Abstract
Most anaplastic large cell lymphomas (ALCL) express oncogenic fusion proteins derived from chromosomal translocations or inversions of the anaplastic lymphoma kinase (ALK) gene. Frequently ALCL carry the t(2;5) translocation, which fuses the ALK gene to the nucleophosmin (NPM1) gene. The transforming activity mediated by NPM-ALK fusion induces different pathways that control proliferation and survival of lymphoma cells. Grb2 is an adaptor protein thought to play an important role in ALK-mediated transformation, but its interaction with NPM-ALK, as well as its function in regulating ALCL signaling pathways and cell growth, has never been elucidated. Here we show that active NPM-ALK, but not a kinase-dead mutant, bound and induced Grb2 phosphorylation in tyrosine 160. An intact SH3 domain at the C terminus of Grb2 was required for Tyr160 phosphorylation. Furthermore, Grb2 did not bind to a single region but rather to different regions of NPM-ALK, mainly Tyr152–156, Tyr567, and a proline-rich region, Pro415–417. Finally, shRNA knockdown experiments showed that Grb2 regulates primarily the NPM-ALK-mediated phosphorylation of SHP2 and plays a key role in ALCL cell growth.
Keywords: Adaptor Proteins, Leukemia, Oncogene, Receptor Tyrosine Kinase, shRNA, Signal Transduction, Anaplastic Large Cell Lymphoma, Grb2, NPM-ALK
Introduction
Anaplastic large cell lymphoma (ALCL)2 is a non-Hodgkin's lymphoma characterized by the t(2;5)(p23;q35) chromosomal translocation that generates an oncogenic fusion protein, NPM-ALK. The t(2,5) translocation represents the more frequent alteration in the ALCLs and is detected in 75–80% of the ALCL patients. The NPM-ALK fusion protein, which includes the NPM oligomerization motif and the ALK catalytic domain, is constitutively activated through autophosphorylation (1, 2).
NPM-ALK-induced signaling pathways control cellular proliferation, inhibit apoptosis, and modify cell-cell adhesion and migration (3–6) and cytoskeleton reorganization and transformation (6, 7). NPM-ALK transforming activity is mediated by different cellular pathways through several downstream adaptors or scaffolding molecules with Src homology 2 (SH2) or phosphotyrosine binding domains that bind to the autophosphorylated NPM-ALK tyrosine residues (2). Many studies have identified several NPM-ALK-interacting molecules such as PLC-γ, IRS-1, Hsp90, Grb2 (growth factor receptor-bound protein 2), ShcC, Jak2, Jak3, PI3K, Stat3, and Stat5 (8). Adaptor proteins Shc (SH2 domain-containing transforming protein), IRS-1 (insulin receptor substrate 1), and Grb2 bind NPM-ALK directly or in complexes, but their specific role in NPM-ALK-mediated lymphomagenesis is still unclear (2, 8). Shc and IRS-1 are tyrosine-phosphorylated and bind NPM-ALK in defined regions (Tyr567 and Tyr152–156 of NPM-ALK, respectively); however, they are dispensable for NPM-ALK-mediated transformation of NIH3T3 cells. In contrast, the interaction between NPM-ALK and Grb2 is still poorly characterized (9, 10).
Grb2 is an ubiquitously expressed adapter protein involved in the signal transduction pathways of protein-tyrosine kinase (11). Grb2 is a 26-kDa protein without catalytic activity and consists of a single SH2 domain flanked by two SH3 domains (12, 13). The SH2 domain of Grb2 specifically binds the tyrosine-phosphorylated peptide sequences of receptor tyrosine kinases such as the EGF receptor (14), PDGF receptor (12), and T-cell receptor (15) and of oncogenic fusions such as BCR-ABL (16) and TPR-MET (17). The SH3 domains of Grb2 bind to proline-rich motifs on the guanine nucleotide-releasing factor Sos (Son of sevenless), leading to the activation of the Ras/MAPK pathway. Two phosphorylation sites on Grb2 have been identified thus far at position Tyr209 in BCR-ABL-expressing cells (16) and Tyr160 by pp60c-src (18).
In this study, we have demonstrated that Grb2 binds to active NPM-ALK and is phosphorylated in human ALCL cells. We identified Tyr160 as a major phosphorylation site of Grb2 by NPM-ALK. Remarkably, we found that Tyr160 of Grb2 also is phosphorylated by other oncogenic fusion tyrosine kinases such as TPR-MET, BCR-ABL, and TEL-JAK2, as well as by wild-type receptor tyrosine kinases such as ALK and MET. Further, we show that NPM-ALK combined mutations in Tyr152–156, Tyr567, and Pro415–417 almost completely abrogated Grb2 binding and Tyr160 phosphorylation. Finally, we show that Grb2 is essential for the activation of SHP2 in ALCL and is required for sustained ALCL cell growth.
MATERIALS AND METHODS
Cell and Culture Conditions
Human embryonic kidney (HEK) cells HEK-293T and 293T-Rex Tet-On (Invitrogen) were cultured in DMEM supplemented with 10% FBS. In HEK-293T tetracycline-inducible systems, tetracycline was added to the medium at 1 μg/ml.
SU-DHL1 and TS (a subclone of SUP-M2) NPM-ALK-positive human ALCL cell lines were obtained from New York University and maintained in RPMI 1640 medium supplemented with 10% FBS. Inducible ALK shRNA SU-DHL-1 and TS (SU-DHL-1 TTA A5 and TS TTA A5, respectively) cells were obtained by co-transduction with pLV-tTRKRAB (TTA) (19) vector and pLVTHM vector containing the H1 promoter ALK-shRNA (A5) cassette (20).
Inducible Grb2 shRNA TS (TS TTA A, B, C, and control sequence) cells were obtained by co-transduction with pLV-tTRKRAB (TTA) vector (19) and pLVTHM vector containing the H1 promoter Grb2 shRNA cassette (20). These cells undergo NPM-ALK or Grb2 silencing when 1 μg/ml doxycycline is added to the medium for at least 72 h. For co-culture experiments, cells were mixed in a 1:1 ratio and cultured under standard conditions for 2 weeks. GFP expression was checked over time by FACS. For proliferation studies, cells were seeded 3 × 103 cells/ml in duplicate in 12-well plates. Viable cells were determined by trypan blue exclusion by using a CountessTM automated cell counter (Invitrogen), and cell growth was analyzed using the ATPlite assay system (PerkinElmer Life Sciences).
Cell Transfection and Reagents
Cells were transfected by Effectene reagent as described by the manufacturer (Qiagen, Valencia, CA). The amount of DNA for each transfection was kept constant at 2 μg by adding an appropriate amount of inert carrier DNA (pBluescript SK plasmid, Stratagene).
ALK inhibitor CEP-14083 (Cephalon) was added at a working concentration of 300 nm. The PP2 Src family kinase inhibitor was purchased from Calbiochem and used at 30 μm.
Cell Lysis, Coimmunoprecipitation, and Immunoblotting Analysis
Whole cell extracts were prepared by resuspending the pelleted cells in lysis buffer containing 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, 0,1% Triton X-100, 1 mm PMSF, 10 mm NaF, 1 mm 3Na3VO4, and protease inhibitors (Roche Applied Science) with incubation at 4 °C for 30 min. Cell lysates were collected by centrifugation at 15,000 × g. Supernatants were analyzed for protein concentration with a Bio-Rad DC protein assay kit (Bio-Rad Laboratories) and stored at −80 °C. Twenty micrograms of protein were run on SDS-PAGE under reducing conditions.
For phosphate digestion, cells were lysed in 20 mm Tris (pH 7.4), 150 mm NaCl, 5 mm EDTA, and 0.1% Triton X-100 in the presence of protease and in the presence/absence of phosphatase inhibitors. 10 μg of whole cell lysates were treated with 800 units of λPPase (New England Biolabs) for 1 h at 30 °C.
For immunoprecipitation experiments, 0.5 mg of whole cell extracts was precleared with 30 μl of protein G-Sepharose beads (Amersham Biosciences) for 1 h at 4 °C and then incubated with 1–5 μg of monoclonal antibody or 5–10 μg of polyclonal antibody at 4 °C for 4 h. Immune complexes were collected on 30 μl of protein G-Sepharose beads, and the beads were washed three times with lysis buffer. Bound proteins were recovered by boiling in Laemmli sample buffer and resolved on SDS-PAGE.
For immunoblotting, proteins were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with the specific antibody. Immune complexes were detected with sheep-anti mouse Ig antibody conjugated to horseradish peroxidase (Amersham Biosciences) and visualized by enhanced chemiluminescence reagent (Amersham Biosciences) according to the manufacturer's instructions. The following antibodies were used: monoclonal anti-ALK (1:2000; Zymed Laboratories Inc., Seattle, WA), monoclonal anti-phospho-Tyr (PY100, 1:1000; Cell Signaling Technology), polyclonal anti-Grb2 (1:2000; Santa Cruz Biotechnology), monoclonal anti-actin (1:2000; Chemicon), polyclonal anti-phospho-Shp2 (Tyr542, 1:1000; Cell Signaling Technology), polyclonal anti-phospho-Stat3 (Tyr705, 1:1000; Cell Signaling Technology), polyclonal anti-Stat3 (1:1000; Cell Signaling Technology), polyclonal anti-phospho-Erk1/2 (1:500; Cell Signaling Technology), polyclonal anti-Erk1/2 (1:1000; Santa Cruz Biotechnology), monoclonal anti-JunB (1:2000; Santa Cruz Biotechnology), polyclonal anti-phospho-Shc (1:1000; Cell Signaling Technology), polyclonal anti-Shc (1:1000; Santa Cruz Biotechnology), polyclonal anti-GFP (1:2000; Invitrogen), polyclonal anti-Src (1:1000; Santa Cruz Biotechnology), polyclonal anti-phospho-Met (1:1000; Cell Signaling Technology), monoclonal anti-human-Met (1:1000; Zymed Laboratories Inc.), monoclonal anti-AKT1 (1:1000; Cell Signaling Technology), monoclonal anti-phospho-AKT1 (Ser473, 1:1000; Cell Signaling Technology), monoclonal anti-JAK2 (1:1000; Cell Signaling Technology), and monoclonal anti-JAK3 (1:1000; Santa Cruz Biotechnology). Secondary anti-mouse or anti-rabbit peroxidase-conjugated antibodies were purchased from Amersham Biosciences.
DNA Constructs and Mutagenesis
Wild-type NPM-ALK or the kinase-dead mutant NPM-ALKK210R was cloned into pcDNA5TO vector (Invitrogen) at HindIII/XhoI sites and stably transfected into 293T-Rex Tet-On cells using Effectene reagent (Qiagen) as described previously (6). Single cell-derived clones were selected for NPM-ALK or NPM-ALKK210R expression levels. Pallino vectors containing NPM-ALK, NPM-ALKK210R, or ATIC-ALK were described previously (21, 22). Pallino CD8-ALK was obtained by cloning the extracellular and intramembranous portion of the human CD8 gene in-frame with the cytoplasmic portion of the ALK gene by recombinant PCR. TPR-MET and the MET expression plasmid were kindly provided by Dr. C. Ponzetto (23). BCR-ABL expression vector was kindly provided by Dr. Saglio. TEL-JAK2 and TEL-JAK3 were kindly provided by Dr. O. Bernard (24). Human ALK full-length (ALK-FL) was cloned in pcDNA3 at HindIII/XhoI sites and hALKF1174L variant was generated by PCR-based mutagenesis (Stratagene, La Jolla, CA).
Grb2WT, the dominant-negative Grb2P49L (mutated in the N-terminal SH3 domain) and Grb2R86K (mutated in the SH2 domain) cloned in the pRK5 vector were kindly provided by Dr. A. Pellicer, New York University. Grb2 tyrosine-mutated forms were generated by PCR-based mutagenesis. All mutations were confirmed by DNA sequencing.
shRNA Sequences, Lentivirus Production, and Cell Infection
Human Grb2-specific shRNA sequences were from the Open Biosystems TRC lentiviral shRNA library. The sense strand of Grb2 shRNA that we used is the following: A, 5′-CAGATATTCCTGCGGGACATA-3′; B, 5′-CGGCTTCATTCCCAAGAACTA-3′; C, 5′-GATCTACATCTGTCTCCAGAA-3′; control, 5′-CCTCTCTGTCAAGTTTGGAAA-3′.
The puromycin resistance gene was removed from the pLKO.1 vector. Lentivirus shGrb2 vectors were constructed by subcloning the H1 promoter-shALK cassette into the EcoRI-ClaI sites of the pLVTHM vector (kindly provided by D. Trono, University of Geneva, Switzerland) (19) as described previously (20).
shRNA Grb2 lentiviruses were obtained by co-transfection of the human HEK-293T cell line (Invitrogen) as described (19). After 72 h the percentages of transduced cells were analyzed for GFP expression by FACS. Inducible Grb2 shRNA interference TS cells were obtained by co-transduction with pLVTHM vector containing the H1 promoter Grb2 shRNA cassette and pLV-tTRKRAB vector. To generate Grb2 shRNA-resistant constructs, wild-type or Y160F Grb2 was mutated in four bases in the sequence corresponding to the shRNA (Grb2WTINT3/4 or Grb2Y160FINT3/4).
Statistical Analysis
The difference in cell growth between the different shGrb2 constructs was evaluated using the Kruskal-Wallis rank test, and the difference between shGrb2 and the relative INT3/4 was evaluated using Student's t test. A p value of ≤0.05 was considered statistically significant.
RESULTS
Grb2 Binds NPM-ALK and Is Tyrosine-phosphorylated
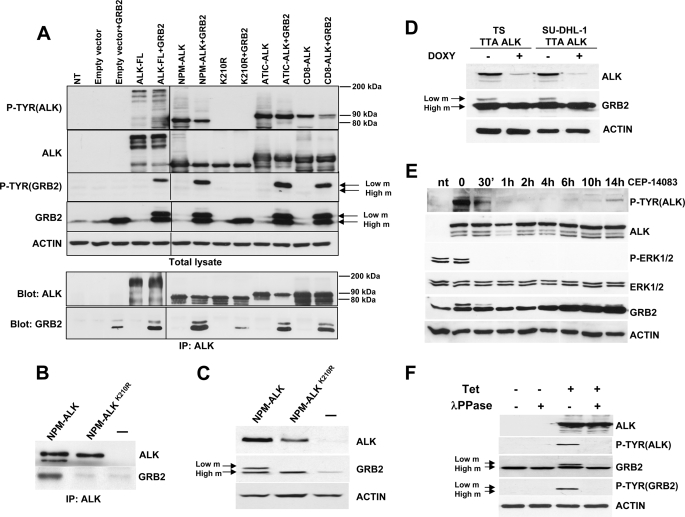
Grb2 has been shown to bind NPM-ALK and ATIC-ALK in previous works (25, 26). First we wanted to test whether Grb2 could bind different ALK fusions regardless of their fusion partner or cellular localization. To this end we took advantage of three different ALK fusions that localize in different cellular compartments. We transfected human HEK-293T with ALK-FL, with NPM-ALK (which is both cytoplasmic and nuclear (27)), with its kinase-dead mutant (K210R), with ATIC-ALK (which is cytoplasmic), and with a chimeric fusion protein between CD8 and ALK (CD8-ALK) that localizes to the cytoplasmic membrane. Immunoprecipitation with an antibody that recognizes the ALK portion of these proteins showed that ALK fusions bound Grb2 regardless of their fusion partner or cellular localization (Fig. 1A).
FIGURE 1.
Phosphorylation of Grb2 in NPM-ALK-expressing cells. A, Grb2 low mobility (m) band and binding do not depend on the type or localization of the ALK fusion proteins. Anti-ALK immunoprecipitations were performed on HEK-293T cells transiently transfected with the indicated ALK full-length and ALK fusion proteins with or without Grb2. Western blots of total lysates and anti-ALK-immunoprecipitated (IP) proteins were blotted with the indicated antibodies. B, HEK-293T cells transiently transfected with NPM-ALK or the kinase-dead mutant were immunoprecipitated with anti-ALK antibodies and blotted with ALK and Grb2 antibodies. C, Grb2 low mobility band is detected only in the presence of the active NPM-ALK but not the kinase-dead NPM-ALKK210R in HEK-293T cells transfected with Grb2 and NPM-ALK or NPM-ALKK210R and blotted with the indicated antibodies. D, Grb2 lower mobility band is detected in ALCL cells. The TS and SU-DHL-1 cell lines were transduced with an inducible anti-ALK shRNA lentivirus (TTA ALK) as described under “Material and Methods.” Cells were cultivated in presence of 1 μg/ml doxycycline (DOXY) for 72 h to knock down NPM-ALK expression, and cell lysates were analyzed by Western blot. E, HEK-293T co-transfected with NPM-ALK and Grb2 were treated with CEP-14083 inhibitor (300 μm) 24 h after transfection for the indicated time points. The effect on NPM-ALK, Grb2, and Erk1/2 phosphorylation was followed over time on total cell extracts by Western blot (nt, not transfected). F, NPM-ALK expression was induced in HEK-293T-Rex by the addiction of 1 μg/ml tetracycline (Tet) to the culture for 24 h. Cell lysates were incubated with or without λPPase. Western blots were blotted with the indicated antibodies.
Unexpectedly, Grb2 was detected on a gel as two bands with different levels of mobility: one band with higher mobility migrating at the expected size of 26 kDa and another with lower mobility. To assess whether this lower mobility band was related to NPM-ALK tyrosine kinase activity, we transfected human HEK-293T cells with either NPM-ALK or its kinase-dead mutant, NPM-ALKK210R. Direct Western blot and Grb2 immunoprecipitation showed that both the binding and the lower mobility species appeared only in the presence of active NPM-ALK (Fig. 1, B and C, and supplemental Fig. 1). To verify that those bands with different mobility were not an artifact of NPM-ALK ectopic expression in HEK-293T cells, we analyzed Grb2 bands as detected in NPM-ALK-positive human lymphoma cells. To this end, we took advantage of a cellular system developed previously by our group based on a doxycycline-dependent inducible anti-ALK shRNA that abrogates NPM-ALK expression in ALCL (20). Under these conditions, the Grb2 band with lower mobility disappeared after NPM-ALK knockdown (Fig. 1D) in TS and SU-DHL-1 cells. Similarly, inhibition of NPM-ALK activity via a specific small molecule, CEP-14083 (28), correlated with the synchronous disappearance of the lower mobility band of Grb2 (Fig. 1E).
Because ALK is a tyrosine kinase, we reasoned that the lower mobility band would correspond to a phosphorylated form of Grb2. Thus, we treated cell extracts from human HEK-293T cells with λPPase, which dephosphorylates phosphoresidue on proteins. After λPPase treatment, the lower mobility band of Grb2 disappeared completely. Furthermore, an antibody specific for phosphotyrosines highlighted a band corresponding to the lower mobility band of Grb2 only in lysates not treated with λPPase (Fig. 1F). Therefore, we concluded that Grb2 is tyrosine-phosphorylated in the presence of active NPM-ALK.
Grb2 Tyr160 Is Phosphorylated in the Presence of NPM-ALK
Seven tyrosine residues are present in the human Grb2 protein. Three of them are located in the N-terminal SH3 domain (Tyr7, Tyr 37, and Tyr 52), two in the SH2 domain (Tyr 118 and Tyr 134), and two in the C-terminal SH3 domain (Tyr 160 and Tyr 209) (Fig. 2A) as described in UniProt, a freely accessible resource of protein sequence and functional information.
FIGURE 2.
Grb2 Tyr160 is phosphorylated in the presence of active NPM-ALK. A, schematic representation of Grb2 structure and tyrosine residues localization. B, HEK-293T cells were co-transfected with Grb2 WT or Grb2 tyrosine mutants together with the active form of NPM-ALK or the NPM-ALK kinase-dead mutant and blotted with the indicated antibodies. m, mobility. C, HEK-293T cells were co-transfected with Grb2WT, Grb2Y160F, or Grb2Y209F mutants and NPM-ALK. Total cells extracts were analyzed by Western blot for Grb2, ALK, and p-Tyr. D, anti-Grb2 immunoprecipitation was performed on HEK-293T cells transiently co-transfected with NPM-ALK or NPM-ALKK210R and Grb2WT or Grb2Y160F. Western blots of total lysates and anti-Grb2-immunoprecipitated (IP) proteins were blotted with the indicated antibodies.
To identify NPM-ALK-induced Grb2 phosphorylation site(s), the Grb2 coding region was mutated by means of site-directed mutagenesis on each tyrosine (Tyr to Phe) and then co-expressed with active NPM-ALK in HEK-293T cells. Previous studies had demonstrated the importance of Tyr209 in Grb2 phosphorylation after binding with the protein kinase BCR-ABL (16). We first analyzed the Y209F mutant, which showed no alteration in the Grb2 migration pattern as compared with the wild-type form in cells expressing active NPM-ALK (Fig. 2B). Next, we mutated all of the other Grb2 tyrosine residues. As shown in Fig. 2B, all of these Grb2 mutants exhibited a similar migration pattern of the two bands except for the Grb2Y160F mutant. Indeed, Grb2 phosphorylation was completely abrogated in the Grb2Y160F mutant as revealed by the disappearance of the phosphorylated low mobility band by a phosphotyrosine antibody (Fig. 2C). This observation was confirmed by immunoprecipitation of Grb2 protein as shown in Fig. 2D.
Tyrosine Phosphorylation of Grb2 at Tyr160 Is a Common Feature of Other Oncogenic and Non-oncogenic Kinases
Next we asked whether the phosphorylation of Grb2 in Tyr160 is limited to NPM-ALK or is shared by other oncogenic tyrosine kinases known to interact with Grb2, such as BCR-ABL (29), TPR-MET (30), and TEL-JAK2 (31). HEK-293T cells were co-transfected with Grb2 wild type or the Y160F mutant and the oncogenic kinases NPM-ALK, BCR-ABL, TPR-MET, TEL-JAK2, and TEL-JAK3, and total cells extracts were analyzed by Western blot. BCR-ABL, TPR-MET, and TEL-JAK2 induced a Grb2 phosphorylation similar to that of NPM-ALK, indicating that other relevant oncogenic kinase fusions have the ability to induce phosphorylation of Grb2 in Tyr160 (Fig. 3). Notably, we showed that a remarkable level of phosphorylation is induced by BCR-ABL on Grb2 Tyr 160, which was not recognized in a previous work (16). Interestingly, TEL-JAK3 oncogenic fusion kinase, which has not been reported to bind Grb2, was unable to phosphorylate Grb2 (Fig. 3C). These results indicate that Grb2 phosphorylation is a common event in many, but not all, oncogenic fusion kinases.
FIGURE 3.
Phosphorylation site at Tyr160 is shared by several oncogenic kinase. A, HEK-293T cells were co-transfected with Grb2WT or the indicated mutants and BCR-ABL. Total cells extracts were analyzed by Western blot for BCR-ABL, Grb2, and p-Tyr. m, mobility. B, HEK-293T cells were co-transfected with Grb2WT or Grb2Y160F and NPM-ALK or TPR-MET. Total cells extracts were analyzed by Western blot for Grb2, ALK, and MET. C, HEK-293T cells were co-transfected with Grb2WT or Grb2Y160F and NPM-ALK, NPM-ALKK210R, TEL-JAK2, or TEL-JAK3. Total cells extracts were analyzed by Western blot for Grb2, Jak2, Jak3, ALK, and p-Tyr.
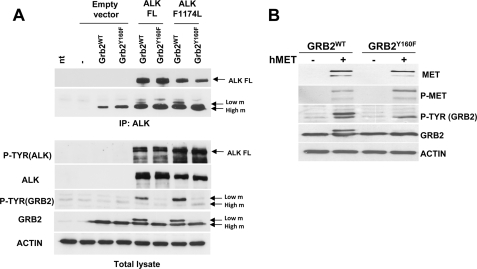
Oncogenic fusion kinases are characterized by strong and continuous kinase activity that alters the intensity and quality of the downstream signaling as compared with their non-oncogenic counterparts (32). Therefore, we asked whether Grb2 phosphorylation is detectable also when non-fusion receptor tyrosine kinases are activated. HEK-293T cells were transfected with wild-type ALK or MET receptors. Remarkably, Grb2 phosphorylation on Tyr160 was observed in the presence of both ALK and MET receptors (Fig. 4). Furthermore, an activating point mutation of the full-length ALK receptor recently described in neuroblastomas (33) was able to phosphorylate Grb2 as well. Altogether, these data indicate that phosphorylation of Grb2 is a common event shared by both wild-type and oncogenic tyrosine kinases.
FIGURE 4.
Non-oncogenic tyrosine kinases phosphorylate Tyr160 on Grb2. A, HEK-293T cells were transiently co-transfected with the human WT ALK receptor tyrosine kinase or its activating mutant (ALKF1174L) combined with Grb2WT or Grb2Y160F. Anti-ALK-immunoprecipitated proteins (IP) and total cell lysates were analyzed by Western blot with p-Tyr, ALK, and Grb2 antibodies. FL, full length; nt, not transfected; m, mobility. B, HEK-293T cells were transiently co-transfected with human MET (hMET) receptor tyrosine kinase and Grb2WT or Grb2Y160F. Cell lysates were analyzed by Western blot. p-Tyr and Grb2 antibodies show that the Y160F mutation abolishes Grb2 phosphorylation in the lower mobility band.
The C-terminal SH3 Domain Is Required for Grb2 Tyr160 Phosphorylation
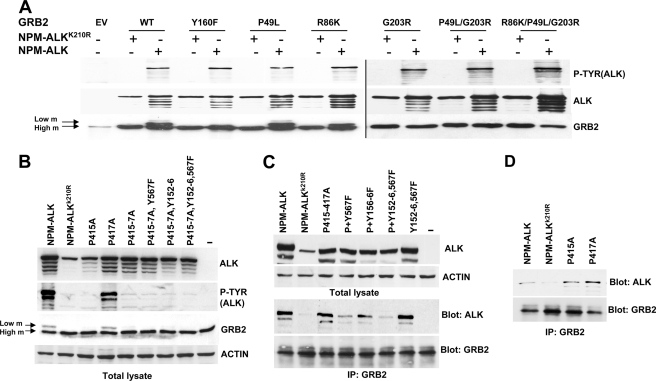
Grb2 acts as an adaptor through its single SH2 and flanked SH3 domains. The backbone dynamics of Grb2 from NMR relaxation data show Grb2 as a flexible protein in which the SH3-SH2 linker regions are flexible (34). The Grb2 crystal structure shows a compact dimer structure with intramolecular contact between two SH3 domains (35). Usually, SH2 domains bind pYXNX phosphotyrosine sequences and SH3 domain proline-rich sequences. To test whether functional Grb2 SH2 and SH3 domains are required for Grb2 interaction with NPM-ALK or for its phosphorylation, we generated different Grb2 mutants by site-directed mutagenesis, inserting a point mutation in each SH domain. As published previously, the introduction in Grb2 of a single point mutation at position R86K leads to a SH2 domain loss-of-function mutant, whereas P49L and G203R mutants disrupt the N-terminal and C-terminal SH3 domains, respectively (36). Mutations in the SH2 domain (R86K) or the N-terminal SH3 domain (P49L) did not affect Grb2 phosphorylation. Conversely, a single point mutation disrupting the C-terminal SH3 domain (G203R) completely abolished Grb2 phosphorylation (Fig. 5A).
FIGURE 5.
Analysis of the Grb2 domains required for Grb2 phosphorylation. A, G203R mutation abolishes Grb2 phosphorylation. HEK-293T cells co-transfected with NPM-ALK or NPM-ALKK210R in combination with Grb2WT or different Grb2 mutants. Cell lysates were analyzed by Western blot for anti-phosphotyrosine, anti-ALK, and anti-Grb2 antibodies. EV, empty vector; m, mobility. B, HEK-293T cells transiently transfected with the indicated combinations of NPM-ALK, NPM-ALKK210R, and NPM-ALK proline mutants and NPM-ALK tyrosine mutants for binding sites for SHC (Y567F) and/or IRS-1 (Y152F,Y156F (Y152–6)). Protein extracts were analyzed by Western blot with Grb2, p-Tyr, and ALK antibodies. C, immunoprecipitation (IP) for Grb2 of HEK-293T cells transfected with combinations of the constructs indicated in A and B. Total lysate (upper lanes) and the anti-Grb2-immunoprecipitated proteins were analyzed by Western blot with anti-ALK and anti-Grb2 antibodies. Data are representative of one of three independent experiments. D, anti-Grb2 immunoprecipitation was performed on HEK-293T cells transiently transfected with NPM-ALK, NPM-ALKK210R, and NPM-ALK proline single and double mutants. Western blots of Anti-Grb2-immunoprecipitated proteins were blotted with the indicated antibodies.
Proline-rich Region and Residues Tyr152–156 and Tyr567 of NPM-ALK Are Involved in Grb2 Binding
NPM-ALK could potentially bind Grb2 directly or through other proteins such as SHP2, Shc, and IRS-1 (7), (26). Therefore, we investigated which region of NPM-ALK was involved in Grb2 binding. Our group recently performed an extensive phosphomapping of the NPM-ALK protein in human lymphoma cell lines, showing that at least 11 NPM-ALK tyrosine residues are phosphorylated in lymphoma cells (37). Because three of these residues (Tyr338, Tyr342, and Tyr343) are located in the NPM-ALK kinase domain and abolish its activity when mutated,3 we decided to mutate all of the other known ALK tyrosine phosphorylation sites sequentially. Lysates from HEK-293T cells transiently transfected with these mutants were analyzed by Western blot for Grb2 phosphorylation. As expected, the sequential addition of mutations to NPM-ALK progressively reduced the ALK phosphorylation status as detected by the phosphotyrosine antibody. Remarkably, the decrease in NPM-ALK phosphorylation correlated with a diminished Grb2 phosphorylation (supplemental Fig. 2). These data suggest that, in contrast to other tyrosine kinases such MET (30, 38), no single tyrosine residue on NPM-ALK is responsible for Grb2 phosphorylation. Rather, Grb2 is recruited and thereby phosphorylated through multiple tyrosine residues directly to NPM-ALK or via other interacting proteins.
Recently, a proline-rich region in NPM-ALK has been shown to regulate the interaction with the SH3 domain of PI3K (39). Therefore, we asked whether the proline-rich region of NPM-ALK was involved in Grb2 binding and/or phosphorylation. To this end, we mutated the proline-rich sequence of NPM-ALK (P415A, P417A, and P415A,P417A). Western blots showed that the P415A, but not the P417A, completely abolished Grb2 phosphorylation by NPM-ALK (Fig. 5B), but Grb2 binding to NPM-ALK was not abolished by such mutations (Fig. 5, C and D). Finally, because IRS-1 and Shc bind NPM-ALK and in turn can recruit Grb2 to NPM-ALK, we sequentially added to the proline-rich mutations other mutations in tyrosine residues Tyr152–156 and Tyr567 known to abolish NPM-ALK binding to IRS-1 and Shc, respectively. Although Tyr152–156 and Tyr567 mutations had little effect (Fig. 5C, lane 7), the combination of such mutations with mutations in the proline-rich domain almost completely abolished Grb2 binding to NPM-ALK (Fig. 5C, lane 6). Overall these data indicated that the binding and phosphorylation of Grb2 by NPM-ALK have different requirements; Grb2 phosphorylation depends mainly on the integrity of NPM-ALK proline-rich region, whereas it's binding to NPM-ALK relies on the same proline-rich region but also on tyrosine residues Tyr152–156 and Tyr567.
Finally, because Src has been described as phosphorylating Grb2 (18), we treated HEK-293T cells with a Src inhibitor after NPM-ALK transfection. Whereas Src phosphorylation was almost completely abrogated after 1 h of inhibition, Grb2 phosphorylation remained unaffected (supplemental Fig. 3), thus showing that NPM-ALK-mediated Grb2 phosphorylation is independent of Src.
Grb2 Regulates ALCL Lymphoma Cell Signaling and Proliferation
To understand the role of Grb2 in ALCL cell signaling and growth, we knocked down Grb2 in ALCL cells through a specific doxycycline-inducible Grb2 shRNA silencing. Three different Grb2 shRNA sequences induced an efficient knockdown with more than 80% reduction of Grb2 protein levels, whereas a fourth sequence did not change Grb2 levels and was used as a control (Fig. 6A). Knockdown of Grb2 protein strongly affected the phosphorylation levels of SHP2 and slightly affected that of Shc, whereas Erk1/2 phosphorylation was not affected (Fig. 6, A and B). To exclude the possibility of off-target effects, we generated a Grb2 cDNA mutated in the shRNA recognition sites and therefore resistant to shRNA knockdown (Grb2INT3/4). When Grb2INT3/4 was expressed in ALCL cells, normal levels of SHP2 and Shc phosphorylation were restored (Fig. 6C).
FIGURE 6.
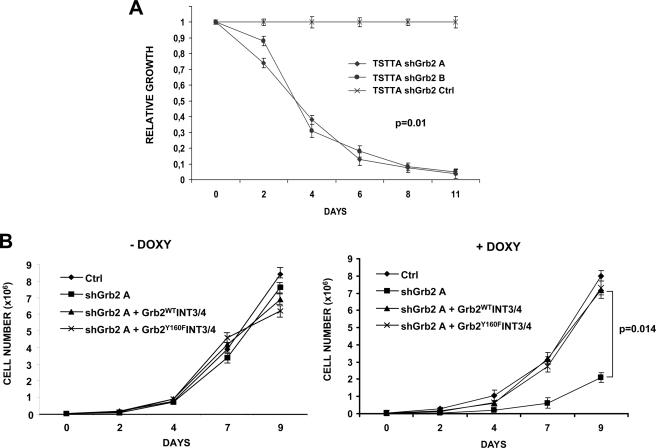
Grb2 knockdown affects downstream effectors activation. A, TS cells were transduced with lentiviral particles for different sequences (A, B, C, and control (Ctrl)) of doxycycline-inducible (DOXY) Grb2 shRNA (shGrb2). After 96 h of induction, Grb2 knockdown and its effects on the downstream effectors were tested by Western blot with the indicated antibodies. B, densitometry analysis of blots from four independent experiments as described in A was performed to quantify the variation of expression of Grb2 and of downstream proteins phosphorylation. Histograms in the left panel indicate the relative expression level of Grb2 as percentage of the expression levels detected in non-transduced cells. The difference in relative expression between shGrb2A or -B and control was evaluated using Student's t test. Grb2 expression was significantly reduced by both shRNA (*, p < 0.01). The right panel shows the relative phosphorylation levels of the indicated proteins as percentages of the phosphorylation levels detected in non-transduced cells. The difference in relative phosphorylation between both shGrb2 and control was evaluated using Student's t test. SHC and SHP2 phosphorylation was significantly reduced by shRNA (*, p < 0.005; **, p < 0.001). In contrast, ERK phosphorylation was not significantly affected by shRNA. C, TS TTA (doxycycline-inducible) cells were infected with Grb2 shRNA (A or B) alone or together with shRNA-resistant Grb2 constructs (Grb2WTINT3/4 or Grb2Y160FINT3/4). Cells were cultured for 96 h with (+) or without (−) 1 μg/ml doxycycline (DOXY). Cell lysates were analyzed by Western blot with the indicate antibodies.
Finally, we asked whether down-regulation of Grb2 could affect lymphoma cell proliferation. ALCL cells transduced with Grb2 shRNA showed a significant disadvantage in co-culture experiments as compared with control infected cells (p = 0.01) (Fig. 7A). Again, the proliferation defect was not due to off-target effects of the shRNA sequences, because it was completely restored by the expression of the shRNA-resistant Grb2INT3/4 cDNA. Tyr160 mutated constructs were equally efficient in restoring ALCL proliferation (Fig. 7B and supplemental Fig. 4). Overall, these data showed that Grb2 is fundamental for the phosphorylation of important molecules in NPM-ALK signaling and for ALCL cell proliferation.
FIGURE 7.
Grb2 knockdown affects lymphoma cells proliferation. A, GFP-negative TS TTA cells were mixed in a 1:1 ratio with GFP-positive TS TTA cells infected with Grb2 shRNA (A or B) or control shRNA. The ratio between the GFP-negative and -positive cells was checked over time by flow cytometry and is indicated as relative growth. Grb2 shRNA showed a significant disadvantage in co-culture experiments as compared with control (Ctrl) infected cells (p = 0.01). Results are from three independent experiments. Statistical analysis was performed by Kruskal-Wallis rank test (hypothesis: three samples from the same population chi-squared = 8.7). The Wilcoxon rank sum test on sample shGrb2A and -B with Bonferroni correction was not significant. B, TS TTA cells transduced with the indicated constructs were plated to a concentration of 30,000 cells/ml and cultured in presence of 1 μg/ml doxycycline (DOXY). Cells were counted at the indicated time points. The effects of the shRNA sequences were completely restored by the expression of shRNA-resistant Grb2INT3/4 cDNA (p = 0.014). Differences in cell growth without doxycycline treatment are not significant. Results are from four independent experiments. The difference in cell numbers between shGrb2A and shGrb2A+INT3/4 was evaluated using Student's t test.
DISCUSSION
In the present work, we have characterized the interaction of the adaptor protein Grb2 with NPM-ALK, the fusion protein involved in the pathogenesis of the anaplastic large cell lymphoma. In particular we have focused on three aspects: the binding of Grb2 to NPM-ALK in cells, the phosphorylation of Grb2 by NPM-ALK, and the role of Grb2 in regulating the signaling pathways and proliferation of ALCL cells.
Grb2 has been shown to bind to some receptor tyrosine kinases or oncogenic tyrosine kinases in specific amino acid residues. For instance, Met receptor binds Grb2 in a well described region of its C-terminal tail characterized by the sequence 1349YVHVNATY1356VNV (30). Similarly, oncogenic BCR-ABL binds Grb2 within the BCR first exon through the residue Tyr177. Mutation of Tyr177 to phenylalanine (Y177F) abolishes Grb-2 binding and abrogates BCR-ABL-induced Ras activation (40). In the case of NPM-ALK, the binding of Grb2 could not be mapped to a single phosphotyrosine residue. In contrast to what has been described for BCR-ABL, where Grb2 binds to BCR, which is the fusion partner of the oncogenic kinase ABL, NPM was dispensable for Grb2 binding, because both a different fusion construct (ATIC-ALK) or a chimeric transmembrane construct (CD8-ALK) was able to equally bind Grb2. Furthermore, extensive mutational analysis showed that not a single phosphotyrosine residue on NPM-ALK was responsible for Grb2 binding. Therefore, we concluded that in cells Grb2 binding to NPM-ALK occurs in multiple regions, directly or through the interaction with other adaptor proteins. This is not totally surprising, because in contrast to MET or BCR-ABL, multiple binding site of Grb2 have been described for its interaction with other tyrosine kinases such as the EGF receptor (40). The Tyr152–156 and Tyr567 NPM-ALK mutants were still able to bind Grb2, as demonstrated previously (9), and to phosphorylate it (data not shown). In contrast, we showed that mutations in a proline-rich region of NPM-ALK previously shown to be important for the binding of the p85 subunit of PI3K (39) were effective in abrogating the phosphorylation of Grb2 in Tyr160 and only slightly diminished its binding to NPM-ALK. However, when we combined the mutations of the proline-rich region with phosphotyrosine residues known to impair the binding of NPM-ALK to IRS-1 and Shc (Tyr152–156 and Tyr567, respectively), we could almost completely abolish Grb2 binding to NPM-ALK.
These data led us to conclude that NPM-ALK binds Grb2 through three regions, a proline-rich region, Pro415–417, and the two phosphotyrosine residues Tyr152–162 and Tyr567, possibly via IRS-1 and Shc. Accordingly, we showed that the C terminal SH3 domain of Grb2 was essential for Grb2 Tyr160 phosphorylation but not for its binding to NPM-ALK. Therefore, we propose a model in which Grb2 binds to NPM-ALK through multiple regions of NPM-ALK but in which Grb2 Tyr160 phosphorylation requires an interaction between the C terminal SH3 domain of Grb2 and the Pro415–417 proline-rich region of NPM-ALK.
Regarding Grb2 phosphorylation, initial work showed that Grb2 was not tyrosine-phosphorylated in response to growth factor stimulation, at least by EGF (12, 41). More recently, however, tyrosine phosphorylation of Grb2 in BCR-ABL-transformed cells on residues Tyr7, Tyr37, Tyr52, and Tyr209 in the SH3 domains has been reported and shown to negatively regulate the Ras/MAPK pathway. This effect has been explained by the fact that Grb2 tyrosine phosphorylation impairs SH3-dependent binding to Sos, thereby blocking Ras activation (16). Furthermore, a recent report showed a role for tyrosine phosphorylation of Grb2 in prolactin receptor/Jak2 signaling (42). In this contest, tyrosine phosphorylation of Grb2 was shown to mediate the inhibitory signals of prolactin on the Ras/MAPK pathway, thereby allowing for the prolactin antagonism of EGF-induced cell proliferation in mammary epithelial cells (43).
In our work we show that, in contrast to BCR-ABL and prolactin, NPM-ALK phosphorylates Grb2 mainly in Tyr160. In fact, the strongest phosphorylated band of Grb2 disappears with the Tyr160 mutant (Fig. 2). In the blots with an antibody that recognizes phosphotyrosine residues, a weakly phosphorylated band with high mobility that runs in the same position of unphosphorylated Grb2 is also detectable despite the Tyr160 mutation in Grb2 (Fig. 2C, lane 3). Therefore, it is possible that other tyrosine residues could be phosphorylated by NPM-ALK although to a much lower level that Tyr160. Interestingly, Tyr160 phosphorylation induces a slower migration of Grb2 in acrylamide gel, whereas Grb2 phosphorylation in other sites does not seem to modify the migration pattern of Grb2. In previous work on BCR-ABL-transformed cells, the high mobility Grb2 band was found to be phosphorylated in Tyr7, Tyr37, Tyr52, and Tyr209 residues, but Grb2 Tyr160 phosphorylation was not detected (16). Our data show that BCR-ABL also phosphorylates Grb2 in Tyr160. In contrast to NPM-ALK and MET, BCR-ABL-mediated phosphorylation of Grb2 seems to be equally distributed between position Tyr160 and other sites. In fact, by quantifying the intensity of the phosphorylated bands, we found that the ratio of Tyr160 (low mobility) to Tyr7, Tyr37, Tyr52, and Tyr209 (high mobility) was 19.2 for NPM-ALK and 8.9 for MET as compared with 0.87 in BCR-ABL-transfected cells.
Previous reports suggested an inhibitory role of Grb2 Tyr7, Tyr37, Tyr52, and Tyr209 phosphorylation in receptor tyrosine kinase signaling (16) (43). Instead, in our system Grb2 Tyr160 mutation was not show to have a role in ALCL proliferation. Further studies are needed to address whether other biological effects, such as cell migration and gene expression, are regulated by Grb2 Tyr160 phosphorylation in ALCL cells and to study the effects of the combination of Tyr160 with other phosphorylation sites.
Finally, we have shown that the lack of Grb2 affects NPM-ALK signaling, in particular Shc and SHP2 activation, without affecting Erk1/2 activation. Even if Grb2 is a key adaptor molecule involved in the activation of the Ras/MAPK cascade, in its absence NPM-ALK induces other pathways such as PI3K that in turn could activate the Ras/MAPK pathway (44). Despite it limited biochemical effect on the Ras/MAPK pathways, the down-modulation of Grb2 in ALCL cells strongly impaired cell proliferation, thus suggesting that Grb2 is fundamental for the full activation of a signaling cascade that involves Shc and SHP2 and assuring an optimal proliferation of lymphoma cells. Thus, Grb2 could represent a potential target for controlling cell proliferation in NPM-ALK-mediated lymphomas.
Supplementary Material
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Compagnia di San Paolo, Torino (Progetto Oncologia), FP7 ERC-2009-StG (Proposal 242965-“LUNELY”) (to R. C.) and the Regione Piemonte (Ricerca Sanitaria Finalizzata and Ricerca Scientifica).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
R. Chiarle, unpublished observations.
- ALCL
- anaplastic large cell lymphoma
- SH
- Src homology
- NPM
- nucleophosmin
- ALK
- anaplastic lymphoma kinase
- λPPase
- λ protein phosphatase.
REFERENCES
- 1.Morris S. W., Kirstein M. N., Valentine M. B., Dittmer K., Shapiro D. N., Look A. T., Saltman D. L. (1995) Science 267, 316–317 [DOI] [PubMed] [Google Scholar]
- 2.Chiarle R., Voena C., Ambrogio C., Piva R., Inghirami G. (2008) Nat. Rev. Cancer 8, 11–23 [DOI] [PubMed] [Google Scholar]
- 3.Wellmann A., Doseeva V., Butscher W., Raffeld M., Fukushima P., Stetler-Stevenson M., Gardner K. (1997) FASEB J. 11, 965–972 [DOI] [PubMed] [Google Scholar]
- 4.Kuefer M. U., Look A. T., Pulford K., Behm F. G., Pattengale P. K., Mason D. Y., Morris S. W. (1997) Blood 90, 2901–2910 [PubMed] [Google Scholar]
- 5.Chiarle R., Gong J. Z., Guasparri I., Pesci A., Cai J., Liu J., Simmons W. J., Dhall G., Howes J., Piva R., Inghirami G. (2003) Blood 101, 1919–1927 [DOI] [PubMed] [Google Scholar]
- 6.Ambrogio C., Voena C., Manazza A. D., Piva R., Riera L., Barberis L., Costa C., Tarone G., Defilippi P., Hirsch E., Boeri Erba E., Mohammed S., Jensen O. N., Palestro G., Inghirami G., Chiarle R. (2005) Blood 106, 3907–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voena C., Conte C., Ambrogio C., Boeri Erba E., Boccalatte F., Mohammed S., Jensen O. N., Palestro G., Inghirami G., Chiarle R. (2007) Cancer Res. 67, 4278–4286 [DOI] [PubMed] [Google Scholar]
- 8.Amin H. M., Lai R. (2007) Blood 110, 2259–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimoto J., Shiota M., Iwahara T., Seki N., Satoh H., Mori S., Yamamoto T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4181–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulford K., Morris S. W., Turturro F. (2004) J. Cell. Physiol. 199, 330–358 [DOI] [PubMed] [Google Scholar]
- 11.Dharmawardana P. G., Peruzzi B., Giubellino A., Burke T. R., Jr., Bottaro D. P. (2006) Anticancer Drugs 17, 13–20 [DOI] [PubMed] [Google Scholar]
- 12.Lowenstein E. J., Daly R. J., Batzer A. G., Li W., Margolis B., Lammers R., Ullrich A., Skolnik E. Y., Bar-Sagi D., Schlessinger J. (1992) Cell 70, 431–442 [DOI] [PubMed] [Google Scholar]
- 13.Songyang Z., Shoelson S. E., McGlade J., Olivier P., Pawson T., Bustelo X. R., Barbacid M., Sabe H., Hanafusa H., Yi T., Baltimore D., Ratnofsky R., Feldman R. A., Contlees L. C. (1994) Mol. Cell. Biol. 14, 2777–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garbay C., Liu W. Q., Vidal M., Roques B. P. (2000) Biochem. Pharmacol. 60, 1165–1169 [DOI] [PubMed] [Google Scholar]
- 15.Vihinen M., Smith C. I. (1996) Crit. Rev. Immunol. 16, 251–275 [DOI] [PubMed] [Google Scholar]
- 16.Li S., Couvillon A. D., Brasher B. B., Van Etten R. A. (2001) EMBO J. 20, 6793–6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponzetto C., Bardelli A., Zhen Z., Maina F., dalla Zonca P., Giordano S., Graziani A., Panayotou G., Comoglio P. M. (1994) Cell 77, 261–271 [DOI] [PubMed] [Google Scholar]
- 18.Jones D. A., Benjamin C. W. (1997) Arch. Biochem. Biophys. 337, 143–148 [DOI] [PubMed] [Google Scholar]
- 19.Wiznerowicz M., Trono D. (2003) J. Virol. 77, 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piva R., Chiarle R., Manazza A. D., Taulli R., Simmons W., Ambrogio C., D'Escamard V., Pellegrino E., Ponzetto C., Palestro G., Inghirami G. (2006) Blood 107, 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raz R., Lee C. K., Cannizzaro L. A., d'Eustachio P., Levy D. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2846–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piva R., Liu J., Chiarle R., Podda A., Pagano M., Inghirami G. (2002) Mol. Cell. Biol. 22, 8375–8387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taulli R., Accornero P., Follenzi A., Mangano T., Morotti A., Scuoppo C., Forni P. E., Bersani F., Crepaldi T., Chiarle R., Naldini L., Ponzetto C. (2005) Cancer Gene Ther. 12, 456–463 [DOI] [PubMed] [Google Scholar]
- 24.Lacronique V., Boureux A., Valle V. D., Poirel H., Quang C. T., Mauchauffé M., Berthou C., Lessard M., Berger R., Ghysdael J., Bernard O. A. (1997) Science 278, 1309–1312 [DOI] [PubMed] [Google Scholar]
- 25.Crockett D. K., Lin Z., Elenitoba-Johnson K. S., Lim M. S. (2004) Oncogene 23, 2617–2629 [DOI] [PubMed] [Google Scholar]
- 26.Trinei M., Lanfrancone L., Campo E., Pulford K., Mason D. Y., Pelicci P. G., Falini B. (2000) Cancer Res. 60, 793–798 [PubMed] [Google Scholar]
- 27.Bischof D., Pulford K., Mason D. Y., Morris S. W. (1997) Mol. Cell. Biol. 17, 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan W., Albom M. S., Lu L., Quail M. R., Becknell N. C., Weinberg L. R., Reddy D. R., Holskin B. P., Angeles T. S., Underiner T. L., Meyer S. L., Hudkins R. L., Dorsey B. D., Ator M. A., Ruggeri B. A., Cheng M. (2006) Blood 107, 1617–1623 [DOI] [PubMed] [Google Scholar]
- 29.Gishizky M. L., Cortez D., Pendergast A. M. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10889–10893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponzetto C., Zhen Z., Audero E., Maina F., Bardelli A., Basile M. L., Giordano S., Narsimhan R., Comoglio P. (1996) J. Biol. Chem. 271, 14119–14123 [DOI] [PubMed] [Google Scholar]
- 31.Ho J. M., Nguyen M. H., Dierov J. K., Badger K. M., Beattie B. K., Tartaro P., Haq R., Zanke B. W., Carroll M. P., Barber D. L. (2002) Blood 100, 1438–1448 [PubMed] [Google Scholar]
- 32.Matsumura I., Mizuki M., Kanakura Y. (2008) Cancer Sci 99, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George R. E., Sanda T., Hanna M., Fröhling S., Luther W., 2nd, Zhang J., Ahn Y., Zhou W., London W. B., McGrady P., Xue L., Zozulya S., Gregor V. E., Webb T. R., Gray N. S., Gilliland D. G., Diller L., Greulich H., Morris S. W., Meyerson M., Look A. T. (2008) Nature 455, 975–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuzawa S., Yokochi M., Hatanaka H., Ogura K., Kataoka M., Miura K., Mandiyan V., Schlessinger J., Inagaki F. (2001) J. Mol. Biol. 306, 527–537 [DOI] [PubMed] [Google Scholar]
- 35.Maignan S., Guilloteau J. P., Fromage N., Arnoux B., Becquart J., Ducruix A. (1995) Science 268, 291–293 [DOI] [PubMed] [Google Scholar]
- 36.Hanafusa H., Torii S., Yasunaga T., Nishida E. (2002) Nat. Cell Biol. 4, 850–858 [DOI] [PubMed] [Google Scholar]
- 37.Boccalatte F. E., Voena C., Riganti C., Bosia A., D'Amico L., Riera L., Cheng M., Ruggeri B., Jensen O. N., Goss V. L., Lee K., Nardone J., Rush J., Polakiewicz R. D., Comb M. J., Chiarle R., Inghirami G. (2009) Blood 113, 2776–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fixman E. D., Fournier T. M., Kamikura D. M., Naujokas M. A., Park M. (1996) J. Biol. Chem. 271, 13116–13122 [DOI] [PubMed] [Google Scholar]
- 39.Polgar D., Leisser C., Maier S., Strasser S., Rüger B., Dettke M., Khorchide M., Simonitsch I., Cerni C., Krupitza G. (2005) Mutat. Res. 570, 9–15 [DOI] [PubMed] [Google Scholar]
- 40.Pendergast A. M., Quilliam L. A., Cripe L. D., Bassing C. H., Dai Z., Li N., Batzer A., Rabun K. M., Der C. J., Schlessinger J., Gishizky M. L. (1993) Cell 75, 175–185 [PubMed] [Google Scholar]
- 41.Rozakis-Adcock M., McGlade J., Mbamalu G., Pelicci G., Daly R., Li W., Batzer A., Thomas S., Brugge J., Pelicci P. G., Schlessinger J., Pawson T. (1992) Nature 360, 689–692 [DOI] [PubMed] [Google Scholar]
- 42.Minoo P., Chughtai N., Campiglio M., Stein-Gerlach M., Lebrun J. J., Ullrich A., Ali S. (2003) Cell. Signal. 15, 319–326 [DOI] [PubMed] [Google Scholar]
- 43.Haines E., Minoo P., Feng Z., Resalatpanah N., Nie X. M., Campiglio M., Alvarez L., Cocolakis E., Ridha M., Di Fulvio M., Gomez-Cambronero J., Lebrun J. J., Ali S. (2009) Mol. Cell. Biol. 29, 2505–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wasik M. A., Zhang Q., Marzec M., Kasprzycka M., Wang H. Y., Liu X. (2009) Semin. Oncol. 36, Suppl. 1, S27–S35 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.