Abstract
Although the induction of genome integration-free induced pluripotent stem cells (iPSCs) has been reported, c-Myc was still required for the efficient generation of these cells. Herein, we report mouse strain-dependent differences in the c-Myc dependence for iPSC generation and the successful generation of genome integration-free iPSCs without c-Myc transduction using C57BL/6 mouse embryonic fibroblasts. We performed 49 independent experiments and obtained a total of 24 iPSC clones, including 18 genome integration-free iPSC clones. These iPSCs were indistinguishable from embryonic stem cells and from iPSCs generated using other methods. Furthermore, the generation of three-factor iPSCs free of virus vectors revealed the contribution of c-Myc to the genomic integration of external genes. C57BL/6 is an inbred mouse strain with substantial advantages for use in genetic and molecular biological studies due to its use in the whole mouse genome sequencing project. Thus, the present series of C57BL/6 iPSCs generated by various procedures will serve as a valuable resource for future genetic studies of iPSC generation.
Keywords: Gene Expression, Gene Regulation, Induced Pluripotent Stem (iPS) Cell, Myc, Stem Cell, Genome Integration-free, Pluripotency, Reprogramming
Introduction
The generation of an induced pluripotent stem cell (iPSC)3 was first accomplished via the ectopic expression of the Oct3/4, Sox2, Klf4, and c-Myc genes (hereafter referred to as 4F) in mouse embryonic fibroblasts (MEFs) using a retrovirus-mediated gene transduction system (1, 2). Although such somatic cell reprogramming suggests the possibility of generating patient-specific pluripotent stem cells (3), several issues remain to be addressed, including the requirement of an oncogene for transduced gene expression, and the use of genome-integrating vectors, which can cause insertional mutations. Indeed, a considerable number of mice derived from 4F iPSCs develop various tumors as a result of c-Myc gene reactivation (4). Although it was subsequently demonstrated that Oct3/4, Sox2, and Klf4 (hereafter referred to as 3F) could also generate iPSCs from MEFs, the exclusion of c-Myc yielded less than 10% of the number of iPSCs when compared with 4F infection and took twice as long (4, 5). These results indicate that although c-Myc is not essential for iPSC generation, it does play a critical role in this process. However, the molecular mechanisms underlying the contribution of c-Myc transduction to iPSC generation is not yet clearly understood.
Thus far, several other combinations of defined factors have been reported to generate iPSCs in the absence of c-Myc. However, these experiments either used tissue stem cells, which are typically difficult to prepare, as the recipient cells or incorporated small compounds that have the potential to cause genetic mutations (6–10). Procedures for generating iPSCs using genome integration-free vector systems, including adenovirus, plasmid, and EBNA-1/Ori-P systems, have also been demonstrated. However, the frequency of iPSC generation using these techniques is extremely low (approximately one per million MEFs), which limits the feasibility of generating patient-specific iPSCs from a small starting population of cells (11–13). In addition, all such methods required at least four defined factors, including c-Myc. In our current study, we report for the first time the successful generation of iPSCs from MEFs using a 2A plasmid vector that contains only three factors and excludes the c-Myc gene (12).
EXPERIMENTAL PROCEDURES
Mouse Strains and MEFs
The following mouse strains were used: C57BL/6, C.B-17/lcr-+/+ Jcl (Crea Japan, Shizuoka, Japan); C3H, 129, C57BL/6-Tg(CAG-EGFP) (Japan SLC, Hamamatsu, Japan); and Tg mice homozygous for Nanog-GFP (RBRC02290; RIKEN BioResource Center). MEFs were prepared from embryonic day 13.5 embryos in each case and used within three cell passages.
Generation of iPSCs Using a Retrovirus Gene Transduction System
Retroviral infections were performed as described previously (1). Briefly, Moloney-based retroviral vectors (pMXs) containing murine cDNA inserts for Oct3/4, Sox2, c-Myc, and Klf4 were obtained from Addgene (Cambridge, MA). These plasmids were transfected into PlatE cells using FuGENE 6 transfection reagent (Roche Applied Science, Basel, Switzerland). The medium was collected at 48 h after transfection, filtered, supplemented with 4 μg/ml polybrene and added to MEFs that had been plated the previous day at 1.25 × 105 cells/3.5-cm diameter dish. At 16–18 h after infection, the medium was replaced with Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum non-essential amino acids, β-mercaptoethanol and leukemia inhibitory factor. At day 4 after infection, the 15% fetal bovine serum supplement was replaced with 15% KnockOut Serum Replacement (Invitrogen). To accurately assess the iPSC incidence, we performed our experiments without passaging the cells to avoid dispersal of the iPSC colonies that generate independently. Furthermore, we quantified the incidence using a 96-well plate. Fibroblasts were seeded onto the plate at 250 cells/well and infected with the four defined factors. The number of iPSC-positive wells was counted after 2 weeks of culture.
Generation of iPSCs Using a Plasmid System
The pCX-OKS-2A and pCX-c-Myc plasmids were purchased from Addgene, and transfections were performed as described previously (12). At 16–18 h after transfection, the medium was replaced with Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum and leukemia inhibitory factor. At day 4 after transfection, the fetal bovine serum was replaced with 15% KnockOut Serum Replacement (Invitrogen). At day 9 after transfection, some cells were harvested with trypsin and reseeded onto 10-cm dishes with either feeder cells or gelatin.
RT-PCR
Total RNA was prepared using the RNeasy mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. RT-PCR was performed using the QuantiTect SYBR Green RT-PCR kit (Qiagen) and the StepOnePlus real-time PCR system (Applied Biosystems, Carlsbad, CA). The reaction conditions were 50 °C for 30 min and 95 °C for 15 min for cDNA synthesis followed by an amplification protocol of 50 cycles at 94 °C for 10 s, 56 °C for 30 s, and 72 °C for 40 s. The data were normalized to the expression of the glyceraldehyde-3-phosphate dehydrogenase gene. The primer sequences were as follows: 5′-CAG GTG TTT GAG GGT AGC TC-3′ and 5′-CGG TTC ATC ATG GTA CAG TC-3′ for Nanog; 5′-GTT CTT CGT GGC TAT GAG CA-3′ and 5′-GTG GGT TAC CTT CAT GGT AG-3′ for Fgf4; 5′-ACG AGT GGC AGT TTC TTC TTG GGA-3′ and 5′-TAT GAC TCA CTT CCA GGG GGC ACT-3′ for Rex1; 5′-TCT TTC CAC CAG GCC CCC GGC TC-3′ and 5′-TGC GGG CGG ACA TGG GGA GAT CC-3′ for endogenous Oct3/4; 5′-TAG AGC TAG ACT CCG GGC GAT GA-3′ and 5′-TTG CCT TAA ACA AGA CCA CGA AA-3′ for endogenous Sox2; and 5′-GAG GCC GGT GCT GAG TAT GTC GT-3′ and 5′-GGT GGT GCA GGA TGC ATT GCT G-3′ for the glyceraldehyde-3-phosphate dehydrogenase gene.
Methylation Analysis
Genomic DNA was prepared using a DNeasy blood and tissue kit (Qiagen) and then subjected to bisulfite analysis using the EpiTect bisulfite kit (Qiagen). The resultant PCR products were cloned using the pGEM-T Easy kit (Promega, Madison, WI) and sequenced. A total of 24 or 48 colonies were sequenced for each sample using the primers 5′-GGT TTT TTA GAG GAT GGT TGA GTG-3′ and 5′-TCC AAC CCT ACT AAC CCA TCA CC-3′ for Oct3/4 and 5′-GAT TTT GTA GGT GGG ATT AAT TGT GAA TTT-3′ and 5′-ACC AAA AAA ACC CAC ACT CAT ATC AAT ATA-3′ for Nanog (14).
Generation of Teratomas and Chimera Mice
Teratomas were produced by injecting cells (1 × 106/injection) subcutaneously into the flanks of nude mice (1). Four weeks after injection, tumors were surgically dissected, fixed, and embedded in paraffin. Tumor sections were stained with hematoxylin and eosin prior to histopathological analysis. Chimeric mice were produced via the aggregation method using ICR eight-cell embryos and subsequently mated with BALB/c female mice for germline transmission (15).
RESULTS
iPSC Generation from C57BL/6 Embryonic Fibroblasts, a High Yield in the Absence of c-Myc Transduction
It is well known that many biological phenomena, particularly tumorigenesis, occur with variable frequency in laboratory mouse strains (16–18). We thus hypothesized that the frequency of iPSC generation might also vary among mouse strains and examined the C57BL/6 strain in this regard. C57BL/6 is an inbred mouse strain that has been used in a large number of laboratory investigations, including longitudinal studies. In addition, this strain has the distinct advantage of having been used in many previous genetic studies, including single nucleotide polymorphism analyses and molecular biological studies, as it was selected for the whole mouse genome sequencing project at the Sanger Institute.
We first attempted to generate iPSCs from MEFs using a retrovirus-mediated gene transduction system. We obtained similar numbers of alkaline phosphatase-positive ES-like colonies by 4F infection when compared with Nanog-Tg MEFs, which have been employed previously for a large number of iPS studies. We unexpectedly found, however, that 3F infection could generate either an equivalent or a greater number of iPSCs when compared with 4F. Because such efficient iPSC generation by 3F infection in C57BL/6 mice is quite unusual, we collected these ES-like colonies, established 13 clones from C57BL/6 MEFs (hereafter designated as R-3FiPS clones, i.e. generated by retroviral vector), and then subjected these clones to further examination.
These colonies were found to be indistinguishable from iPS colonies generated by 4F infection (R-4FiPS) and also from ES colonies, which have sharp and defined borders. RT-PCR, bisulfite analysis of Oct3/4 promoter regions, and immunostaining for Oct3/4 also demonstrated that the R-3FiPS clones were very similar to ES and R-4FiPS clones (supplemental Fig. S1, A–C). In addition, the pluripotency of the R-3FiPS cells was verified by their successful generation of subcutaneous teratomas, including three germ layers, and chimeric mice (supplemental Fig. S1, D and E). Germline transmission was also observed (supplemental Fig. S1, F and G). Notably, this is the first report to demonstrate the germline transmission of R-3FiPS cells derived from MEFs. Hence, we conclude that R-3FiPS cells established from C57BL/6 MEFs are identical to the iPSCs that have been reported thus far. This indicates that 3F iPSCs are generated efficiently from C57BL/6 mice.
iPSC Generation via a Plasmid Vector Bearing the Oct3/4, Sox2, and Klf4 Genes
The efficient iPSC generation we obtained by 3F introduction of C57BL/6 mice using a retrovirus-mediated gene transduction system prompted us to examine whether iPSCs could be induced from C57BL/6 MEFs using a genome integration-free vector with 3F, i.e. with the exclusion of the c-Myc gene. We used an expression plasmid construct reported previously by Okita et al. (12) in which the three defined factors were linked to form a single open reading frame with the 2A peptide.
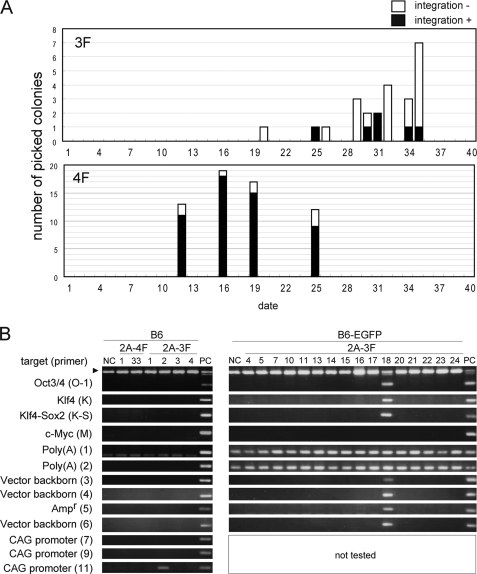
We performed 49 independent experiments, which yielded a total of 24 iPS clones (hereafter referred to as 2A-3FiPS; supplemental Table S1). In contrast, 13 independent 4F gene transfections of the same MEF preparation using 3F in the 2A plasmid vector and c-Myc in another vector (12) generated 134 iPS clones (hereafter referred to as 2A-4FiPS; supplemental Table S1). Notably, the generation of colonies by 3F transfection was slower when compared with 4F transfection, i.e. colonies generated from 4F and 3F transfections appeared at ∼16 and 30 days after transfection, respectively (Fig. 1A).
FIGURE 1.
Genome integration-free iPS clones generated by the transfection of C57BL/6 MEFs with the 3F plasmid. A, appearance of 2A-3FiPS cell colonies. B, PCR analysis using previously designed primers (12). The arrowhead indicates bands corresponding to the endogenous Oct3/4 gene. We used two strains and ES cells from C57BL/6 (B6) mouse and the tails of B6-EGFP Tg mouse for negative controls (NC). 2A-4F-25, in which the genomic integration of 4F genes was confirmed, was used as a positive control (PC) for genome integration. Note that 2A-3FiPS clone 18, which was generated from B6-EGFP Tg, was positive for integration and that no c-Myc signal was detected. PCR was not performed for the 2A-3FiPS clones from B6-EGFP Tg MEFs using the primers for the CAG promoter because the genome of this strain contains the same promoter. The rabbit β-globin poly(A) signal region is also present in the genome of B6-EGFP Tg. Hence, signals corresponding to these sequences were detected in all clones generated from this mouse strain, B6-EGFP 2A-3F clones 4–24. C, the regions used to generate Southern hybridization probes for each gene are indicated below each panel. Genomic DNA (4 μg) was digested with EcoRI and BamHI, and 2A-4F-25 was used as a positive control (PC) for genome integration. 2A-plasmid (V), which was the 3F vector used in the transfections, was loaded at an amount corresponding to two copies/cell. The closed and open arrowheads indicate bands from the endogenous gene allele and pseudo gene allele, respectively. The asterisk indicates the integration of an exogenous Oct3/4 gene into the genome of clone 2A-3F-2 in which we could not detect the integration of any other exogenous gene. D, summary of the results obtained from the transfection experiments.
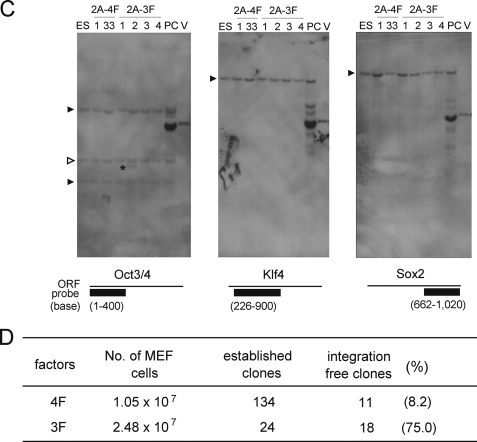
Genome Integration-free iPSCs, Correlation of c-Myc Transduction with the Genomic Integration of Exogenous Genes
PCR analysis revealed that 18 of our 24 2A-3FiPS clones were genome integration-free, with subsequent Southern hybridization analysis of four 2A-3FiPS clones confirming these results (Fig. 1, B–D). In contrast, only 11 of the 134 2A-4FiPS clones were found to be genome integration-free (Fig. 1, B–D). Thus, there was a clear difference in the incidence of genome integration-free iPSCs between the 3F and 4F transfections (18/24 (75%) versus 11/134 (8.2%); Fig. 1D). Also, notably, in the genome integration-positive clone 2A-3FiPS, only a small vector region of ∼1 kb covering the 5′ region of the Oct3/4 expression vector had integrated (Fig. 1, B and C). All of the remaining genome integration-positive 2A-3FiPS clones contained all of the regions investigated (supplemental Fig. S2).
Characterization of the Genome Integration-free 2A-3FiPS Clones
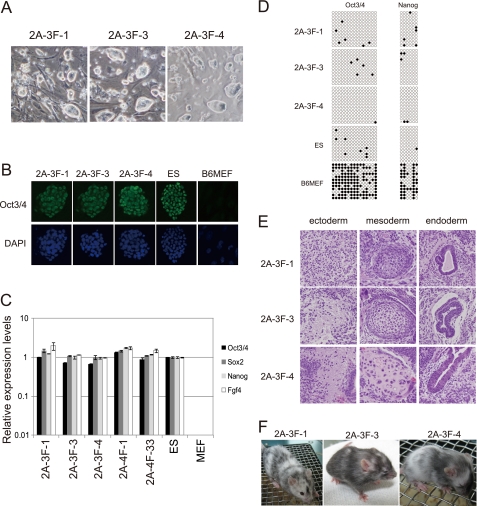
The genome integration-free colonies were found to be morphologically indistinguishable from ES and 4FiPS cells (Fig. 2A). Various investigations, including the immunostaining of Oct3/4, RT-PCR for stem cell marker genes, bisulfite analysis of the promoter regions in Oct3/4 and Nanog genes, and gene expression profiling, indicated that these cells were nearly identical to mouse ES cells or 4FiPS cells generated by a retrovirus or 2A plasmid (Fig. 2, B–D, supplemental Fig. S3 and supplemental Table S2). Furthermore, the developmental potential of the 2A-3FiPS clones, 2A-3F-1, -3, and -4, was demonstrated by their formation of subcutaneous teratomas, including three germ layers in nude mice and generation of chimeric mice (Fig. 2, E and F).
FIGURE 2.
Characterization of the genome integration-free 2A-3FiPS clones. A, morphology of three genome integration-free 2A-3FiPS colonies. B, immunocytochemical detection of Oct3/4 using the h-134 antibody (Santa Cruz Biotechnology). 4′,6-Diamidino-2-phenylindole (DAPI) was used as a nuclear counter stain. C, RT-PCR analysis of ES-marker genes. Error bars indicate standard deviations (n = 3). D, methylation analysis of the Oct3/4 and Nanog gene promoter regions using bisulfite genomic sequencing. Open and closed circles indicate unmethylated and methylated CpGs, respectively. E, teratomas derived from the 3 2A-3FiPS clones. F, a chimeric mouse generated from 2A-3FiPS-1, -3, and -4.
Differences in the c-Myc Dependence of iPSC Generation between Mouse Strains
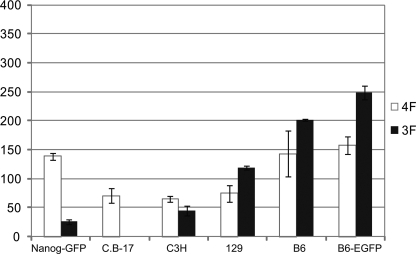
Because we observed unique characteristics of iPSC generation in the C57BL/6 mouse strain, we examined this process in four additional mouse strains, including the inbred strains C3H and 129 (Fig. 3). Nanog-GFP Tg mice, which possess the DBA, 129, and C57BL/6 genetic backgrounds, were used as controls (2). B6-EGFP is a Tg strain derived from the C57BL/6 strain (19). Using MEFs from each strain or C57BL/6, we compared the incidence of iPS formation following infection with 3F or 4F using the retroviral system. To avoid an overestimation of the frequency at which iPSCs arise, our experiment was performed without passaging the cells as this can lead to frequent separation of iPSC colonies and the production of multiple additional colonies. This in turn causes an overestimation of the iPSC incidence (20, 21).
FIGURE 3.
iPSC formation in various mouse strains. The graph indicates the number of ES-like colonies at day 26 after infection. 4F, Oct3/4, Sox2, Klf4, and c-Myc; 3F, Oct3/4, Sox2, and Klf4. B6 denotes the C57BL/6 mouse strain. Error bars indicate the standard deviations (n = 2).
In addition, we randomly picked 48 colonies generated from C57BL/6 with three factors to determine whether these were genuine iPSCs. It has been suggested that clones arising in an iPSC generation assay are not always true iPSC clones. We found, however, that 44 out of the 48 isolated colonies grew, and by RT-PCR analysis, we found that all colonies expressed each of the stem cell marker genes investigated (supplemental Fig. S4). Taken together, we conclude from our observations shown in supplemental Figs. S1A and S4 that almost all of the colonies detected in our assay were iPSCs.
The results showed that in 129 and C57BL/6 mice, the 3F infections induced more iPSCs than 4F. There was little difference in iPS cell formation following 3F or 4F infection in the C3H strain. In contrast 4F produced iPSCs more efficiently than 3F in the Nanog-GFP Tg and C.B-17 strains, consistent with previous reports (1, 5). Also, notably, in the C.B-17 strain, the ratio of 4F:3F was 71:1.
In addition, we quantified the iPSC generation from C57BL/6 MEFs and Nanog-Tg MEFs using another assay, using a 96-well plate. Fibroblasts infected with the four defined factor genes were cultured in each well of a 96-well plate (250 cells/well; ∼4.8 × 104 cells in total). To avoid an overestimation of the frequency at which iPSCs arise, a small number of fibroblasts, ∼25% of the number of fibroblasts that must be required for iPSC generation, were seeded in each well. After 14 days of culture, the iPSC-positive wells were counted, and an efficient iPSC generation from C57BL/6 MEFs by 3F infection was observed (supplemental Fig. S5), consistent with the findings in culture dishes (Fig. 3).
DISCUSSION
All previously reported genome integration-free transfection methods have been found to require c-Myc transduction, and there are no previous reports of iPSC generation using integration-free methods without the incorporation of the c-Myc gene or its products (13, 22–24). Indeed, several genome integration-free methods require additional genes or small, potentially mutagenic compounds in addition to the four established factors. The EBNA-1/ori-P system requires seven factors (13), and an additional system in which the gene products are directly introduced into fibroblasts requires six factors or additional small compounds (23, 24). In our present experiments, we used MEFs from C57BL/6 mice and successfully succeeded in generating genome integration-free iPSCs with a plasmid bearing only three defined transcription factors. Hence, our current demonstration of iPSC generation from fibroblasts using a genome integration-free plasmid vector that does not contain the c-Myc gene represents an important advance in the possible future clinical application of iPSCs.
The success of our integration-free 3F iPSC generation system allowed us to further elucidate the effects of c-Myc on genome integration events. Interestingly, the formation frequency for genome integration-free 2A-3FiPS colonies was found to be ∼10-fold higher than that for 2A-4FiPS colonies. It was also observed that the yield of genome integration-free colonies was similar for 2A-3F (0.73 × 10−6) and 2A-4F (1.05 × 10−6), although the frequency at which 2A-3FiPS colonies arose, 0.97 × 10−6, was less than that of 2A-4FiPS colonies (1.24 × 10−5) (Fig. 1D). These data indicate that c-Myc transduction enhances the genome integration of external genes, which is an important consideration for the design of future improvements to iPSC generation methodologies.
In addition, it is noteworthy that the time to 2A-3FiPS colony appearance following transfection (at around day 30) differed from that of 2A-4FiPS colonies (around day 16). Importantly, this time difference was not dependent upon c-Myc integration (Fig. 1A) as the 2A-4FiPS colonies all appeared at around day 16, including the clones in which c-Myc had integrated.
Further examination of iPS generation from six different mouse strains demonstrated strain-dependent differences in this process and further revealed iPSC generation-sensitive strains. When we examined the quantity of endogenous c-Myc transcripts, little difference was observed between C57BL/6 and Nanog-GFP Tg MEFs, suggesting that strain differences in iPSC generation were not due to the quantity of endogenous c-Myc products (supplemental Fig. S6A). Moreover, Nanog Tg × C57L/6 F1 mice exhibited the Nanog Tg iPSC generation phenotype (supplemental Fig. S6B). Another unexpected observation was that the C57BL/6 phenotype following 3F infection to efficiently generate iPSCs was not observed when a plasmid vector was used (Fig. 3 and supplemental Fig. S7A), although the frequency of iPSC formation was still high.
Although the basis for our current observations seemed initially to be quite complex, we speculated that these findings could be explained more simply by the presence of factors that negatively influence iPSC generation. These factors should be inactivated or suppressed by c-Myc transduction and which exhibit variable expression levels in different mouse strains (supplemental Fig. S7B). In this hypothesis, it is assumed that the concentration of such inhibitors in C57BL/6 MEFs is lower than in Nanog Tg MEFs. It is not surprising that the quantity of promoters, Oct3/4, Sox2, and Klf4, expressed by the retrovirus system is greater than the 2A plasmid system. The molecules controlled by p53 are strong candidates in this regard (20, 25).
The C57BL/6 strain will likely be highly useful for future iPSC studies. Different cell types prepared from these mice will also allow us to conduct more in-depth and fruitful investigations on the mechanisms of iPSC generation. This is because C57BL/6 is an inbred strain and is one of the most popular variants used in previous studies including longitudinal analyses. This mouse strain will also be useful for performing further genetic studies based on the complete genomic DNA sequence. Hence, the series of C57BL/6 iPSC generated by the various means reported herein, including the retrovirus or 2A plasmid methodologies and 4F or 3F transfections, will also be valuable cellular reagents for future precise genetic studies of iPSC generation.
Finally, our current results suggest that the requirement for c-Myc transduction in iPSC generation will also be variable among individual humans because the observed differences among mouse strains may also be reflected in human genetic polymorphisms. If the putative iPSC generation suppressor molecule is identified, it will be possible to survey human populations and identify individuals whose fibroblasts may be more suitable for generating integration-free 3F iPSCs.
Supplementary Material
Acknowledgments
We are grateful to Shinya Yamanaka (University of Kyoto, Kyoto, Japan) for generously providing the Nanog-GFP mice through RIKEN BRC and also the 2A and retrovirus vectors through Addgene. We also thank Toshio Kitamura (University of Tokyo, Tokyo, Japan) for providing PlatE cells and the retrovirus vector pMXs and Akira Nifuji for helpful discussions. We further thank Masahiro Uda, Ryoko Watanabe, Yuko Noda, Misato Sunayama, Akemi Ishibashi, Kinuko Nishikawa, Hana Yoshida, and Matsuko Nakamura for technical assistance.
This work was supported in part by a research grant from Precursory Research for Embryonic Science and Technology (PRESTO), Japan Science and Technology Agency (JST).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1 and S2.
- iPSC
- induced pluripotent stem cells
- iPS
- induced pluripotent stem
- ES
- embryonic stem
- MEF
- mouse embryonic fibroblast
- Tg
- transgenic
- GFP
- green fluorescent protein
- EGFP
- enhanced GFP
- RT-PCR
- reverse transcription-PCR.
REFERENCES
- 1.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 2.Okita K., Ichisaka T., Yamanaka S. (2007) Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 3.Hanna J., Wernig M., Markoulaki S., Sun C. W., Meissner A., Cassady J. P., Beard C., Brambrink T., Wu L. C., Townes T. M., Jaenisch R. (2007) Science 318, 1920–1923 [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 5.Wernig M., Meissner A., Cassady J. P., Jaenisch R. (2008) Cell Stem Cell 2, 10–12 [DOI] [PubMed] [Google Scholar]
- 6.Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huangfu D., Osafune K., Maehr R., Guo W., Eijkelenboom A., Chen S., Muhlestein W., Melton D. A. (2008) Nat. Biotechnol. 26, 1269–1275 [DOI] [PubMed] [Google Scholar]
- 8.Kim J. B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Araúzo-Bravo M. J., Ruau D., Han D. W., Zenke M., Schöler H. R. (2008) Nature 454, 646–650 [DOI] [PubMed] [Google Scholar]
- 9.Kim J. B., Sebastiano V., Wu G., Araúzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B. K., Zaehres H., Schöler H. R. (2009) Cell 136, 411–419 [DOI] [PubMed] [Google Scholar]
- 10.Shi Y., Do J. T., Desponts C., Hahm H. S., Schöler H. R., Ding S. (2008) Cell Stem Cell 2, 525–528 [DOI] [PubMed] [Google Scholar]
- 11.Stadtfeld M., Nagaya M., Utikal J., Weir G., Hochedlinger K. (2008) Science 322, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. (2008) Science 322, 949–953 [DOI] [PubMed] [Google Scholar]
- 13.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I. I., Thomson J. A. (2009) Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura M., Miura K., Iwabuchi K., Ichisaka T., Nakagawa M., Lee J., Kanatsu-Shinohara M., Shinohara T., Yamanaka S. (2006) BMC Dev. Biol. 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens L. C., Little C. C. (1954) Proc. Natl. Acad. Sci. U.S.A. 40, 1080–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens L. C., Varnum D. S. (1974) Dev. Biol. 37, 369–380 [DOI] [PubMed] [Google Scholar]
- 18.Smith G. S., Walford R. L., Mickey M. R. (1973) J. Natl. Cancer Inst. 50, 1195–1213 [DOI] [PubMed] [Google Scholar]
- 19.Okabe M., Ikawa M., Kominami K., Nakanishi T., Nishimune Y. (1997) FEBS Lett. 407, 313–319 [DOI] [PubMed] [Google Scholar]
- 20.Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. (2009) Nature 460, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith Z. D., Nachman I., Regev A., Meissner A. (2010) Nat. Biotechnol. 28, 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woltjen K., Michael I. P., Mohseni P., Desai R., Mileikovsky M., Hämäläinen R., Cowling R., Wang W., Liu P., Gertsenstein M., Kaji K., Sung H. K., Nagy A. (2009) Nature 458, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D., Kim C. H., Moon J. I., Chung Y. G., Chang M. Y., Han B. S., Ko S., Yang E., Cha K. Y., Lanza R., Kim K. S. (2009) Cell Stem Cell 4, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H., Wu S., Joo J. Y., Zhu S., Han D. W., Lin T., Trauger S., Bien G., Yao S., Zhu Y., Siuzdak G., Schöler H. R., Duan L., Ding S. (2009) Cell Stem Cell 4, 381–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marión R. M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M. A. (2009) Nature 460, 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.