assay and anti-GFP antibody. C, TDP-43 decreases the aggregation of expanded huntingtin protein in a FRET assay. HeLa cells were co-transfected with HttQ72-CFP/YFP coding plasmids along with plasmids coding for TDP-43, profilin, or empty vector (pcDNA3.1) and assayed for FRET after 48 h. Calculated FRET was normalized to the donor levels and normalized to pcDNA3.1-transfected cells. Averages from four independent experiments performed in quadruplicate are shown with S.E. as error bars. **, p < 0.01; ***, p < 0.001, paired t test. d, C-terminal (221–414) region of TDP-43 decreases polyglutamine solubility. HeLa cells were co-transfected with Q80-CFP and N-terminal (1–265) or C-terminal (221–414) Cherry-tagged TDP-43 proteins, and filter trap assay was performed with 20 or 5 μg lysates. Right panel, similar level of expression of Q80-CFP in lysates was verified by Western blot. E, TDP-43 fragment containing the Q/N-rich domain decreases aggregation of expanded huntingtin protein. HeLa cells were co-transfected with HttQ72-CFP/YFP along with the indicated mCherry-tagged TDP-43 plasmids and then assayed for FRET after 48 h. Calculated FRET was normalized to the donor levels and represented as % of mCherry-transfected cells. Averages from four independent experiments performed in quadruplicate are shown with S.E. as error bars. ***, p < 0.01, paired t test. F, TDP-43 C-terminal fragment (221–414) reduced inclusion formation, whereas the N-terminal (1–265) fragment did not. HeLa cells were co-transfected with Q80-CFP and either mCherry or the indicated mCherry-tagged TDP-43 constructs. Cells were cultured for 24 h and fixed prior to counting inclusions in at least 2,000 cells from 10 fields per well. The y axis represents the % of transfected cells containing aggregates. **, p < 0.001, t test.
As a consequence of Q/N-rich proteins becoming incorporated into polyQ aggregates, they can alter the dynamics of polyglutamine aggregate formation (21, 25). We observed that overexpression of TDP-43 led to significantly less Q80-CFP retention on the membrane, indicating that increased levels of TDP-43 could suppress polyglutamine aggregation (Fig. 5, A and B). The decreased Q80-CFP aggregation observed was not due to cell death induced by TDP-43 overexpression, which was minimal at this early time point (supplemental Fig. S2). To quantitatively assess the influence of TDP-43 overexpression on polyglutamine aggregation, we utilized a fluorescence resonance energy transfer (FRET)-based assay to measure aggregation of HttQ72-YFP and HttQ72-CFP in HeLa cells. This assay was previously used to identify modifiers of HttQ72 aggregation (26). Overexpression of TDP-43 suppressed HttQ72 aggregation as measured by HttQ72-CFP:HttQ72-YFP FRET similar in degree to profilin, an actin regulatory factor that binds the polyproline region of Htt and strongly inhibits its aggregation (Fig. 5C) (45).
We mapped the domain of TDP-43 required for polyglutamine aggregate suppression by expressing various truncation/deletion mutants; an N-terminal fragment of TDP-43(1–265), which retains the RNA-binding motifs and normal nuclear localization (Fig. 3E), or a C-terminal fragment (221–414) carrying the Q/N domain (Fig. 3D). On both the filter trap assay and the Q80-CFP:Q80-YFP FRET assay, the C-terminal region of TDP-43 suppressed polyglutamine aggregation, whereas the N-terminal region did not (Fig. 5, D and E). Additionally, using fluorescence microscopy, we observed that the TDP-43 C-terminal domain strongly suppressed the percentage of Q80-CFP-transfected cells, which formed polyglutamine inclusions, whereas the N-terminal region had no effect (Fig. 5F).
These data indicate that TDP-43, like other Q/N-rich domain-containing proteins, is sequestered by polyglutamine aggregates and itself becomes detergent-insoluble. Furthermore, increasing levels of TDP-43 inhibits polyglutamine-dependent protein aggregation, in a manner dependent on the C-terminal Q/N-rich region of TDP-43.
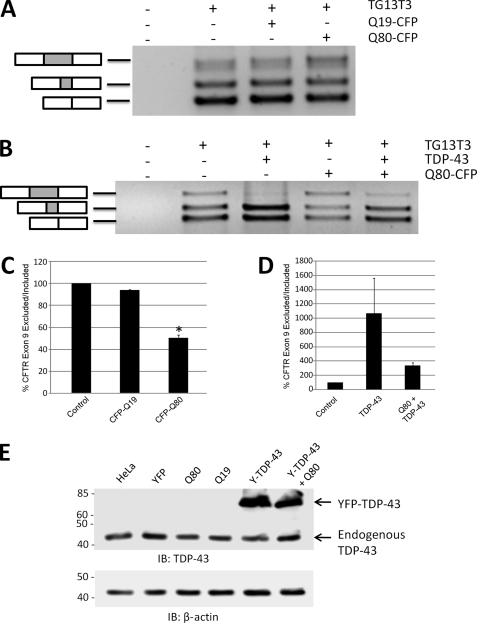
Sequestration of Endogenous TDP-43 into Polyglutamine Aggregates Suppresses TDP-43-mediated Splicing
The prior experiments demonstrated that polyglutamine inclusions sequester endogenous TDP-43 into insoluble aggregates, which could disrupt normal TDP-43 function. We tested this idea by assessing the alternative splicing of a minigene construct (“TG13T3”) containing a known TDP-43 splicing target, exon 9 of the CFTR (30). TDP-43 binds directly to the UG13 repeat region upstream of CFTR exon 9 and suppresses inclusion of this exon (30, 46). We transfected HeLa cells with the CFTR exon 9 reporter construct TG13T3 and performed RT-PCR to assess splicing. Endogenous levels of TDP-43 led predominantly to exclusion of CFTR exon 9 (bottom two bands), and expression of Q19-CFP did not alter this splicing pattern (Fig. 6A). In contrast, expression of Q80-CFP with TG13T3 increased exon 9 inclusion (top band), with decreased exon 9 skipping (Fig. 6, A and C, bottom two bands). These findings are consistent with a loss of TDP-43-mediated exon 9 skipping, presumably from sequestration of TDP-43 into Q80-CFP aggregates. Although polyglutamine aggregates sequester nearly all endogenous TDP-43 (Fig. 1B), overexpression of TDP-43 led to retention of a significant amount of TDP-43 in the nucleus (Fig. 3A). Therefore, we examined whether overexpression of TDP-43 could rescue the suppression of CFTR exon 9 skipping induced by Q80-CFP. Overexpression of TDP-43 alone led to complete loss of exon 9 inclusion (top band), and an increase in exon 9 skipping (bottom two bands), similar to previous reports (Fig. 6B) (30, 36, 46). Interestingly, co-expression of both Q80-CFP and TDP-43 returned the splicing pattern of TG13T3 similar to that seen in control cells (Fig. 6, B and D). This implied that the effect of Q80-CFP on alternative splicing of CFTR exon 9 was directly through sequestration of TDP-43. Thus, sequestration of TDP-43 by polyglutamine aggregates alters the normal function of TDP-43, suggesting the possibility that secondary loss of TDP-43 function may play a role in polyglutamine toxicity.
FIGURE 6.
Sequestration of nuclear TDP-43 into polyglutamine aggregates suppresses TDP-43-mediated splicing. To assess whether sequestration of TDP-43 into polyglutamine aggregates had an effect on TDP-43 function, HeLa cells were transfected with a CFTR minigene construct (TG13T3) as a reporter of TDP-43-mediated splicing. In the presence of endogenous TDP-43 levels in HeLa cells, three bands are observed, corresponding to exon 9 inclusion (upper band) or skipping of exon 9 (lower two bands). The middle band is due to the adoption of a cryptic splice acceptor site in exon 9, and the bottom band is from complete exon 9 skipping. A, co-transfection of TG13T3 with Q80-CFP increased exon 9 inclusion, consistent with a loss of basal levels of TDP-43-mediated exon 9 skipping by sequestration into polyQ aggregates. B, overexpression of TDP-43 strongly suppressed exon 9 inclusion (upper band) and enhanced exclusion (lower bands). The alteration in splicing of the CFTR exon 9 minigene induced by Q80-CFP was largely normalized by overexpression of TDP-43. C and d,, mean ratio of exon 9 exclusion/inclusion from three independent experiments, normalized to basal level in HeLa cells (control). *, p < 0.05, paired t test, control versus Q80-CFP. E, immunoblot (IB) of HeLa cell lysates with antibodies to TDP-43, showing that total TDP-43 levels were not affected by transfection with the polyQ-CFP constructs or the YFP-TDP-43 (Y-TDP-43) fusion protein. β-Actin immunoblot is shown below as a loading control.
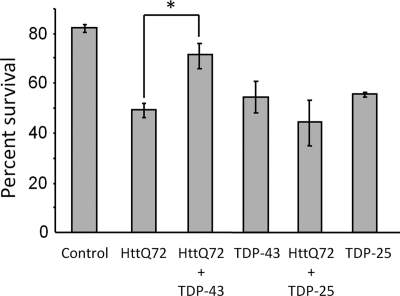
Increased Expression of TDP-43 Suppresses Polyglutamine Aggregate Toxicity
Sequestration of nuclear proteins has been shown to mediate the toxicity of polyglutamine aggregates in cultured cells (40). Given that polyglutamine aggregates sequester nuclear TDP-43 which can alter TDP-43-mediated splicing in the nucleus, we investigated whether increasing levels of TDP-43 could rescue the cytotoxicity of the huntingtin exon 1 fragment with a polyglutamine expansion (HttQ72) (Fig. 7). 48 h after transfection, minimal cell death was observed in HttQ72-expressing COS7 cells, similar to what was observed in HeLa cells (supplemental Fig. S2). However, 72 h after transfection, HttQ72 induced significant cell death, with ∼49% survival for HttQ72 versus ∼82% survival for cells transfected with pCherry alone. Similar to a previous report, overexpression of full-length TDP-43 alone showed some cytotoxicity (36). Interestingly, co-transfection of HttQ72 together with full-length TDP-43 significantly rescued HttQ72 toxicity. The observed rescue of HttQ72 toxicity by TDP-43 overexpression could either be from repletion of TDP-43 function in the nucleus or from direct suppression of HttQ72 aggregation as described above (Fig. 5). To distinguish between these possibilities, we examined the effect of the TDP-43 C-terminal fragment (221–414) containing the Q/N domain, which suppresses HttQ72 aggregation but remains in the cytoplasm and does not enter the nucleus. Transfection of the TDP-43 C-terminal fragment (221–414) alone induced cell death similar to full-length TDP-43 at 72 h. However co-transfection of HttQ72 together with TDP-43(221–414), which suppresses HttQ72 aggregation, did not rescue the cytotoxicity of HttQ72. These data indicate that increased expression of full-length TDP-43 can significantly rescue the cytotoxicity of HttQ72 and suggest that this is due to repletion of TDP-43 in the nucleus, rather than through suppression of HttQ72 aggregation.
FIGURE 7.
Increased TDP-43 expression rescues cell death induced by polyglutamine toxicity. COS7 cells were transfected with mCherry alone (Control) or together with the indicated constructs. mCherry positive cells were counted in the same fields at 24 and 72 h after transfection, and percent survival is shown (number of mCherry positive cells at 24 h/number of mCherry positive cells at 72 h) for >200 cells in duplicate wells. HttQ72-induced significant cell death is compared with mCherry alone. However, co-transfection of TDP-43 together with HttQ72 significantly improved cell survival compared with HttQ72 alone (71% versus 49% survival; *, p < 0.05, paired t test). By contrast, expression of the C-terminal fragment of TDP-43(221–414) (TDP-25), which suppresses HttQ72 aggregation but does not localize to the nucleus, did not rescue HttQ72 toxicity. Similar to previous reports, full-length TDP-43 and TDP-25 were also toxic and induced a similar amount of cell death.
DISCUSSION
Here, we report that TDP-43 is sequestered into polyglutamine aggregates and that this binding requires the presence of a previously uncharacterized Q/N-rich region in the C-terminal domain of TDP-43. Recruitment into polyglutamine aggregates led TDP-43 to become detergent-insoluble, and overexpression of TDP-43 altered the dynamics of polyglutamine aggregate formation. Finally, we found that sequestration into polyglutamine aggregates disrupted TDP-43-mediated splicing in the nucleus, which could be counteracted by overexpression of TDP-43. These data suggest that, like other polyglutamine aggregate interacting proteins, sequestration and loss of normal TDP-43 function may play a role in the pathogenesis of polyglutamine diseases. Furthermore, the identification of a Q/N-rich prion-related domain in TDP-43 may have important implications for understanding normal and pathologic functions of TDP-43.
Aggregate Interacting Proteins in the Pathogenesis of Polyglutamine Diseases
Polyglutamine aggregate interacting proteins are myriad; some bind to specific nonpolyglutamine regions such as huntingtin-associated protein 1 (47); some are involved in the normal recognition and degradation of misfolded proteins, including heat shock proteins and ubiquitin proteosome components (43, 44, 48, 49); and some aberrantly incorporate into polyglutamine aggregates via either a polyQ or Q/N-rich region (20, 21). These aberrant protein-protein interactions provide a potential molecular mechanism by which expanded polyglutamine proteins develop a toxic gain of function, disrupt normal cellular function, and ultimately induce neurodegeneration (18).
Elongated stretches of pure polyglutamine spontaneously aggregate to form amyloid fibrils in vitro and in vivo (50), possibly via hydrogen-bonded polar zipper formation between polyglutamine molecules (51). Based on this model, proteins with nonpathogenic polyglutamine stretches are predicted to incorporate into polyglutamine aggregates (20), and this has been demonstrated for several nonpathogenic length polyglutamine proteins, including cAMP-response element-binding protein-binding protein, and TATA box-binding protein (40, 41, 52, 53). In these cases, the unexpanded polyglutamine proteins were depleted from their normal cellular localization and incorporated into detergent-insoluble polyglutamine aggregates, leading to loss of their normal function, similar to TDP-43. It is notable that repletion of TDP-43 was only able to partially rescue HttQ72 toxicity, consistent with the idea that sequestration of multiple different proteins likely plays a role in polyglutamine toxicity.
Although TDP-43 does not contain a polyQ sequence, our deletion analysis identified a Q/N-rich region in the C terminus of TDP-43 that is required for its sequestration into polyglutamine aggregates. This is noteworthy, as several polyglutamine aggregate interacting proteins have recently been identified that contain similar Q/N-rich domains, including the transcription factor nuclear transcription factor Y (54), and the DNA/RNA-binding proteins TIA-1 and FUS/TLS (21, 25, 42). We hypothesize that polyglutamine aggregates seed aggregation of Q/N-rich proteins via a similar mechanism to the recruitment of pure polyQ proteins. It has been proposed that this “cross-seeding” of numerous Q/N proteins may in part be responsible for the pathologic diversity in polyglutamine diseases (21). As TDP-43 has been observed to co-localize with polyglutamine aggregates in Huntington disease patients (10), TDP-43 could take part in this type of a cross-seeding reaction between polyQ fibrils and Q/N proteins and possibly influence cellular dysfunction and neurodegeneration.
Loss of TDP-43 Functions as a Potential Mechanism in Neurodegeneration
In cases of ALS and FTLD, the loss of normal nuclear staining in cells, which contain cytoplasmic TDP-43 aggregates, is striking (1). Thus, TDP-43 aggregation might lead to loss of normal nuclear TDP-43 function, either by direct sequestration as shown here for polyglutamine aggregates or by an indirect mechanism (55). Although it is still uncertain whether cytoplasmic aggregates of the C-terminal region of TDP-43 sequester endogenous TDP-43 and alter its function (36, 37), the loss of nuclear TDP-43 was a common feature prior to degeneration of susceptible neurons in two recently published TDP-43 transgenic mouse models (56, 57). Additionally, depletion of TDP-43 in cultured cells produced dysregulation of cell cycle, altered cell morphology, and cell death (58, 59). Finally, Drosophila models also support that loss of TDP-43 is toxic to motor neurons (60), and recent studies of TDP-43 null alleles in mice confirmed that TDP-43 is required for early mouse development (61, 62). These studies clearly indicate that acquired loss of TDP-43 could be toxic and support the concept that loss of normal TDP-43 function may play a role not only in ALS and FTLD but possibly also in other neurodegenerative diseases. Our finding that repletion of TDP-43 in the nucleus by overexpression had a protective effect against polyglutamine aggregate toxicity in vitro suggests that strategies to maintain TDP-43 nuclear function might be considered as a therapeutic approach in ALS as well as polyglutamine diseases (55).
Structure and Function of the C-terminal Domain of TDP-43
A better understanding of the structure and function of the C-terminal domain of TDP-43 is of particular importance for ALS research, as all but one of the ALS-associated missense mutations in TDP-43 occur within this region (55). TDP-43 structurally resembles hnRNP proteins, and when it was originally identified based on binding to the TAR DNA sequence of HIV-1, it was noted to have a glycine-rich domain (residues 274–314), similar to other RBD-Gly family proteins, including hnRNPA1 (35, 63). Subsequently, some have referred to the entire C-terminal region as a glycine-rich domain (64, 65). However, unlike hnRNPA1, the C-terminal domain of TDP-43 has not been implicated in binding to DNA or RNA or nuclear shuttling (63, 66, 67). Instead, the C terminus of TDP-43 is required for it to function as a silencer at several splicing targets (68, 69) and for TDP-43 to act as a transcriptional insulator for the mouse sp-10 gene (70), presumably through the regulation of protein-protein interactions.
Our deletion analysis suggests that the glycine-rich domain (274–314) (63) is dispensable for binding to polyglutamine aggregates. Rather the adjacent Q/N-rich region (320–367) is required. This region is similar in Q/N content (31%) to other polyQ-binding proteins such as TIA-1 (31%), FUS/TLS (26%), and nuclear transcription factor Y (33%) (21, 25, 54). By comparison, the C-terminal domain of hnRNPA1, which does not bind to polyglutamine aggregates, is significantly more glycine-rich than TDP-43, and it has much lower Q/N content throughout (Fig. 4). The Q/N-rich region in TDP-43 overlaps almost exactly with the region recently found to be necessary for interaction between TDP-43 and hnRNPA2 (71). This is not surprising as Q/N-rich domains are common in DNA- and RNA-binding proteins and are hypothesized to be involved in both protein-protein interactions and in aggregation (72, 73).
Q/N-rich domains were first characterized in yeast, where they are responsible for protein aggregation and prion-like inheritance of the non-Mendelian factors, including PSI+ (Sup35) and URE3 (Ure2p). Thus, they are sometimes referred to as prion-related domains (22). In yeast, although prion-like protein aggregation sometimes inactivates the relevant protein, in other cases it can activate or alter the function of the protein, indicating that regulated protein aggregation may be utilized to serve normal cellular functions (22). Furthermore, Q/N domains in higher organisms also appear to use regulated self-aggregation to alter protein function in response to various stimuli. For example, the Q/N-rich prion related domain of TIA-1 is critical for TIA-1 aggregation and the assembly of cytoplasmic stress granules (74), and Q/N-rich domains have been shown to regulate protein aggregation and synaptic function in both aplysia (75) and Drosophila (76).
Identification of a Q/N-rich region in the C-terminal domain of TDP-43 is of particular importance because of the known tendency of C-terminal fragments of TDP-43 to aggregate both in cell culture models (36–38, 77) and in patients with ALS and FTLD (1). Although not all Q/N domain-containing proteins are capable of amyloid formation and prion-like propagation in yeast, most have a strong tendency to aggregate (72). Therefore, it is likely that the Q/N domain required for binding to polyglutamine aggregates also influences the tendency of the C-terminal domain of TDP-43 to self-associate and form protein aggregates. Future experiments will be required to determine how the Q/N domain influences pathologic aggregation of TDP-43 and what role it may play in normal TDP-43 function.
Supplementary Material
Acknowledgments
We thank Francisco Baralle for generously providing the TG13T3 plasmid and Heather True and Tim Miller for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants NS055980 and R01NS069669 (to R. H. B.). This work was also supported by the Neuroscience Blueprint Core Grant NS057105 (to Washington University), the Hope Center for Neurological Disorders, the McDonnell Center for Cellular and Molecular Neurobiology, the Muscular Dystrophy Association Grant 135428, and Children's Discovery Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- FTLD
- frontotemporal lobar degeneration
- Q/N
- glutamine/asparagine
- FUS/TLS
- fused in sarcoma, translocated in liposarcoma
- Htt
- huntingtin
- hnRNPA1
- heterogeneous nuclear ribonucleoprotein A1
- YFP
- yellow fluorescent protein
- CFP
- cyan fluorescent protein
- CFTR
- cystic fibrosis transmembrane receptor
- RBD-Gly
- RNA binding domain-glycine-rich domain.
REFERENCES
- 1.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 2.Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., Hatanpaa K. J., White C. L., 3rd, Bigio E. H., Caselli R., Baker M., Al-Lozi M. T., Morris J. C., Pestronk A., Rademakers R., Goate A. M., Cairns N. J. (2008) Ann. Neurol. 63, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., Okamoto K., Nishizawa M., Takahashi H., Onodera O. (2008) Ann. Neurol. 63, 538–542 [DOI] [PubMed] [Google Scholar]
- 4.Van Deerlin V. M., Leverenz J. B., Bekris L. M., Bird T. D., Yuan W., Elman L. B., Clay D., Wood E. M., Chen-Plotkin A. S., Martinez-Lage M., Steinbart E., McCluskey L., Grossman M., Neumann M., Wu I. L., Yang W. S., Kalb R., Galasko D. R., Montine T. J., Trojanowski J. Q., Lee V. M., Schellenberg G. D., Yu C. E. (2008) Lancet Neurol. 7, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., Miller C. C., Nicholson G., Shaw C. E. (2008) Science 319, 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., Rouleau G. A. (2008) Nat. Genet. 40, 572–574 [DOI] [PubMed] [Google Scholar]
- 7.Josephs K. A., Whitwell J. L., Knopman D. S., Hu W. T., Stroh D. A., Baker M., Rademakers R., Boeve B. F., Parisi J. E., Smith G. E., Ivnik R. J., Petersen R. C., Jack C. R., Jr., Dickson D. W. (2008) Neurology 70, 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakashima-Yasuda H., Uryu K., Robinson J., Xie S. X., Hurtig H., Duda J. E., Arnold S. E., Siderowf A., Grossman M., Leverenz J. B., Woltjer R., Lopez O. L., Hamilton R., Tsuang D. W., Galasko D., Masliah E., Kaye J., Clark C. M., Montine T. J., Lee V. M., Trojanowski J. Q. (2007) Acta Neuropathol. 114, 221–229 [DOI] [PubMed] [Google Scholar]
- 9.King A., Sweeney F., Bodi I., Troakes C., Maekawa S., Al-Sarraj S.Neuropathology, in press [DOI] [PubMed] [Google Scholar]
- 10.Schwab C., Arai T., Hasegawa M., Yu S., McGeer P. L. (2008) J. Neuropathol. Exp. Neurol. 67, 1159–1165 [DOI] [PubMed] [Google Scholar]
- 11.Weihl C. C., Temiz P., Miller S. E., Watts G., Smith C., Forman M., Hanson P. I., Kimonis V., Pestronk A. (2008) J. Neurol. Neurosurg. Psychiatry 79, 1186–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moisse K., Volkening K., Leystra-Lantz C., Welch I., Hill T., Strong M. J. (2009) Brain Res. 1249, 202–211 [DOI] [PubMed] [Google Scholar]
- 13.Ross C. A. (2002) Neuron 35, 819–822 [DOI] [PubMed] [Google Scholar]
- 14.Gatchel J. R., Zoghbi H. Y. (2005) Nat. Rev. Genet. 6, 743–755 [DOI] [PubMed] [Google Scholar]
- 15.Shao J., Diamond M. I. (2007) Hum. Mol. Genet. 16, R115–R123 [DOI] [PubMed] [Google Scholar]
- 16.Taylor J. P., Tanaka F., Robitschek J., Sandoval C. M., Taye A., Markovic-Plese S., Fischbeck K. H. (2003) Hum. Mol. Genet. 12, 749–757 [DOI] [PubMed] [Google Scholar]
- 17.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 18.Williams A. J., Paulson H. L. (2008) Trends Neurosci. 31, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 20.Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11404–11409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa Y., Kaneko K., Matsumoto G., Kurosawa M., Nukina N. (2009) J. Neurosci. 29, 5153–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickner R. B., Edskes H. K., Roberts B. T., Baxa U., Pierce M. M., Ross E. D., Brachmann A. (2004) Genes Dev. 18, 470–485 [DOI] [PubMed] [Google Scholar]
- 23.Kwiatkowski T. J., Jr., Bosco D. A., Leclerc A. L., Tamrazian E., Vanderburg C. R., Russ C., Davis A., Gilchrist J., Kasarskis E. J., Munsat T., Valdmanis P., Rouleau G. A., Hosler B. A., Cortelli P., de Jong P. J., Yoshinaga Y., Haines J. L., Pericak-Vance M. A., Yan J., Ticozzi N., Siddique T., McKenna-Yasek D., Sapp P. C., Horvitz H. R., Landers J. E., Brown R. H., Jr. (2009) Science 323, 1205–1208 [DOI] [PubMed] [Google Scholar]
- 24.Vance C., Rogelj B., Hortobágyi T., De Vos K. J., Nishimura A. L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., Ganesalingam J., Williams K. L., Tripathi V., Al-Saraj S., Al-Chalabi A., Leigh P. N., Blair I. P., Nicholson G., de Belleroche J., Gallo J. M., Miller C. C., Shaw C. E. (2009) Science 323, 1208–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doi H., Okamura K., Bauer P. O., Furukawa Y., Shimizu H., Kurosawa M., Machida Y., Miyazaki H., Mitsui K., Kuroiwa Y., Nukina N. (2008) J. Biol. Chem. 283, 6489–6500 [DOI] [PubMed] [Google Scholar]
- 26.Pollitt S. K., Pallos J., Shao J., Desai U. A., Ma A. A., Thompson L. M., Marsh J. L., Diamond M. I. (2003) Neuron 40, 685–694 [DOI] [PubMed] [Google Scholar]
- 27.Ju J. S., Miller S. E., Hanson P. I., Weihl C. C. (2008) J. Biol. Chem. 283, 30289–30299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju J. S., Fuentealba R. A., Miller S. E., Jackson E., Piwnica-Worms D., Baloh R. H., Weihl C. C. (2009) J. Cell Biol. 187, 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaufele F., Carbonell X., Guerbadot M., Borngraeber S., Chapman M. S., Ma A. A., Miner J. N., Diamond M. I. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9802–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagani F., Buratti E., Stuani C., Romano M., Zuccato E., Niksic M., Giglio L., Faraguna D., Baralle F. E. (2000) J. Biol. Chem. 275, 21041–21047 [DOI] [PubMed] [Google Scholar]
- 31.Galbiati F., Volonte D., Minetti C., Bregman D. B., Lisanti M. P. (2000) J. Biol. Chem. 275, 37702–37711 [DOI] [PubMed] [Google Scholar]
- 32.Levy J. R., Sumner C. J., Caviston J. P., Tokito M. K., Ranganathan S., Ligon L. A., Wallace K. E., LaMonte B. H., Harmison G. G., Puls I., Fischbeck K. H., Holzbaur E. L. (2006) J. Cell Biol. 172, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allemand E., Guil S., Myers M., Moscat J., Cáceres J. F., Krainer A. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3605–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garee J. P., Oesterreich S. (2010) J. Cell. Biochem. 109, 312–319 [DOI] [PubMed] [Google Scholar]
- 35.He Y., Smith R. (2009) Cell. Mol. Life Sci. 66, 1239–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y. J., Xu Y. F., Cook C., Gendron T. F., Roettges P., Link C. D., Lin W. L., Tong J., Castanedes-Casey M., Ash P., Gass J., Rangachari V., Buratti E., Baralle F., Golde T. E., Dickson D. W., Petrucelli L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igaz L. M., Kwong L. K., Chen-Plotkin A., Winton M. J., Unger T. L., Xu Y., Neumann M., Trojanowski J. Q., Lee V. M. (2009) J. Biol. Chem. 284, 8516–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. (2009) Hum. Mol. Genet. 18, 3353–3364 [DOI] [PubMed] [Google Scholar]
- 39.Wang H. Y., Wang I. F., Bose J., Shen C. K. (2004) Genomics 83, 130–139 [DOI] [PubMed] [Google Scholar]
- 40.Nucifora F. C., Jr., Sasaki M., Peters M. F., Huang H., Cooper J. K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V. L., Dawson T. M., Ross C. A. (2001) Science 291, 2423–2428 [DOI] [PubMed] [Google Scholar]
- 41.Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F. U. (2004) Mol. Cell 15, 95–105 [DOI] [PubMed] [Google Scholar]
- 42.Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., Wanker E. E. (2001) Mol. Biol. Cell 12, 1393–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S., Nollen E. A., Kitagawa K., Bindokas V. P., Morimoto R. I. (2002) Nat. Cell Biol. 4, 826–831 [DOI] [PubMed] [Google Scholar]
- 44.Mitsui K., Nakayama H., Akagi T., Nekooki M., Ohtawa K., Takio K., Hashikawa T., Nukina N. (2002) J. Neurosci. 22, 9267–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao J., Welch W. J., Diprospero N. A., Diamond M. I. (2008) Mol. Cell. Biol. 28, 5196–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., Baralle F. E. (2001) EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X. J., Li S. H., Sharp A. H., Nucifora F. C., Jr., Schilling G., Lanahan A., Worley P., Snyder S. H., Ross C. A. (1995) Nature 378, 398–402 [DOI] [PubMed] [Google Scholar]
- 48.Chai Y., Koppenhafer S. L., Shoesmith S. J., Perez M. K., Paulson H. L. (1999) Hum. Mol. Genet. 8, 673–682 [DOI] [PubMed] [Google Scholar]
- 49.Jana N. R., Zemskov E. A., Wang Gh., Nukina N. (2001) Hum. Mol. Genet. 10, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 50.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 51.Perutz M. F., Johnson T., Suzuki M., Finch J. T. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5355–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C. C., Faber P. W., Persichetti F., Mittal V., Vonsattel J. P., MacDonald M. E., Gusella J. F. (1998) Somat. Cell Mol. Genet. 24, 217–233 [DOI] [PubMed] [Google Scholar]
- 53.Busch A., Engemann S., Lurz R., Okazawa H., Lehrach H., Wanker E. E. (2003) J. Biol. Chem. 278, 41452–41461 [DOI] [PubMed] [Google Scholar]
- 54.Yamanaka T., Miyazaki H., Oyama F., Kurosawa M., Washizu C., Doi H., Nukina N. (2008) EMBO J. 27, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buratti E., Baralle F. E. (2009) Adv. Genet. 66, 1–34 [DOI] [PubMed] [Google Scholar]
- 56.Wegorzewska I., Bell S., Cairns N. J., Miller T. M., Baloh R. H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18809–18814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wils H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Groote C. C., Van Broeckhoven C., Kumar-Singh S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3858–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ayala Y. M., Misteli T., Baralle F. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iguchi Y., Katsuno M., Niwa J., Yamada S., Sone J., Waza M., Adachi H., Tanaka F., Nagata K., Arimura N., Watanabe T., Kaibuchi K., Sobue G. (2009) J. Biol. Chem. 284, 22059–22066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feiguin F., Godena V. K., Romano G., D'Ambrogio A., Klima R., Baralle F. E. (2009) FEBS Lett. 583, 1586–1592 [DOI] [PubMed] [Google Scholar]
- 61.Sephton C. F., Good S. K., Atkin S., Dewey C. M., Mayer P., 3rd, Herz J., Yu G. (2010) J. Biol. Chem. 285, 6826–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu L. S., Cheng W. C., Hou S. C., Yan Y. T., Jiang S. T., Shen C. K. (2010) Genesis 48, 56–62 [DOI] [PubMed] [Google Scholar]
- 63.Ou S. H., Wu F., Harrich D., García-Martínez L. F., Gaynor R. B. (1995) J. Virol. 69, 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lagier-Tourenne C., Cleveland D. W. (2009) Cell 136, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pesiridis G. S., Lee V. M., Trojanowski J. Q. (2009) Hum. Mol. Genet. 18, R156–R162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buratti E., Baralle F. E. (2001) J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 67.Winton M. J., Igaz L. M., Wong M. M., Kwong L. K., Trojanowski J. Q., Lee V. M. (2008) J. Biol. Chem. 283, 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y. M., Baralle F. E. (2005) J. Biol. Chem. 280, 37572–37584 [DOI] [PubMed] [Google Scholar]
- 69.Ayala Y. M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F. E. (2005) J. Mol. Biol. 348, 575–588 [DOI] [PubMed] [Google Scholar]
- 70.Abhyankar M. M., Urekar C., Reddi P. P. (2007) J. Biol. Chem. 282, 36143–36154 [DOI] [PubMed] [Google Scholar]
- 71.D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y. M., Baralle F. E. (2009) Nucleic Acids Res. 37, 4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Michelitsch M. D., Weissman J. S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11910–11915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrison P. M., Gerstein M. (2003) Genome Biol. 4, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L. M., Anderson P. (2004) Mol. Biol. Cell 15, 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Si K., Choi Y. B., White-Grindley E., Majumdar A., Kandel E. R. (2010) Cell 140, 421–435 [DOI] [PubMed] [Google Scholar]
- 76.Salazar A. M., Silverman E. J., Menon K. P., Zinn K. (2010) J. Neurosci. 30, 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson B. S., Snead D., Lee J. J., McCaffery J. M., Shorter J., Gitler A. D. (2009) J. Biol. Chem. 284, 20329–20339 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.