Abstract
HMG-CoA lyase (HMGCL) is crucial to ketogenesis, and inherited human mutations are potentially lethal. Detailed understanding of the HMGCL reaction mechanism and the molecular basis for correlating human mutations with enzyme deficiency have been limited by the lack of structural information for enzyme liganded to an acyl-CoA substrate or inhibitor. Crystal structures of ternary complexes of WT HMGCL with the competitive inhibitor 3-hydroxyglutaryl-CoA and of the catalytically deficient HMGCL R41M mutant with substrate HMG-CoA have been determined to 2.4 and 2.2 Å, respectively. Comparison of these β/α-barrel structures with those of unliganded HMGCL and R41M reveals substantial differences for Mg2+ coordination and positioning of the flexible loop containing the conserved HMGCL “signature” sequence. In the R41M-Mg2+-substrate ternary complex, loop residue Cys266 (implicated in active-site function by mechanistic and mutagenesis observations) is more closely juxtaposed to the catalytic site than in the case of unliganded enzyme or the WT enzyme-Mg2+-3-hydroxyglutaryl-CoA inhibitor complex. In both ternary complexes, the S-stereoisomer of substrate or inhibitor is specifically bound, in accord with the observed Mg2+ liganding of both C3 hydroxyl and C5 carboxyl oxygens. In addition to His233 and His235 imidazoles, other Mg2+ ligands are the Asp42 carboxyl oxygen and an ordered water molecule. This water, positioned between Asp42 and the C3 hydroxyl of bound substrate/inhibitor, may function as a proton shuttle. The observed interaction of Arg41 with the acyl-CoA C1 carbonyl oxygen explains the effects of Arg41 mutation on reaction product enolization and explains why human Arg41 mutations cause drastic enzyme deficiency.
Keywords: Acetoacetate, Acetyl Coenzyme A, Coenzyme A, Crystal Structure, Enzyme Mechanisms, Fatty Acid Metabolism, Ketogenesis, Mitochondria
Introduction
HMG-CoA lyase (HMGCL4 ; EC 4.1.3.4) catalyzes a cation-dependent cleavage of substrate into acetyl-CoA and acetoacetate (Scheme 1) (1). This reaction is a key step in ketogenesis, the products of which support anaplerotic metabolism in bacteria (2) and energy production in nonhepatic animal tissues (3). Ketogenesis is particularly important to human metabolism during the prenatal period and during fasting or starvation. In accordance with these physiological roles, it is not surprising that gene knock-out in mice results in embryonic lethality (4). The physiological importance of the enzyme in humans is underscored by the observation that mutations that diminish HMGCL activity correlate with inherited metabolic disease that can be lethal if uncontrolled (5).
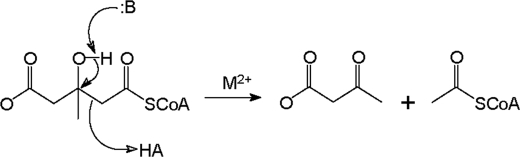
SCHEME 1.
HMG-CoA lyase reaction scheme.
A variety of human mutations, including many point mutations in protein-coding exons of the gene, have been documented (6). A computational modeling approach was used to explain the molecular basis for some mutations linked to inherited disease (7). This led to the prediction that HMGCL adopts a β/α-barrel fold and a proposal that the acyl-S-pantetheine moiety of the bound substrate passes through the barrel lumen. Initial structural work on human HMGCL liganded to cation and hydroxyglutarate (8) demonstrated that the folding prediction was reasonable but that the substrate binding proposal was unlikely to be correct. The positions of bound cation and hydroxyglutarate (from hydrolyzed hydroxyglutaryl-CoA) indicated the catalytic site to be positioned at the C-terminal end of the barrel, but the absence of a full acyl-CoA molecule in the experimentally determined structure limited detailed insight into the substrate-binding site.
To more fully address questions regarding the conformation of bound substrate, activator cation liganding, details concerning reaction chemistry and specificity, as well as the molecular basis for certain inherited HMGCL deficiencies, new structural information on enzyme bound to an intact acyl-CoA molecule is required. To remedy this need, complexes of the WT enzyme with the competitive inhibitor 3-hydroxyglutaryl-CoA and also of catalytically deficient R41M enzyme with the authentic substrate HMG-CoA have been supplemented with the activator cation Mg2+, and crystallization of the desired ternary complexes has been accomplished. Diffraction quality crystals have been produced, supporting three-dimensional structural determinations for ternary complexes of enzyme, cation, and either the acyl-CoA substrate or inhibitory analog. These findings are now described and interpreted to address the questions outlined above.
EXPERIMENTAL PROCEDURES
Expression and Isolation of HMGCL Proteins
The WT and R41M mutant HMGCL proteins were expressed using plasmid pTrc-HL-1 and purified using the protocol developed for the WT enzyme (9). Cloning, expression, and purification of the R41M mutant were carried out as described previously (10). For investigation of the functional consequences of Lys48 mutations, N-terminally His6-tagged proteins were produced by ligation of the NcoI/BamHI coding fragment (derived from pTrc-HL-1) into comparably restricted pET30b. The QuikChange mutagenesis protocol (Stratagene) was used to introduce mutations encoding K48N and K48Q. After DNA sequencing confirmed the desired mutations and the absence of artifacts in the coding sequence, the plasmids were used for protein expression. Active enzyme was expressed, and the efficient isolation of homogeneous N-terminally His-tagged proteins was facilitated by nickel-agarose chromatography. The WT enzyme (Vmax = 136 units/mg; Km for HMG-CoA = 26 μm) was comparable with the pTrc-HL-1-expressed enzyme. The protein concentration was determined by the method of Bradford (11). The enzyme activity was determined by a citrate synthase-coupled assay (1), modified as described previously (12).
Crystallization, Data Collection, and Structure Determination
WT crystals were obtained by the vapor diffusion method in the presence of 1 mm inhibitor, 3-hydroxyglutaryl-CoA (HG-CoA), as described previously (8). The R41M mutant crystals were obtained under similar conditions, except that the substrate (HMG-CoA) was used in place of the inhibitor (HG-CoA). Small needles formed immediately after the enzyme (6 mg/ml in 20 mm potassium phosphate buffer, pH 7.8) was mixed 1:1 with equilibration buffer consisting of 0.1 m Hepes, pH 7.5, 60 mm MgCl2, 5% glycerol, and 15–18% PEG 8000. These needles required 3–4 weeks to grow into large enough crystals for data collection.
Crystals of the WT lyase in complex with the inhibitor HG-CoA were generated by soaking WT crystals in mother liquor supplemented with additional 5 mm HG-CoA for 4 h. Generation of the complexed crystals of the R41M mutant lyase with the substrate HMG-CoA followed the identical procedure, i.e. the mutant crystals were soaked in mother liquor containing 5 mm HMG-CoA. All crystals were soaked in cryoprotectant consisting of mother liquor with an additional 20% glycerol prior to flash-freezing in liquid nitrogen. X-ray data for the WT lyase crystal soaked with the inhibitor HG-CoA (hereafter referred to as WT/HG-CoA) and for the R41M mutant were collected at −180 °C to resolutions of 2.4 and 2.2 Å, respectively, using an in-house R-AXIS IV++ detector coupled to a Rigaku Micromax 007 x-ray generator operating at 40 kV and 20 mA. For R41M crystals soaked with the substrate HMG-CoA (hereafter referred to as R41M/HMG-CoA), 2.25-Å resolution data were collected using synchrotron radiation at BioCARS beamline 14-BM-C at the Advanced Photon Source, Argonne National Laboratory. Data processing was carried out using the HKL2000 program package. All crystals belong to the monoclinic space group C2, with six monomers in the asymmetric unit and with Matthews coefficients of Vm = 2.2 Å3/Da, corresponding to 42% solvent content.
The crystal structure of WT/HG-CoA was determined and refined using the WT human lyase structure (Protein Data Bank code 2CW6) (8) as the starting model. Bound HG-CoA was located using the difference Fourier map calculated with the |Fo| − |Fc| coefficient. The R41M mutant crystal structure was determined by molecular replacement with MOLREP within the CCP4 program suite (13) using the monomer structure of WT lyase (Protein Data Bank code 2CW6) as the search model. There is an ∼2-Å difference in the a axis between the WT/HG-CoA lyase and R41M mutant enzyme crystals. The structure of R41M/HMG-CoA was also solved using the R41M structure by the difference Fourier method. Refinements were carried out using the program CNS (14) and together with manual model adjustments using the program TURBO-FRODO (15) and COOT (16). Water molecules were added at electron densities >3σ in the |Fo| − |Fc| map after several cycles of model building and structure refinement. The final models gave the following crystallographic R-factor/Rfree values: WT/HG-CoA, 0.211/0.284; R41M, 0.219/0.263; and R41M/HMG-CoA, 0.222/0.274. The data collection and refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics
PDB, Protein Data Bank; r.m.s.d., root mean square deviation.
| Crystal |
|||
|---|---|---|---|
| WT/HG-CoA (PDB code 3MP3) | R41M (PDB code 3MP4) | R41M/HMG-CoA (PDB code 3MP5) | |
| Diffraction data | |||
| Resolution range (Å) | 30–2.40/2.44–2.40 | 30–2.2/2.28–2.2 | 20–2.25/2.33–2.25 |
| No. total reflections | 247,687 | 466,437 | 430,579 |
| No. unique reflections | 66,824/2315 | 86,402/5896 | 86,171/8582 |
| Completeness (%) | 92.3/64.6 | 94.4/64.4 | 100/99.9 |
| Redundancy (%) | 3.7/2.2 | 5.4/3.6 | 5.0/4.6 |
| I/σ(I) | 8.8/2.0 | 28.9/2.6 | 15.9/2.1 |
| Unit cell dimensions | a = 197.4, b = 117.0, c = 86.9 Å; β = 112.3° | a = 196.5, b = 116.6, c = 87.0 Å; β = 112.8° | a = 198.0, b = 116.7, c = 86.5 Å; β = 112.2° |
| Space group | C2 | C2 | C2 |
| Rsym | 0.100/0.323 | 0.051/0.455 | 0.101/0.674 |
| Vm (Å3/Da)/solvent content (%) | 2.2/42.0 | 2.2/42.0 | 2.2/42.0 |
| Monomers in asymmetric unit | 6 | 6 | 6 |
| Refinement | |||
| Rcrystal/Rfree | 0.212/0.282 | 0.219/0.261 | 0.221/0.274 |
| r.m.s.d. bond length (Å)/bond angle | 0.007/1.25° | 0.006/1.20° | 0.007/1.24° |
| No. protein atoms | 13,107 | 13,049 | 13,034 |
| No. water molecules | 437 | 382 | 344 |
| No. bound ligand atoms | 67a | 58b | |
| No. metal ions | 6 | 2 | |
| Average B-factor | |||
| Main chain atoms (Å2) | 31.0 | 45.4 | 36.4 |
| Side chain atoms (Å2) | 32.6 | 47.4 | 38.3 |
| All protein atoms (Å2) | 31.8 | 46.3 | 37.3 |
| Water molecules (Å2) | 27.5 | 40.6 | 31.3 |
| Ligands (Å2) | 66.5 | 73.3 | |
| Metal ions (Å2)c | 21.9 | 38.7 | |
a Includes 57 atoms for HG-CoA and 10 atoms for HGA.
b For HMG-CoA.
c Six Mg2+ ions in the WT crystal and two ions in R41M/HMG-CoA.
RESULTS
Overall Structure and the Ligand-binding Site
We have determined structures of human HMGCL in three different crystal forms: the WT enzyme crystallized in the presence of a competitive inhibitor, HG-CoA (17), and soaked with HG-CoA (WT/HG-CoA); the R41M mutant crystallized in the presence of the substrate HMG-CoA but with no ligand bound (R41M); and R41M soaked with HMG-CoA (R41M/HMG-CoA). In each crystal form, there are six monomers (three dimers) in the asymmetric unit, and not all of the six protomers contain bound ligand. In fact, in the WT/HG-CoA crystal, only one monomer contains HG-CoA, and another monomer contains 3-hydroxyglutaric acid (HGA; presumably the hydrolysis product of HG-CoA). The R41M crystal has no Mg2+ ion bound in any of the six monomers. In the R41M/HMG-CoA crystal, only one monomer contains bound HMG-CoA and Mg2+, one contains Mg2+ only, and the other four monomers contain no Mg2+ and no ligand. Thus, we have a total of 18 different monomer structures, including the WT with Mg2+ bound (WT-Mg2+), R41M with apo structure (i.e. no Mg2+ ion) (R41M), R41M binary complex with Mg2+ ion (R41M-Mg2+), and three different ternary complexes (the WT with HG-CoA and Mg2+, i.e. WT-Mg2+-HG-CoA; the WT with HGA and Mg2+, i.e. WT-Mg2+-HGA; and R41M with Mg2+ and HMG-CoA, i.e. R41M-Mg2+-HMG-CoA). Because the structures of R41M and R41M-Mg2+ are the same, R41M will be used for both structures. Also, as all ternary complexes include Mg2+, they will be referred to as simply WT-HG-CoA, WT-HGA, and R41M-HMG-CoA. With the exception of some residues (positions 265–271) in the glycine-rich loop that are disordered in some monomer structures, the entire polypeptide starting from the N terminus of the mature protein (Thr28) to Cys323 (sequence alignment in Fig. 1) is observed in all 18 monomers in the three crystal forms. The last two C-terminal residues, Lys324 and Leu325, are not observed in any of the 18 structures. Because there are no significant differences among the structures of apo and binary complexes of both the WT and R41M with Mg2+ ion (root mean square deviations between pairs of <0.6 Å for the entire visible 296 Cα atoms), they will be referred to as WT and R41M. Furthermore, apart from the two regions Ser131–Gln148 and Gly264–Gly274, the latter of which is disordered in some monomer structures, the overall polypeptide folds are the same among all 18 monomers with root mean square differences ranging from 0.41 to 0.72 Å for 274 Cα atoms (Fig. 2A). The structural and functional significance of the movements/disorderedness of these two regions will be discussed below. It is interesting to note that not all 18 monomers contain bound Mg2+, even though the crystallization media for all lyase crystals contained 60 mm MgCl2. Although all of the WT enzyme monomers contain Mg2+, only two of the 12 monomers in R41M mutant crystals contain the metal ion, including the one that has bound HMG-CoA. Whether the mutant has a lower affinity for Mg2+ is not known. However, the Kd value of the mutant for Mn2+ (which binds more tightly than Mg2+) is almost the same as that of the WT (2.1 versus 1.8 μm) (10). Each monomer of human lyase is composed of nine β-strands and 12 α-helices and forms a (βα)8-triosephosphate isomerase (TIM) barrel with an additional helical region, residues 290–325, as reported previously (8). A structure of the physiological dimer designated on the basis of the extensive intersubunit contact area (8) is shown in Fig. 2B; one subunit is rotated by ∼90° with respect to the other.
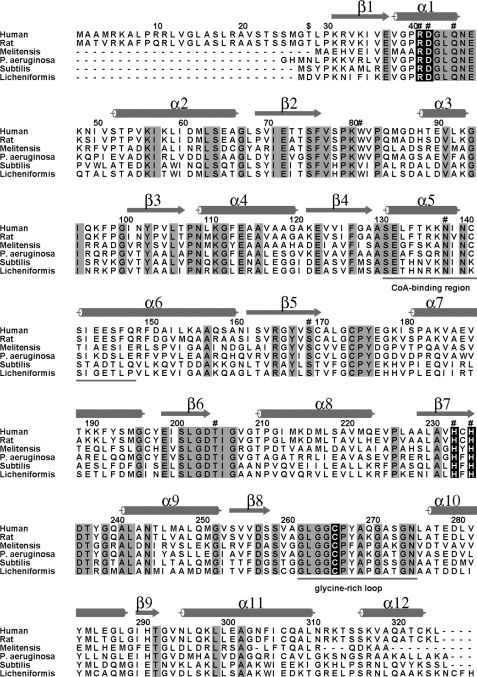
FIGURE 1.
Sequence alignment of HMGCLs from Homo sapiens (human), Rattus norvegicus (rat), Brucella melitensis, Pseudomonas aeruginosa, Bacillus subtilis, and Bacillus licheniformis. The dollar sign indicates the start of the mature protein at Thr28. Identical residues in all six organisms are highlighted in gray. Secondary structural elements of human HMGCL are shown as cylinders (12 α-helices) and arrows (nine β-strands). Catalytic residues and residues that coordinate Mg2+ ion are marked highlighted in black. Residues interacting with the CoA ligand are indicated (#). The underlined helical CoA-binding region displays some mobility upon ligand binding. The underlined glycine-rich loop delineates the signature sequence for the HMGCL family of enzymes. Residues after position 290 are largely helical, and this part of the polypeptide is external to the TIM barrel.
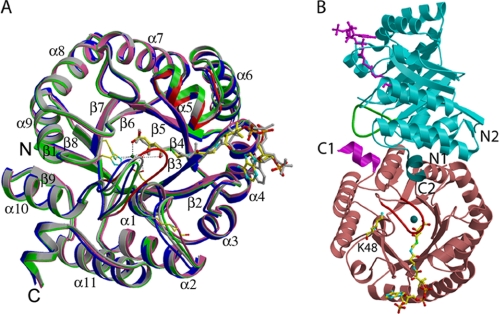
FIGURE 2.
Ribbon diagrams of the human HMGCL structure. A, an overlay of four monomer structures is shown. Light gray, WT lyase; blue, WT lyase complexed with the inhibitor HG-CoA; green, R41M; pink, R41M-HMG-CoA. Mg2+ is shown as a blue ball; the inhibitor HG-CoA is shown with atomic coloring; and the substrate HMG-CoA is displayed in gray. Hydrogen bonds with protein or bonds to Mg2+ are indicated by dotted lines. Significant differences between the overlaid monomers are marked in red. The glycine-rich loop containing Leu263–Asn275 shows significant movement when the R41M-HMG-CoA complex is compared with the other three monomers. Ala129–Cys141 also shows mobility when CoA ligands are bound in the L-shaped conformation. B, the dimer structure of mutant R41M complexed with the substrate HMG-CoA and Mg2+. The tan lower monomer shows bound HMG-CoA with atomic coloring. The cyan upper monomer displays the apo-R41M structure, and a modeled magenta HMG-CoA is shown only to indicate the active site. The barrel of the cyan monomer is rotated by ∼90° relative to the tan lower monomer. The magenta C terminus of the lower monomer is interacting with the green Leu263–Asn275 loop of the cyan upper monomer. The dark cyan C terminus of the upper monomer is no longer interacting with the red Leu263–Asn275 loop of the bottom monomer due to the loop movement upon ligand binding. Lys48 is located far from the active site and does not interact with the bound CoA ligand.
Structures of Ternary Complexes
Electron densities corresponding to the bound ligands are displayed in Fig. 3. Overall, the electron densities are well defined with the exception of that for the adenine ring in the ternary structure of R41M with HMG-CoA (Fig. 3E).
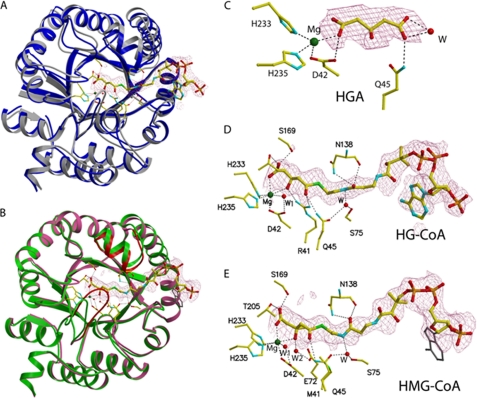
FIGURE 3.
Active site of human HMGCL. A, overlay of monomer structures of WT lyase with (blue) and without (gray) the bound inhibitor HG-CoA. Atom coloring is used for all ligands, and bond interactions are indicated by dotted lines. B, monomer overlay of mutant R41M in green and R41M with the substrate HMG-CoA in pink. Red indicates the mobility of the Cys266-containing loop residues Leu263–Asn275. This loop moves closer into the active site when the substrate is bound. C–E, electron density maps showing bound ligands: HGA (a hydrolysis product of the inhibitor HG-CoA; C), the inhibitor HG-CoA (D), and the substrate HMG-CoA (E) fitted into densities from |Fo| − |Fc| maps at the 2.5σ level. The adenine ring of HMG-CoA (gray) is disordered.
To trap the ternary complex of the WT, the competitive inhibitor HG-CoA was used for the WT lyase. Because our previous co-crystallization attempts using HG-CoA resulted in the lyase complex with a hydrolysis product (HGA) of the inhibitor (8), both co-crystallization and soaking methods were employed. Preformed WT lyase crystals that were obtained in the presence of HG-CoA were soaked further with a solution containing HG-CoA. However, of six monomers (three dimers) in the asymmetric unit, only one monomer of the WT enzyme has a bound inhibitor (Fig. 3, A and D), and the other monomer of the same dimer has bound HGA (Fig. 3C), as observed previously with co-crystallization with HG-CoA (8). The remaining four monomers contain only Mg2+ ion, but no other ligand. In the structure of the monomer that contains HGA and Mg2+, one of the carboxylates of the hydrolysis product (HGA) is coordinating Mg2+, in contrast to the structure of the HGA complex obtained by co-crystallization only. In the latter case, the HGA molecule is >6 Å away from the Mg2+ ion (8). Thus, the HGA binding mode in the active-site cavity is not unique and is quite different from that of the glutaryl moiety of HG-CoA or HMG-CoA (see below). Therefore, model building for the ternary complex structure of a CoA derivative based on the structure of either of these two HGA complexes would not result in a correct binding mode for the substrate or inhibitor. These observations serve as a reminder that one must proceed with caution when modeling a ligand-bound structure on the basis of a reference structure that is missing a significant portion of the ligand.
The inhibitor HG-CoA is bound with the acyl moiety adjacent to the Mg2+ activator cation, which is situated near the base of the C-terminal cavity of the barrel (Fig. 2A and 3A), as was predicted previously from the WT structure (8). Although crystal growth required the presence of HMG-CoA substrate or HG-CoA analog, soaking the crystals with these compounds was required to improve occupancy and detectable electron density. Nonetheless, the orientation of bound ligand satisfies base-line functional expectations because, as indicated below, the acyl group C3 and C5 oxygens are coordinated with the activator cation. Moreover, only the physiologically active S-isomer of the substrate/analog is bound despite the fact that chemically synthesized mixtures of isomers of HMG-CoA substrate or HG-CoA analog were used for co-crystallization and soaking. The bound ligand adopts an L-shaped conformation; the acylpantetheine moiety forms the long arm, and the 3′-AMP forms the short arm, with the pyrophosphate making the right angle turn, resulting in exposure of the 3′-AMP to solvent at the surface of the molecule. This conformation corresponds to that observed in the structures of other acyl-CoA-binding enzymes, including the acyl-CoA dehydrogenase family (18) and malate synthase (19, 20), and contrasts with the compact bent conformation observed for citrate synthase (21) or the completely extended linear shape observed for methylmalonyl-CoA mutase (22).
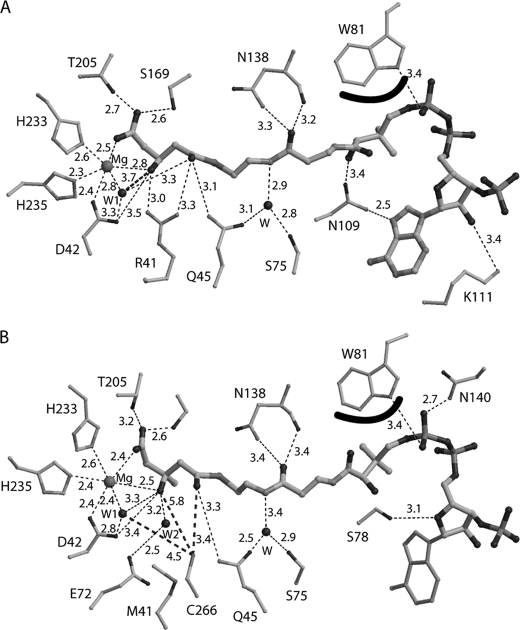
Fig. 3 depicts the overlay of monomers of the WT enzyme with enzyme complexed to the competitive inhibitor HG-CoA and monomers of R41M with the R41M complex containing HMG-CoA, respectively. Upon acyl-CoA liganding, there are minimal structural changes in most of the protein backbone (root mean square deviation of ∼0.5 Å) except for the residues in α-helix 5 (Ser131–Ile139) and the subsequent residues (Asn140–Ser142), which move ∼3 Å closer to the pantetheine portion of CoA. Fig. 4 shows detailed interactions between the bound ligand and the surrounding residues. In the structure of WT-HG-CoA (Fig. 4A), the oxygen atoms of the C5 carboxyl and C3 hydroxyl of the hydroxyglutaryl moiety of the inhibitor are coordinating the activator Mg2+ ion. In addition, there are hydrogen bond interactions from a C5 carboxyl oxygen to hydroxyl oxygens of Ser169 (2.5 Å) and Thr205 (2.7 Å). The Asp42 carboxyl displays an interaction with the C3 oxygen (3.5 Å). The guanidinium nitrogens of Arg41 closely interact with the C3 (3.0 Å) and C1 (3.3 Å) oxygens, and the Gln45 amide nitrogen interacts with the C1 oxygen (3.1 Å). Also, a well ordered water molecule is in close proximity to both the C1 (3.3 Å) and C3 (3.8 Å) oxygens and one of the Asp42 carboxyl oxygens (3.3 Å). In addition to these multiple binding contacts for the acyl group, the CoA moiety makes additional interactions with the polypeptide: the amide nitrogen and the main chain carbonyl oxygen of Asn138 form hydrogen bonds with the oxygen of the amide that links cysteamine and 4-phosphopantetheine moieties; the Trp81 indole nitrogen makes a hydrogen bond with a pyrophosphoryl oxygen; the indole ring also flips to shield the pantetheine portion of the CoA moiety from solvent; Lys111 forms a hydrogen bond with the ribityl 2′-hydroxyl oxygen; and Asn109 makes hydrogen bonds with both the 4-phosphopantetheine-bridging hydroxyl oxygen and N7 of the adenine ring. These latter interactions explain the L-shaped bend observed for binding of the CoA moiety. Furthermore, the interactions involving Asn138 are at least partially responsible for the bending and pulling of the helix-loop-helix region containing Ser131–Gln148 (Fig. 2A). Note that interactions between the polypeptide and the pyrophosphoryl adenosine moiety of CoA observed in the structure of R41M-HMG-CoA are slightly different from those seen in the WT-HG-CoA structure, presumably due to the disordered adenine ring of HMG-CoA.
FIGURE 4.
Interactions between the HMGCL polypeptide and bound acyl-CoA ligands. A, schematic drawing of interactions between WT human lyase and HG-CoA. Hydrogen and coordination bonds are indicated by dashed lines. Thicker lines reflect the interaction of the HG-CoA analog with ordered water (A) or the interactions of HMG-CoA and ordered water with Cys266 (B). The curved line at Trp81 indicates a hydrophobic interaction between the indole ring and bound inhibitor. B, interactions between HMGCL R41M and HMG-CoA. The disordered adenine of HMG-CoA is depicted as thinner lines. Most interactions are similar when WT-HG-CoA and R41M-HMG-CoA are compared. Some minor differences in the binding region of the pyrophosphoryl adenosine moiety of CoA are observed due to the disordered adenine ring in R41M-HMG-CoA. However, the most significant changes are seen where arginine is replaced with methionine.
Structure of R41M in Complex with HMG-CoA
To determine the binding mode of the substrate HMG-CoA, the catalytically deficient R41M mutant was employed. The overall binding mode is essentially the same as that of HG-CoA to the WT enzyme (Fig. 3B); however, there are a few subtle but significant differences. Fig. 4B shows the detailed interactions between the substrate and the surrounding residues. In addition to the water coordinating to the Mg2+ ion (W1), a second ordered water (W2 in Fig. 4B) is situated in close proximity (3.2 Å) to the substrate C3 oxygen and is stabilized by interaction with the Glu72 carboxyl oxygen (2.5 Å). This water (W2) is positioned where the guanidinium group of Arg41 lies in the WT structure. Thus, two ordered water molecules interact with the substrate C3 oxygen; one (Mg2+-coordinated) is positioned by Asp42 (2.8 Å), and the other is situated in the space vacated due to the R41M mutation and stabilized by Glu72. A major difference in the HMG-CoA complex is the position of the glycine-rich loop, which moves closer to the acyl group of bound substrate by ∼6 Å. This brings Cys266, a residue implicated as functionally important by kinetic, mutagenesis, and affinity labeling studies, into the active site. The thiol group of Cys266 is now situated 3.4 Å from the C1 carbonyl oxygen of HMG and 4.5 Å from the water molecule that coordinates Mg2+ (Figs. 3B and 4B).
Divalent Activator Mg2+ Ion-binding Site
When the hydrolysis product HGA is bound to the enzyme, i.e. when the bound ligand lacks the CoA moiety, the ligand binds in two different modes. When the HGA concentration is relatively low, the Mg2+ coordination involves His233, His235, Asp42, Asn275, and two water molecules, and HGA is not within the coordination sphere. Thus, the HGA ligand is away from the bottom of the active-site pocket (8). On the other hand, when the HGA concentration is higher (this study, due to both co-crystallization and soaking with HG-CoA), the Mg2+ ion coordination is tetrahedral, involving His233, His235, Asp42, and one carboxyl oxygen of HGA. Thus, HGA is positioned deeper into the long narrow cavity of the active site, but not deep enough for both the carboxyl and hydroxyl oxygens of HGA to coordinate to the Mg2+ ion. For the enzyme complex with either HG-CoA or HMG-CoA (i.e. when the CoA moiety is present), the Mg2+ ion has nearly perfect octahedral coordination, involving His233, His235, Asp42, C3, and C5 oxygens from the S-3-hydroxyglutaryl moiety (Fig. 4A) or S-3-hydroxy-3-methylglutaryl moiety (Fig. 4B) and a well ordered water molecule. The S-isomer of HG and HMG moieties is required for Mg2+ coordination of the C3 oxygen. This isomer is selectively bound in these complexes, in agreement with the observation (1) of the stereospecificity of this enzyme. All four amino acids involved in activator cation coordination are invariant residues that have been shown to make substantial contributions to HMGCL function. Interestingly, two other members of the HMGCL TIM barrel family of enzymes, 4-hydroxy-2-ketovalerate aldolase (23) and isopropylmalate synthase (24), also utilize for cation coordination two histidines and one aspartate; these residues are conserved in the HMGCL family.
Role of Lys48
Lys48 of HMGCL has been implicated in an inherited K48N mutation (25). In addition, proteomics results have shown that this residue is a site of post-translational acetylation (26). For these reasons, K48N and K48Q mutant proteins (which model the proposed human mutation and mimic Lys48 acetylation, respectively) were engineered, isolated, and kinetically characterized (Table 2). The purified mutant proteins retain substantial activity, exhibiting Vmax values that are 65% (K48N) and 74% (K48Q) of the estimate for the WT enzyme (Vmax = 136.0 units/mg). The Km values for HMG-CoA of K48N (20.9 μm) and K48Q (13.4 μm) do not suggest impaired substrate saturation in comparison with the WT enzyme (Km = 26.5 μm). These experimental results for purified K48N and K48Q mutant proteins do not indicate a critical role for Lys48 in HMGCL function and are compatible with the position of bound acyl-CoA observed in structures of the protein complexes with the analog HG-CoA (Fig. 3A) or with the substrate HMG-CoA (Figs. 2B and 3B). The distances from both the Cα and ϵ-NH2 of Lys48 to the carbonyl oxygen of the thioester are >15 Å, and their closest distances to the bound HMG-CoA are also >14 Å to the amide nitrogen of the 4-phosphopantothenic acid moiety (Fig. 2B). The C terminus of the lyase molecule lies >25 Å from the adenine ring of the substrate, contrary to the results obtained from the modeled structure of the enzyme-ligand complex (25). Instead, the C terminus is interacting with the glycine-rich loop of the other monomer (Fig. 2B).
TABLE 2.
Characterization of HMG-CoA lyase Lys48 mutant proteins
Experiments were performed using homogeneous enzyme preparations. Kinetic parameters are based on assays performed at 30 °C using the spectrophotometric method described under “Experimental Procedures.” Data were fit by nonlinear regression analysis.
| Protein | Vmax | Km for HMG-CoA | t½ a |
|---|---|---|---|
| units/mg | μm | min | |
| WT | 136.0 ± 4.4 | 26.5 ± 3.2 | 36.3 ± 6.7 |
| K48N | 87.8 ± 2.5 | 20.9 ± 2.3 | 27.1 ± 3.7 |
| K48Q | 100.3 ± 2.7 | 13.4 ± 1.4 | 22.2 ± 2.1 |
a t½ measurements were performed at 37 °C. Samples of the purified proteins (in pH 7.8 incubation buffer containing 25 mm sodium phosphate, 300 mm NaCl, 300 mm imidazole, 5 mm mercaptoethanol, and 20% glycerol) were kept on ice until initiation of the experiment by transfer to a 37 °C water bath. At 10-min intervals over a period of 90 min, aliquots of the incubation samples were diluted 500-fold into cuvettes containing the reagents for the standard citrate synthase-coupled spectrophotometric assay (1, 12) for determination of activity. Assays were performed in triplicate. t½ values were calculated by fitting the incubation time-dependent activity data to an exponential decay curve using nonlinear regression analysis (GraphPad Prism 4).
DISCUSSION
Human HMGCL Ternary Complex Structures: Similarities and Contrasts
Because HMGCL is a single substrate enzyme, to prepare enzyme-Mg2+-acyl-CoA complexes for crystallization, it was necessary to employ the competitive inhibitor HG-CoA to avoid turnover in the complex with the WT enzyme. Conversely, for a complex with the substrate HMG-CoA to persist, the catalytically deficient R41M mutant protein was used. As discussed in more detail below, the positioning observed for the flexible loop (Leu263–Asn275) that harbors the signature sequence for HMGCL is substantially different in these two acyl-CoA complexes. However, regardless of which protein or acyl-CoA molecule is used to detect bound acyl-CoA in these two ternary complexes, observed cation liganding is similar, but it differs from observations made for the enzyme-Mg2+-3-hydroxyglutarateternary complex (8). In complexes with either an acyl-CoA substrate or analog, oxygen atoms from both the C5 carboxyl and C3 hydroxyl groups ligand to cation. In the absence of a CoA ligand, Mg2+ is coordinated to a second water molecule and the Asn275 amide nitrogen, both of which replace the ligand oxygens. In all three cases, the four remaining cation ligands are contributed by Asp42, His233, His235, and an ordered water molecule.
The physiological significance of the observed acyl-CoA binding in both ternary complexes is underscored by the fact that, despite incubation of enzymes with chemically synthesized RS-mixtures of HMG-CoA or 3-hydroxyglutaryl-CoA, the only bound species observed in each complex represents the S-isomer. Stegink and Coon (1) experimentally demonstrated that HMGCL is specific for the S-isomer. Cation coordination to substrate/analog contributes to specificity because coordination to both the C3 hydroxyl and C5 carboxyl oxygens of the acyl moiety could not occur for R-acyl-CoA isomers.
Contrasts are also apparent upon comparison of the acyl-CoA-binding site experimentally determined for both of these ternary complexes with the model predicted on the basis of molecular docking (25). Those computational studies relied on structural coordinates determined for the enzyme-cation-HGA complex (8). Although the structural results indicated the approximate location of the catalytic site, some ambiguity persisted due to the presence of two carboxyl functionalities in bound HGA. Based on the available information, the modeling study imposed the constraint that the HMG-CoA acyl chain dock to the protein surface where HG binding had been observed. Without availability of other information on which carboxyl group of bound HG is originally thioesterified to CoA and without details implicating specific residues in CoA binding, the molecular docking model that resulted suggested that CoA binds in an extended fashion, with the adenosine moiety oriented toward the C-terminal α-helical region of the protein. Interaction of the HMG-CoA 3′-phosphoryl with Asn311 and Lys313 was proposed. Frameshift mutations in DNA encoding the human enzyme result in C-terminal deletions. Observation of diminished activity for such mutants was interpreted in the context of this docking model of the protein. Additionally, a missense mutation that produces K48N protein was proposed to correlate with inherited metabolic disease. This residue was modeled to suggest interaction of Lys48 with a pantetheine oxygen of HMG-CoA. Disruption of substrate binding or conformation of bound substrate upon mutation of Lys48 was therefore proposed (25) as the basis for the observation of diminished mutant enzyme activity. Because the ϵ-amino of Lys48 is observed to be located >15 Å from the CoA pantetheine oxygen (Fig. 2B), and the C terminus of the protein is >25 Å from the adenine moiety of the substrate, other explanations (altered expression, folding, stability, etc.) offered to account for the low activity reported for these inherited HMGCL mutants need to be carefully considered. For example, truncation of the C-terminal sequence could disrupt dimerization and activity. Native active enzyme from eukaryotes and prokaryotes (9, 12, 27, 28) has been characterized as a dimer of identical subunits.
Interpretation of Functional Observations in the Context of Ternary Complex Structure
HMGCL functions in eukaryotes and prokaryotes, catalyzing key reactions in ketone body biosynthesis and leucine catabolism. Reaction products support metabolic energy production (acetoacetate) and anaplerotic metabolism (acetyl-CoA). The initial identification of functional residues at the active site of the enzyme (29) was accomplished by affinity labeling of the Pseudomonas mevalonii enzyme using 2-butynoyl-CoA to covalently modify the protein. This reagent exhibits Ki values of 65 and 320 μm for the bacterial and animal enzymes, respectively. The cysteine (Cys237) that was identified in the modified P. mevalonii enzyme maps as Cys266 in the human enzyme; this residue is conserved in all HMGCL proteins. Subsequent mutagenesis of this residue in recombinant forms of human (30) and bacterial (31) enzymes resulted in decreases in catalytic activity of 4 orders of magnitude upon substitution of cysteine with alanine.
Knock-out of the gene in animals (4) causes prenatal lethality. Human disease results from a variety of inherited HMGCL mutations. Consequently, our functional investigations have focused not only on conserved residues identified by protein modification approaches but also on those point mutations that map to invariant residues in HMGCL proteins. We have modeled such mutations in a recombinant form of the human enzyme and characterized the purified mutants in an attempt to test enzymatic function of the mutated residues. A combination of chemical modification results and characterization of proteins mutated at these sites indicated functional roles for invariant residues His233 and His235, which were subsequently identified in initial structural work (8, 32) as activator cation ligands. Other human mutations implicated invariant residues Asp42 and Arg41. Characterization of purified proteins containing mutations of each of these side chains (10, 33) indicated major decreases (4 and 5 orders of magnitude, respectively) in catalytic activity. Moreover, using the acetyldithio-CoA (an analog of the reaction product acetyl-CoA) to measure product enolization, Arg41 mutation was shown to eliminate this partial reaction (10). The ternary complex structure of the WT enzyme with Mg2+ and the inhibitor HG-CoA (Fig. 3A) is particularly informative in demonstrating the proximity (3.3 Å) of the Arg41 guanidinium moiety to the thioester carbonyl oxygen of bound acyl-CoA (Fig. 4A). A similar interaction of arginine with bound acyl-CoA has been suggested in work on the related malate synthase reaction (20). Interestingly, residues corresponding to Arg41, Asp42, His233, and His235 of the human enzyme are conserved in other members of a broader HMGCL family of proteins such as isopropylmalate synthase, homocitrate synthase, and hydroxyketovalerate aldolase. All of these enzymes utilize acetyl-CoA as a substrate or product. In contrast, only HMGCL proteins contain the affinity label-targeted cysteine in a flexible loop (Leu263–Asn275) that harbors a highly conserved “signature” sequence for this protein. Upon substrate binding, this loop moves over to the C-terminal end of the TIM barrel of protein that contains the active site, including the invariant residues identified above.
Another factor to be considered in correlating the tertiary complex structural information with functional observations involves reaction stereochemistry. Cleavage of substrate by HMGCL occurs with incorporation of deuterium from solvent D2O into the methyl group of acetyl-CoA and inversion of stereochemistry at the substrate C3 (34). In this respect, the stereochemistry of this reaction is in accord with other enzyme-catalyzed Claisen condensations or cleavages (35). This observation requires that deprotonation of the substrate C3 alcohol and protonation of the carbanion produced upon C2–C3 bond cleavage occur from the same side of the substrate; this is the side involved in coordination of C3 and C5 oxygens to the activator cation (Fig. 3). An ordered water molecule, as well as two amino acids (Asp42 and Cys266) with side chains shown to have a large influence on reaction rate, would qualify for participation in these steps based on stereochemical considerations and on the orientation of these candidates in the structures of the HMG-CoA- and HG-CoA-containing ternary complexes (Fig. 3).
Candidates for Functional Roles in Substrate Cleavage
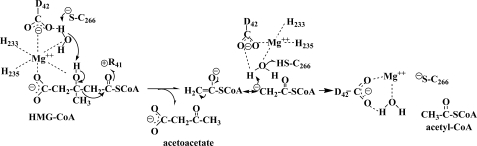
The reaction is critically dependent on the presence of a charge sink for the electron density generated as a result of substrate C2–C3 bond cleavage. Clearly, the interaction of the guanidinium group of Arg41 with the C1 carbonyl oxygen satisfies this requirement. There are several candidates for participation as a general acid or base in HMG-CoA cleavage, so these assignments are more complicated. Candidates include Asp42 and the ordered water molecule situated in closest proximity to C2 and C3 of bound acyl-CoA in the ternary complex structures (Figs. 3 and 4). In these structures, the carboxyl of Asp42 (elimination of which decreases activity by >104-fold) is in close proximity to the bound ligand C3 hydroxyl oxygen (3.5 Å), but so is the ordered water oxygen, which is positioned 3.3 Å (Fig. 4B) or 3.8 Å (Fig. 4A) away from this C3 oxygen in the ternary complex with HMG-CoA or HG-CoA, respectively. This solvent oxygen is closer to the ligand C2 (4.0 Å) than is the Asp42 carboxylate (5.6 Å). The requisite proton abstraction from the substrate and the carbanion quenching by protonation of the cleavage product by either candidate (as well as by Cys266) would be compatible with observed reaction stereochemistry. The participation of ordered water in shuttling a proton between the substrate and an active-site residue side chain has precedent in related aldolase reactions. More specifically, in 2-keto-3-deoxy-6-phosphogluconate aldolase (36), the shuttle involves an ordered water and the carboxyl group of a glutamate implicated in catalysis.
Consideration of candidates for participation as active-site acid/base residues (Fig. 5) should, however, also take into account the measurement of pKa = 8.0 in the pH/rate profile for the WT enzyme (30). No titratable group is detectable when the comparable experiment is performed using C266S, suggesting that the sulfhydryl of Cys266 accounts for the observed pKa, and therefore, its participation in reaction chemistry is important. Moreover, a residue corresponding to this cysteine is covalently modified by the affinity label 2-butynoyl-CoA, demonstrating that, in solution, close juxtapositioning of Cys266 to bound acyl-CoA can occur. Assignment of a role for Cys266 becomes complicated because of the considerable mobility of the Cys266-containing flexible loop that is evident from our structural results (Fig. 2). In the R41M-containing complex with the substrate, Cys266 is in closer proximity to bound acyl-CoA than in structures of WT enzyme complexes with hydroxyglutarate or HG-CoA. Although loop positioning in the R41M mutant structure may not reveal the closest possible proximity of Cys266 to the substrate, it reflects the most relevant experimental structural results available. In the R41M structure (Fig. 4B), Cys266 is somewhat remote from the substrate C3 hydroxyl oxygen (5.8 Å) but closer to the C1 carbonyl oxygen (3.4 Å) and the ordered solvent oxygen (4.5 Å).
FIGURE 5.
Proposed reaction mechanism for HMGCL. This schematic drawing shows functionally important active-site side chains, Mg2+, an ordered water molecule, and the HMGCL substrate and products. The left side of the diagram indicates the six Mg2+ coordination ligands, as found in the R41M-Mg2+-HMG-CoA ternary complex. In the absence of the HMG-CoA or HG-CoA ternary complex ligands, the substrate C5 carboxyl and C3 hydroxyl oxygen ligands are replaced with water and Asn275 oxygens (not shown) (8).
Although both Asp42 and Cys266 are plausible participants in a water-mediated proton shuttle, the simplest model (Fig. 5) that is in accord with both the mechanistic and functional data as well as the ternary complex structural results involves participation of the aforementioned ordered water molecule in a proton shuttle with Cys266 functioning as the partner in the relay. Interactions with both Mg2+ and Asp42 precisely position this solvent molecule both in close proximity to substrate and 4.5 Å from the sulfur atom of Cys266 in the mobile loop. Thus, water-mediated deprotonation of the substrate C3 hydroxyl can be supported by the cysteine thiolate anion. Subsequently, the Cys266 thiolate anion re-forms upon shuttling of a proton from the neutral thiol to the hydroxyl anion that develops after an ordered solvent proton is used to quench the carbanionic form of the acetyl-CoA reaction product.
Acknowledgments
We thank the staff at Advanced Photon Source beamline BioCARS 14-BM-C for excellent assistance in data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants GM29076 (to J.-J. P. K.) and DK21491 (to H. M. M.). Use of the Advanced Photon Source was supported by the United Stated Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38, and Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357. Use of BioCARS Sector 14 was supported by National Center for Research Resources Grant RR07707 from the National Institutes of Health.
The atomic coordinates and structure factors (codes 3MP3, 3MP4, and 3MP5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HMGCL
- HMG-CoA lyase
- HG-CoA
- 3-hydroxyglutaryl-CoA
- HGA
- 3-hydroxyglutaric acid
- TIM
- triosephosphate isomerase.
REFERENCES
- 1.Stegink L. D., Coon M. J. (1968) J. Biol. Chem. 243, 5272–5279 [PubMed] [Google Scholar]
- 2.Siddiqi M. A., Rodwell V. W. (1967) J. Bacteriol. 93, 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson A. M., Williamson D. H. (1980) Physiol. Rev. 60, 143–187 [DOI] [PubMed] [Google Scholar]
- 4.Wang S. P., Marth J. D., Oligny L. L., Vachon M., Robert M. F., Ashmarina L., Mitchell G. A. (1998) Hum. Mol. Genet. 7, 2057–2062 [DOI] [PubMed] [Google Scholar]
- 5.Gibson K. M., Breuer J., Nyhan W. L. (1988) Eur. J. Pediatr. 148, 180–186 [DOI] [PubMed] [Google Scholar]
- 6.Pié J., López-Viñas E., Puisac B., Menao S., Pié A., Casale C., Ramos F. J., Hegardt F. G., Gómez-Puertas P., Casals N. (2007) Mol. Genet. Metab. 92, 198–209 [DOI] [PubMed] [Google Scholar]
- 7.Casals N., Gómez-Puertas P., Pié J., Mir C., Roca R., Puisac B., Aledo R., Clotet J., Menao S., Serra D., Asins G., Till J., Elias-Jones A. C., Cresto J. C., Chamoles N. A., Abdenur J. E., Mayatepek E., Besley G., Valencia A., Hegardt F. G. (2003) J. Biol. Chem. 278, 29016–29023 [DOI] [PubMed] [Google Scholar]
- 8.Fu Z., Runquist J. A., Forouhar F., Hussain M., Hunt J. F., Miziorko H. M., Kim J. J. (2006) J. Biol. Chem. 281, 7526–7532 [DOI] [PubMed] [Google Scholar]
- 9.Roberts J. R., Narasimhan C., Hruz P. W., Mitchell G. A., Miziorko H. M. (1994) J. Biol. Chem. 269, 17841–17846 [PubMed] [Google Scholar]
- 10.Tuinstra R. L., Wang C. Z., Mitchell G. A., Miziorko H. M. (2004) Biochemistry 43, 5287–5295 [DOI] [PubMed] [Google Scholar]
- 11.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 12.Kramer P. R., Miziorko H. M. (1980) J. Biol. Chem. 255, 11023–11028 [PubMed] [Google Scholar]
- 13.CCP4 (1994) Acta Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 14.Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 15.Roussel A., Inisan A. G., Knoop-Mouthuy A., Cambillau C. (1999) TURBO-FRODO, CNRS/Universitite, Marseille, France [Google Scholar]
- 16.Emsley P., Cowtan K. (2004) Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17.Kramer P. R., Miziorko H. M. (1983) Biochemistry 22, 2353–2357 [DOI] [PubMed] [Google Scholar]
- 18.Kim J. J., Wang M., Paschke R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7523–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith C. V., Huang C. C., Miczak A., Russell D. G., Sacchettini J. C., Höner zu Bentrup K. (2003) J. Biol. Chem. 278, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 20.Anstrom D. M., Kallio K., Remington S. J. (2003) Protein Sci. 12, 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remington S., Wiegand G., Huber R. (1982) J. Mol. Biol. 158, 111–152 [DOI] [PubMed] [Google Scholar]
- 22.Mancia F., Keep N. H., Nakagawa A., Leadlay P. F., McSweeney S., Rasmussen B., Bösecke P., Diat O., Evans P. R. (1996) Structure 4, 339–350 [DOI] [PubMed] [Google Scholar]
- 23.Manjasetty B. A., Powlowski J., Vrielink A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6992–6997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koon N., Squire C. J., Baker E. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrasco P., Menao S., López-Viñas E., Santpere G., Clotet J., Sierra A. Y., Gratacós E., Puisac B., Gómez-Puertas P., Hegardt F. G., Pie J., Casals N. (2007) Mol. Genet. Metab. 91, 120–127 [DOI] [PubMed] [Google Scholar]
- 26.Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 27.Narasimhan C., Antholine W. E., Miziorko H. M. (1994) Arch. Biochem. Biophys. 312, 467–473 [DOI] [PubMed] [Google Scholar]
- 28.Tuinstra R. L., Burgner J. W., 2nd, Miziorko H. M. (2002) Arch. Biochem. Biophys. 408, 286–294 [DOI] [PubMed] [Google Scholar]
- 29.Hruz P. W., Miziorko H. M. (1992) Protein Sci. 1, 1144–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts J. R., Narasimhan C., Miziorko H. M. (1995) J. Biol. Chem. 270, 17311–17316 [DOI] [PubMed] [Google Scholar]
- 31.Narasimhan C., Roberts J. R., Miziorko H. M. (1995) Biochemistry 34, 9930–9935 [DOI] [PubMed] [Google Scholar]
- 32.Forouhar F., Hussain M., Farid R., Benach J., Abashidze M., Edstrom W. C., Vorobiev S. M., Xiao R., Acton T. B., Fu Z., Kim J. J., Miziorko H. M., Montelione G. T., Hunt J. F. (2006) J. Biol. Chem. 281, 7533–7545 [DOI] [PubMed] [Google Scholar]
- 33.Tuinstra R. L., Miziorko H. M. (2003) J. Biol. Chem. 278, 37092–37098 [DOI] [PubMed] [Google Scholar]
- 34.Messner B., Eggerer H., Cornforth J. W., Mallaby R. (1975) Eur. J. Biochem. 53, 255–264 [Google Scholar]
- 35.Hanson K. R., Rose I. A. (1975) Acc. Chem. Res. 8, 1–10 [Google Scholar]
- 36.Fullerton S. W., Griffiths J. S., Merkel A. B., Cheriyan M., Wymer N. J., Hutchins M. J., Fierke C. A., Toone E. J., Naismith J. H. (2006) Bioorg. Med. Chem. 14, 3002–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]