Abstract
Recent studies suggest that DNA topoisomerase IIβ (topo IIβ) is involved in transcriptional activation of certain genes, which assumes accurate targeting of the enzyme to its action site. The target selection may be achieved by cooperation with unknown regulatory factors. To seek out such factors, we looked for proteins associated with the enzyme in differentiating cerebellar neurons. Antibody against topo IIβ co-precipitated RNA-binding proteins including PSF, NonO/p54nrb, as well as hnRNP U/SAF-A/SP120. Reconstitution experiments with tag-purified proteins showed that topo IIβ associates stoichiometrically with SP120 in the presence of RNA that was co-purified with SP120. The most effective RNA species for the complex formation was a subset of cellular polyadenylated RNAs. The C-terminal 187-residue domain of SP120 was necessary and sufficient for the association with both topo IIβ and the endogenous RNA. The RNA isolated from the tag-purified SP120 inhibited the relaxation of supercoiled DNA by topo IIβ. When the enzyme associates with SP120, however, the inhibition was abolished and the catalytic property was modulated to more processive mode, which may prolong its residence time at the genomic target site. Furthermore, the presence of SP120 was required for the stable expression of topo IIβ in vivo. Thus, SP120 regulates the enzyme in dual ways.
Keywords: DNA Topoisomerase, Neurodifferentiation, Nuclear Matrix, Nuclear Organization, Ribonuclear Protein (RNP)
Introduction
DNA topoisomerase II (topo II)2 can be described as an enzyme with two different catalytic activities, deoxyribonuclease and DNA ligase. Between the cleavage and religation of one duplex DNA (G-segment), another duplex (T-segment) is transferred through the gate between the cleaved ends that are held through a covalent linkage to the tyrosine residues of two identical subunits (1). Multiple cycles of these events dissipate the supercoiling or intertwining structures in substrate DNA. Thus, the enzyme plays a major role in cellular processes such as DNA replication, transcription, and recombination (2). Topo II orthologs are widely distributed in organisms from bacteria to mammals and two paralogs, named α and β, were first identified in human cells (3). These paralogs (isozymes) that are present in most vertebrates show similar enzymatic properties in vitro but their expression is regulated quite differently. Topo IIα is essential in cell proliferation because it catalyzes the segregation of daughter chromosomes at the mitotic phase. This role is played by a unique topo II in lower eukaryotes. In contrast, the biological function of topo IIβ had been totally ambiguous until we compared the isozyme expression patterns in developing cerebellar cortex (4, 5). As expected, topo IIα is the predominant isozyme expressed in proliferating precursors of granule neurons. When cells begin to differentiate after the final cell division, the isozyme expression pattern switches from topo IIα to topo IIβ. Later studies with cultured granule neurons and topo II-specific inhibitors revealed that the activity of topo IIβ is required for the transcriptional activation of a subset of genes that are induced in terminal differentiation (6). Analyses of topo IIβ-knock-out mice have also shown that the enzyme is necessary for the formation of neuromuscular junction (7), normal development of cerebral cortex (8), and gene expression in embryonic brain (9). Thus, it is now clear that topo IIβ plays significant roles in the regulation of gene expression at the final stage of neural development.
Recently, using DNA microarray techniques we have extended the catalogue of genes that are regulated by topo IIβ (referred to as A1 genes) and analyzed genomic sites targeted by the enzyme (10). Two distinct classes of topo IIβ action sites (termed c1 and c2 toposites) were discriminated. There was a significant correlation between the genomic positions of A1 genes and the toposites. The c2 toposites that are located frequently in AT-rich genomic regions are particularly interesting because they are concentrated around transcription start sites of A1 genes, but not around those of the genes that are transcribed independently of topo IIβ. How topo IIβ is targeted to toposites should be clarified to further understand its regulation mechanism. While topo II itself may have a binding preference to AT-rich sequences, it is likely that other protein factors associated with the enzyme are involved in the target selection.
HnRNP U/SAF-A/SP120 is an abundant nuclear protein that directly binds to nascent hnRNA, first described as protein component “U” of the hnRNP complex isolated from HeLa cells (11). Its primary structure was deduced from cDNA sequencing and a motif in C-terminal domain called RGG box was shown to be essential for RNA binding (12). The same protein was characterized as a DNA-binding protein that selectively binds to SAR in the presence of nonspecific competitor DNA, thus called SAF-A (13). A short stretch of amino acids in the N-terminal region, designated SAF-box or SAP domain, was demonstrated to be responsible for the association with SAR (14). In an independent work (15), we described a nuclear scaffold protein SP120 that selectively binds to SAR or MAR. The protein recognizes AT-rich sequences in most SAR/MAR and later turned out to be a rat homologue of hnRNP U/SAF-A. Because SP120 has a preference to AT-rich sequences (16) and it is co-purified with topo IIβ in the nuclear matrix fraction under certain conditions, we assumed that they interact physically and cooperate together in the determination of genomic action sites of topo IIβ.
In the present study, the extraction and subsequent immunoprecipitation of nuclear proteins under mild conditions revealed that topo IIβ is associated with SP120 in differentiating granule neurons. We have further shown that a certain class of RNA tightly bound to SP120 mediates the association, which is required for the in vivo stability of topo IIβ protein. When complexed with SP120, the catalytic property of topo IIβ was modulated so as to the enzyme might stay on the same genomic site during its action. Taken together, the results represent a novel mechanism for the regulation of topo IIβ in its action site determination.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies used in the study are described in supplemental materials.
Construction of Plasmids
The plasmid pFLAG-topo2b that encodes the full-length rat topo IIβ (1,614 amino acids) with FLAG sequence tagged at the N terminus was constructed. The original topo IIβ cDNA clone (AB262979) was amplified with primers listed in the supplemental materials. A high-fidelity DNA polymerase, KOD-Plus DNA polymerase (Toyobo), was used throughout. The PCR product was inserted in-frame between the EcoRV/SalI sites of pFLAG-CMV-2 expression vector (Sigma). All the resulting constructs were sequenced to verify the absence of any amino acid changes.
For the expression of SP120 and its domain variants, plasmids pMyc-SP120WT, pMyc-SP120ΔN, pMyc-SP120ΔC, and pMyc-SP120C187 were constructed using PCR-generated fragments encoding the amino acid residues 1–798, 205–798, 1–611, and 612–798 of rat SP120 (798 amino acids), respectively. The original SP120 cDNA clone (D14048) was amplified using the forward and reverse primers listed in supplemental materials. The PCR products were inserted in frame between the EcoRI/XhoI sites of the pCMV-Myc expression vector (Clontech). The plasmid pFLAG-SP120WT for the expression of full size SP120 tagged with FLAG sequence at the N terminus was constructed similarly using the pFLAG-CMV vector.
Co-transfection Experiments
pFLAG-topo2b (7.2 μg) was co-transfected either with pMyc-SP120WT, ΔC or ΔN (0.3 μg) into HEK293 cells (106 cells/100 mm dish) by using a conventional calcium phosphate procedure. Two days later, cells were lysed in 1 ml of extraction buffer containing 50 mm HEPES-NaOH (pH 7.4), 1 mm EDTA, 0.3 m NaCl, 0.2% Nonidet P-40, and proteinase inhibitor mixture (PIC, Roche). Lysates were centrifuged at 20,000 × g for 10 min, and the supernatants were subjected to pre-clearing with 10 μl-slurry of Dynabeads Protein G (Invitrogen) at 4 °C for 2 h. After removing the beads by magnetic separation, fresh beads pre-coated with anti-FLAG M2 antibody (5 μg) was added and incubated at 4 °C overnight. After incubation, the beads were washed and then suspended in SDS-PAGE sample buffer.
Protein Expression and Purification
To express epitope-tagged proteins for in vitro reconstitution experiments, HEK293 cells (106 cells/100 mm dish) were transfected with 3 μg of the construct plasmids and cultured for 3 days. Cells were lysed on ice in 1 ml/dish of ice-cold HSB (50 mm HEPES-NaOH: pH 7.4, 1 mm EDTA, 420 mm NaCl, 0.1% Nonidet P-40, PIC). Clear lysates were prepared by repeated centrifugation. In some experiments, lysates were treated with 0.2 mg/ml RNase A at 25 °C for 20 min and re-centrifuged to remove any aggregates. Immunoprecipitation was performed at 4 °C for 4 h with Dynabeads Protein G that had been pre-coated with anti-tag antibodies. After incubation, the beads were washed three times with HSB. FLAG-tagged proteins were eluted on ice for 30 min with 100 μl of 150-μg/ml 3x FLAG peptide (Sigma) in PBS. The elution was repeated once more, and the eluates were combined. Myc-tagged proteins were used without elution still bound on beads.
In Vitro Reconstitution Experiments
Topo IIβ-SP120-RNA ternary complex was reconstituted in vitro by incubating the isolated components on ice for various times in the reconstitution buffer containing 20 mm HEPES-NaOH (pH 8.0), 0.1 m NaCl, 3 mm MgCl2, 2 mm CaCl2, 0.5 mm EDTA, 1 mm DTT, 0.05% Tween20, 1 mm PMSF, PIC, and 10 units/ml placental RNase inhibitor. Depending on experiments, either topo IIβ or SP120 was fixed on magnetic beads, and the binding of its partner protein was measured. Proteins on washed beads were separated by SDS-PAGE and detected by SYPRO Ruby staining or Western blotting.
FLAG-topo IIβ was always purified from RNase-treated cell lysates by immunoprecipitation with anti-FLAG antibody, followed by elution with FLAG peptide. Whereas, SP120 (Myc- or Flag-tagged) was immunopurified from cell lysates that had been incubated with or without RNase. In reconstitution experiments with fractionated RNA preparations, purified RNA (1.0 μg) was mixed with FLAG-topo IIβ and RNase-treated Myc-SP120 that was fixed on magnetic beads.
Stoichiometric binding between topo IIβ and SP120 was tested by mixing FLAG-topo IIβ and FLAG-SP120 (contains endogenous RNA) at various molar ratios. Immunoprecipitation was done with anti-topo IIβ or anti-SP120 peptide antibodies. Proteins bound on beads and unbound in solution were separated and analyzed by SDS-PAGE. Protein bands in gels were quantified by SYPRO Ruby staining followed by densitometry.
RNase Resistance Assay
The ternary complex was first formed by incubating FLAG-topo IIβ and equimolar amounts of FLAG-SP120 with bound RNA. RNase A (0.2 mg/ml) was then added and the mixture was incubated at 25 °C for 20 min. Immunoprecipitation was done on ice for 1 h using anti-topo IIβ peptide antibody. As a control, FLAG-SP120 pretreated with RNase was used. Protein bands were quantified by SYPRO Ruby staining followed by densitometry. Purified RNA recovered from the beads was quantified by RiboGreen (Molecular Probes).
RNA Preparation
Total RNA fractions were purified from HEK cells and Escherichia coli cells by using RNeasy Mini kit (Qiagen). Cellular RNA co-purified with SP120 was purified by phenol-chloroform extraction. Poly(A) RNA fraction was isolated from HEK cells using PolyATract System 1000 (Promega).
Relaxation Assay
FLAG-tagged topo IIβ and SP120 (with or without bound RNA) were mixed and incubated on ice for 20 min as in reconstitution experiments. After dilution, supercoiled pUC18 (50 ng) and 0.5 mm ATP were added to initiate the reaction. Unless otherwise stated, the reaction was carried out at 30 °C for 30 min. After adding the stop solution containing SDS, proteinase K, and loading dye, the mixture was incubated at 55 °C for 1 h, and subjected to gel electrophoresis in 1% agarose. Topoisomer DNA bands were visualized by staining with 0.5 μg/ml ethidium bromide. Fully supercoiled DNA substrates that remained unrelaxed after the reaction were quantified by densitometry.
siRNA Experiments
siRNA against rat SP120 (L-095044–01, Dharmacon), rat topo IIβ (10) and control siRNA (D-001810-10-05, Dharmacon) were transfected to freshly isolated cerebellar granule cells by using a Nucleofector Kit (rat neuron) and an optimized instrument (Amaxa) as described previously (10). Cells were harvested at the 2nd and 5th day in culture and subjected to Western blotting. To normalize the loading volume, nucleic acids concentrations were determined with RiboGreen (Molecular Probes). Total RNA was purified by RNeasy Mini Kit (Qiagen) and stored at −80 °C until use.
Reverse Transcription-Quantitative PCR (RT-qPCR)
cDNA was synthesized from 1 μg of total RNA using QuantiTect Reverse Transcription Kit (Qiagen). Real time PCR reactions were set up with QuantiTect SYBR Green PCR Master Mix (Qiagen) and run on the ABI 5700 real-time PCR system (Applied Biosystems). Gene-specific primer pairs used are listed in supplemental materials.
RESULTS
Analysis of Proteins Associated with Topo IIβ in Cultured Granule Cells
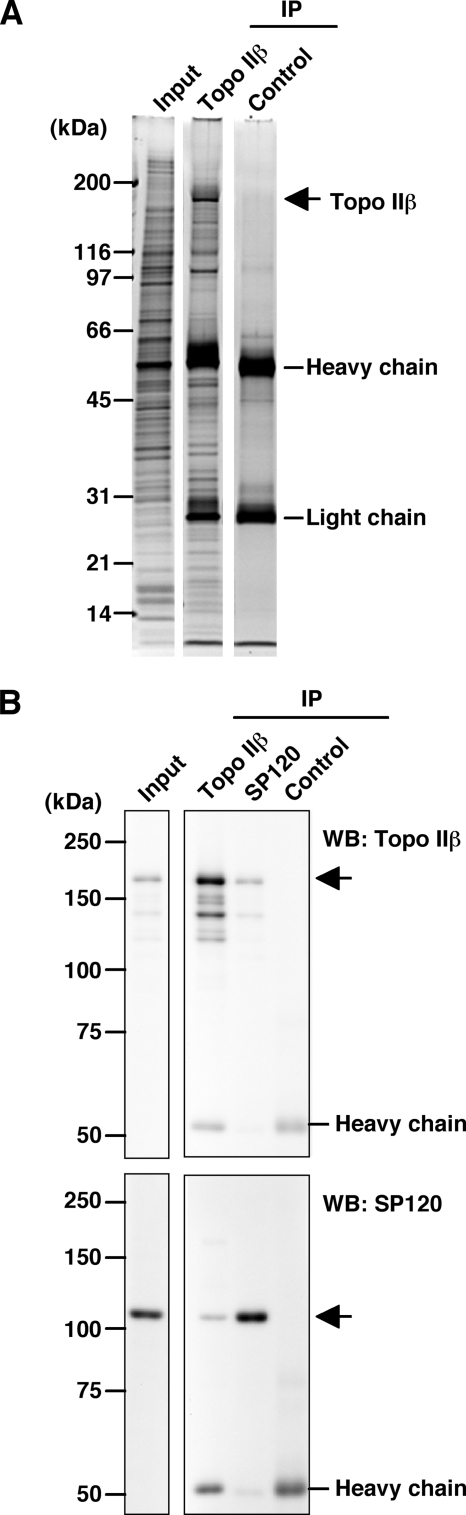
Because topo IIβ has been demonstrated to be involved in neuronal differentiation in developing cerebellar cortex (6), rat cerebellar granule cells cultured in vitro were used for the analysis. Nuclei isolated from the cells at day 3 in culture were treated with micrococcal nuclease (MNase) in 0.1 m NaCl and the solubilized material was recovered by centrifugation. The extract was subjected to immunoprecipitation with monoclonal antibody to topo IIβ (3B6) or preimmune mouse IgG followed by SDS-PAGE (Fig. 1A). Detailed procedure is described in supplemental materials. Many protein bands, including topo IIβ, were detected specifically. Antibodies used in the present study, including 3B6 (6), were quite specific in immunoblotting (supplemental Fig. S1A). When used for immunoprecipitation in high-salt conditions, 3B6 precipitates only topo IIβ (Fig. S1B).
FIGURE 1.
Analysis of proteins co-precipitated with topo IIβ. A, nuclear extract from cultured granule cells was immunoprecipitated using a monoclonal antibody against topo IIβ. Mouse IgG was used as a control. Proteins in immunoprecipitates (IP) were separated by 4–20% gradient SDS-PAGE and stained with SYPRO Ruby. B, nuclear extract was immunoprecipitated with monoclonal antibodies to topo IIβ (3B6) or SP120 (1–67D). IP fractions were subjected to Western blotting to detect topo IIβ (upper panels) or SP120 (lower panels) that are indicated by arrows. Additional bands detected below the topo IIβ band are degradation products of topo IIβ. Detailed procedures are given in supplemental materials.
Co-precipitated proteins were eluted with 0.5 m NaCl and analyzed by LC/MS (Table 1). Among the proteins identified with high statistical confidence, three nuclear RNA-binding proteins (PSF, NonO/p54nrb, and hnRNP U) were noted. Others were all ribosomal proteins, which might originate from ribosome contamination in crude nuclear preparation used here. Most protein bands migrating ahead of the antibody heavy chain probably represent ribosomal proteins. In contrast, PSF and hnRNP U are likely to be genuine binding partners of topo IIβ. PSF is often accompanied by NonO (17). From among these proteins, we focused on hnRNP U/SAF-A/SP120 because it binds to DNA that shares similar feature with topo IIβ target sites (see Introduction). Their co-existence in both immunoprecipitates, as detected by Western blotting, supported the interaction between these proteins (Fig. 1B).
TABLE 1.
List of proteins detected by LC/MS analysis
Only proteins with search scores higher than 50 are shown. Experimental details are given in supplemental materials.
| SwissProt Database Accession# | Symbol | Protein MW (Da) | Distinct Summed MS/MS Search Score | No. of peptide | % AA Coverage | Protein Name | Species |
|---|---|---|---|---|---|---|---|
| P23246 | PSF | 76149.8 | 118.7 | 8 | 14 | Splicing factor, proline- and glutamine-rich | HUMAN |
| P62243 | RPS8 | 24205.3 | 105.47 | 6 | 36 | 40S ribosomal protein S8 | RAT |
| P21533 | RPL6 | 33561.8 | 79.87 | 6 | 23 | 60S ribosomal protein L6 | RAT |
| P62425 | RPL7A | 29995.8 | 76.28 | 6 | 22 | 60S ribosomal protein L7a | RAT |
| P62909 | RPS3 | 26674.4 | 75.45 | 5 | 26 | 40S ribosomal protein S3 | RAT |
| Q5FVM4 | NONO | 54925.6 | 75.42 | 5 | 11 | Non-POU domain-containing octamer-binding protein | RAT |
| P50878 | RPL4 | 47257.1 | 70.95 | 5 | 13 | 60S ribosomal protein L4 | RAT |
| P41123 | RPL13 | 24309.6 | 61.13 | 4 | 20 | 60S ribosomal protein L13 | RAT |
| P62755 | RPS6 | 28680.8 | 60.61 | 4 | 18 | 40S ribosomal protein S6 | RAT |
| P05426 | RPL7 | 30329.4 | 56.39 | 5 | 22 | 60S ribosomal protein L7 | RAT |
| Q00839 | HNRPU | 90513.8 | 51.58 | 3 | 4 | Heterogeneous nuclear ribonucleoprotein U | HUMAN |
| P09895 | RPL5 | 34458.9 | 51.39 | 4 | 16 | 60S ribosomal protein L5 | RAT |
Immunopurified SP120 Contains Tightly Bound RNA
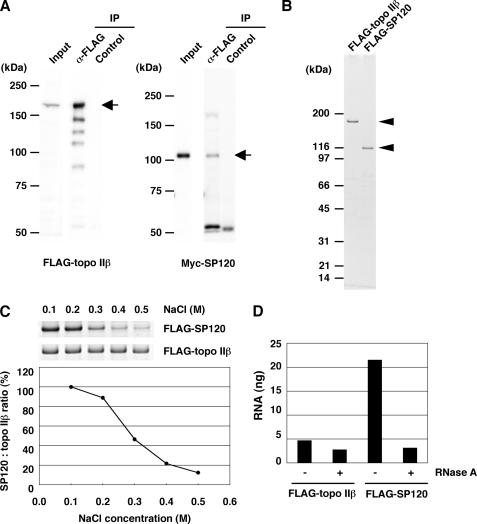
To further investigate the interaction between topo IIβ and SP120, we used human cells transfected with epitope-tagged rat proteins. As shown in Fig. 2A, FLAG-topo IIβ precipitated with anti-FLAG antibody from whole cell extract contained Myc-tagged SP120, corroborating that their interaction also occurs in human cells. However, the proteins were dissociated from each other and purified separately under higher salt conditions (Fig. 2B). After eluting with FLAG peptide, we used these preparations for the reconstitution experiments in vitro. The immunopurified topo IIβ and SP120 were mixed at 0.1 m NaCl and topo IIβ was precipitated with anti-topo IIβ onto magnetic beads. Both proteins were detected on the beads, indicating their efficient interaction (Fig. 2C). After washing the beads with increasing concentrations of NaCl, SP120 remaining on beads decreased significantly. Although the protein complex is stable under physiological ionic conditions, it dissociates in higher salt ranges. This is consistent with the results shown in Fig. 2, A and B.
FIGURE 2.
Topo IIβ-SP120 complex contains RNA. A, HEK293 cells were co-transfected with pFLAG-topo 2b and pMyc-SP120 and the whole cell extract was used for immunoprecipitation by anti-FLAG antibody. The expressed proteins in IP were detected by Western blotting with anti-FLAG (left panel) or anti-Myc (right panel) antibodies. B, FLAG-tagged proteins were expressed separately in HEK cells and immunopurified in high-salt conditions. These preparations were free from cross-contamination as shown by the stained gel (arrowheads). C, immunopurified FLAG-topo IIβ and FLAG-SP120 were mixed in 0.1 m NaCl and immunoprecipitated with anti-topo IIβ peptide antibody. The protein complex formed on beads was treated with increasing concentrations of NaCl as indicated. Proteins remaining on beads were separated by SDS-PAGE and detected by staining (upper panels). Relative amounts of SP120 are plotted in the lower graph. D, high-salt (0.42 m NaCl) cell extracts were treated or untreated with RNase A, and the proteins were immunopurified. Amounts of RNA associated with proteins were quantified by RiboGreen. RNA per 100 ng of protein is shown in the graph.
Because SP120 was first characterized as an RNA-binding protein and topo IIα was also shown to bind RNA (18), we examined the RNA content in topo IIβ and SP120 preparations that were tag-purified in high salt. Only SP120 contained significant amounts of RNA, which is sensitive to RNase A even in its protein-bound state (Fig. 2D). It is worth noting that at high ionic conditions the association between topo IIβ and SP120 is unstable but the RNA is tightly bound to SP120. The SP120-bound RNA consisted of multiple molecular species of heterogeneous sizes (not shown).
C-terminal Domain of SP120 with Bound RNA Is Sufficient for Its Interaction with Topo IIβ
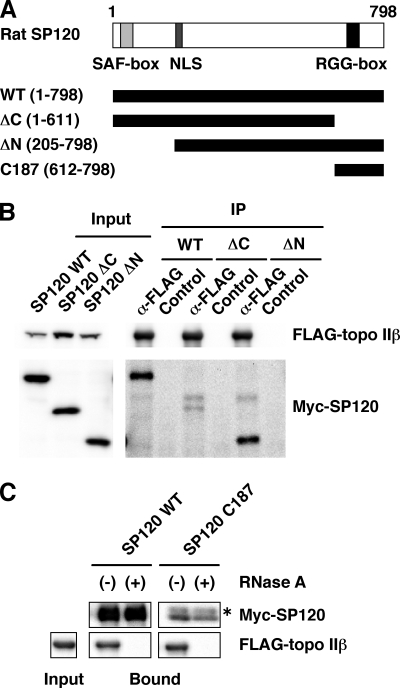
Two functional domains in SP120 molecule had been identified and characterized. The N-terminal domain termed SAF-box or SAP domain is required for DNA binding (14) and the C-terminal domain called RGG-box is essential for the interaction with hnRNA (12) (Fig. 3A). To assess the importance of these domains in the interaction with topo IIβ, FLAG-tagged topo IIβ, and Myc-tagged SP120, either wild type (WT) or truncation mutants (ΔC or ΔN), were co-expressed in HEK293 cells. After immunoprecipitation of cell lysates with FLAG-specific antibody, co-precipitated SP120 was detected with anti-Myc antibody (Fig. 3B). Very little SP120 protein was present in the IP from the cells transfected with ΔC mutant as compared with those with WT or ΔN mutant, suggesting that the C-terminal domain of SP120 contributes to the binding with topo IIβ.
FIGURE 3.
The C-terminal domain of SP120 with tightly bound RNA is involved in the complex formation. A, domain structure of SP120 and its variants used for the analysis. All recombinant proteins had Myc tag on the N terminus. Numbers in parentheses stand for amino acid residue numbers. SAF-box and RGG-box in SP120 have been ascribed as essential motifs for the binding of DNA and RNA, respectively. NLS, nuclear localization signal. B, HEK cells were co-transfected with FLAG-topo IIβ and Myc-SP120 (WT, ΔC, or ΔN). The expressed proteins were detected by anti-tag antibodies (left panel). Cell extracts were immunoprecipitated with anti-FLAG antibody and subjected to Western blotting with anti-tag antibodies (right panel). C, tagged proteins were expressed separately in HEK cells and tag-purified in high-salt extract. Myc-SP120 extracts were either treated (+) or untreated (−) with RNase A. SP120 proteins (WT and C187) fixed on beads through anti-Myc antibody were incubated with FLAG-topo IIβ free in solution. Proteins bound on the beads were detected by Western blotting with anti-tag antibodies. The asterisk indicates the light chain of the antibody used for IP.
The result does not necessarily mean that the site of physical contact with topo IIβ also resides within the deleted domain. To clarify this point, an in vitro binding assay was performed using the C-terminal domain of SP120 (C187), which was deleted in the ΔC mutant (Fig. 3C). Cell lysates containing Myc-tagged SP120 (WT and C187) were either treated or untreated with RNase A and the proteins were subjected to immunoprecipitation with anti-Myc antibody. The FLAG-topo IIβ isolated from RNase-treated lysate was eluted from the beads by FLAG peptide and mixed with the SP120 proteins that were still retained on the beads. As shown in the figure unequivocally, not only WT protein but also C187 bound topo IIβ, while the binding was abolished when the Myc-tagged proteins had been treated with RNase. Immunopurified C187 also contained RNA, whose size distribution was similar to that of WT protein (data not shown). The results demonstrate that the C-terminal 187 residues of SP120 are solely responsible for both the interaction with topo IIβ and the retention of RNA that is essential for the formation of ternary complex.
Polyadenylated RNA Is Involved in the Complex Formation
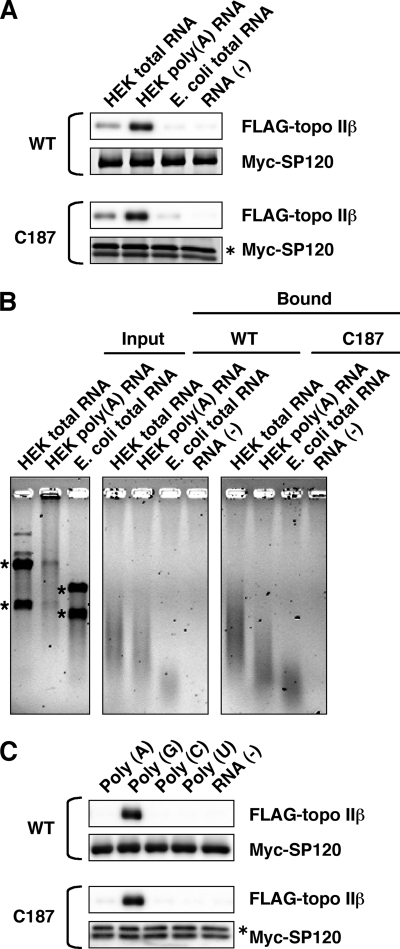
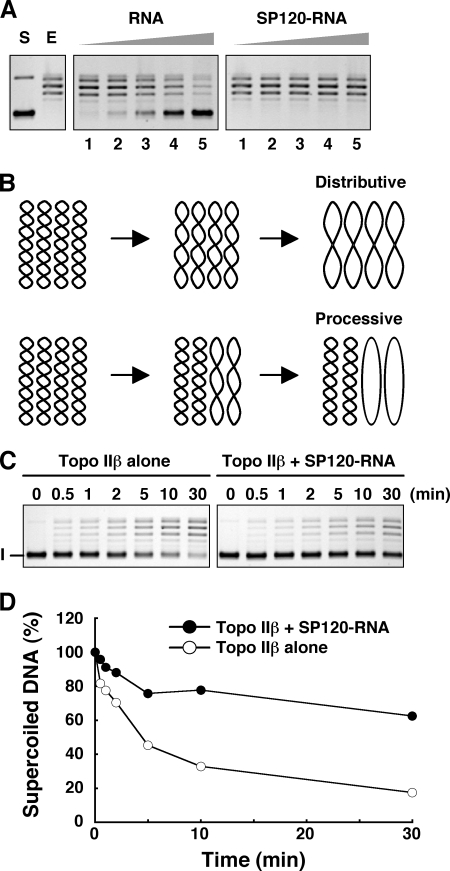
To identify RNA species that is most effective in the complex formation, binding of topo IIβ to SP120 immobilized on magnetic beads was measured as described above, except that endogenous cellular RNA was removed by RNase digestion and the binding was driven by purified RNA fractions. When the same amounts were added to the reaction, poly(A) RNA was most effective for both WT and C187 (Fig. 4A). Total RNA from HEK cells was less effective than poly(A) RNA and that from E. coli cells was almost ineffective. RNA composition of the bound fraction was analyzed by agarose gel electrophoresis under denatured conditions (Fig. 4B). Smearing RNA bands were detected in the protein complex that was formed with RNA preparations isolated from HEK cells. The result also shows a complete absence of ribosomal RNAs in the complex, suggesting that RNA species responsible for the complex formation is a population of polyadenylated mRNA-like hnRNAs. The total RNA of E. coli appears to contain smaller RNA species that does bind to SP120 but is unable to facilitate the interaction with topo IIβ.
FIGURE 4.
Identification of RNA species that are required for the SP120-topo IIβ interaction. A, in vitro binding assay was performed as in Fig. 3C, except that endogenous RNA bound to SP120 was digested with RNase and replaced by the purified RNA preparations as indicated. FLAG-topo IIβ was detected by Western blotting with anti-FLAG antibody, whereas Myc-SP120 recombinants (WT and C187) were visualized directly by SYPRO Ruby staining. Asterisk indicates the antibody's light chain. B, RNA added to the binding reaction (input) and the RNA recovered from the protein complex were separated by 1% agarose gel electrophoresis under denatured conditions and detected by staining with SYBR Green II. Asterisks indicate rRNAs. C, binding experiment was performed as in A with 1 μg of synthetic polyribonucleotides instead of natural RNAs. Proteins on beads were detected by Western blotting with anti-tag antibodies. The asterisk indicates the light chain of the IP antibody.
We next examined whether the poly(A) tail is directly involved in the enhanced interaction between these proteins by using synthetic homoribopolymers (Fig. 4C). Unexpectedly, for both WT and C187 poly(G) was by far the most effective polynucleotide and others including poly(A) showed little promoting activity. To see whether a single species within the heterogeneous RNA population can support the interaction, RNA associated with the complex was randomly cloned and the resulting cDNA clones were transcribed in vitro to test the complex-forming activity. Out of 12 sequenced clones, 9 were identified as mature mRNAs, 2 were unspliced introns, and 1 was a noncoding RNA (supplemental Fig. S2A). Transcripts from full-sized mRNA clones indeed facilitated the interaction between the proteins (supplemental Fig. S2B). Not all clones represent particularly abundant transcripts. Therefore, some common feature in poly(A) RNA, possibly a G-rich stretch, might be involved in the selective interaction as a determinant factor.
Stoichiometry of the Topo IIβ-SP120 Interaction in the Presence of RNA
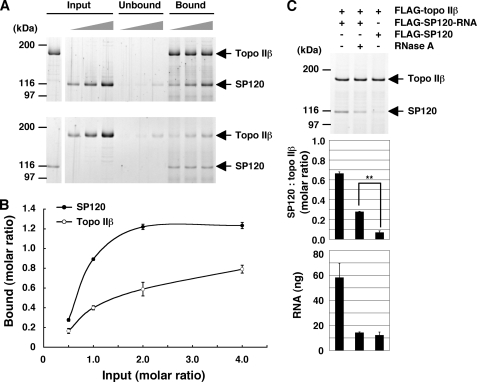
Molecular ratio between topo IIβ and SP120 in the RNA-dependent protein complex was determined more quantitatively using the complex immobilized on magnetic beads in vitro. FLAG-tagged proteins, topo IIβ (Fig. 5A, upper panel) or SP120 (lower panel), were incubated with increasing amounts of corresponding partners and then immunoprecipitated by antibodies against topo IIβ (upper panel) or SP120 (lower panel). Proteins bound on washed beads were separated by SDS-PAGE and quantified by densitometry of stained bands (Fig. 5B). For both proteins, the binding increased as added protein increased until they were detected in unbound fraction (Fig. 5A). Although the saturation levels appear to differ significantly between the two curves, the result suggests that topo IIβ and SP120 form a one-to-one molecular ratio complex when semi-quantitative nature of the method is taken into account.
FIGURE 5.
The topo IIβ-SP120-RNA complex: Stoichiometry and resistance to RNase. A, tag-purified FLAG-topo IIβ and FLAG-SP120 with tightly-bound RNA were mixed at the molar ratio of 0.5, 1.0, 2.0, keeping one of the proteins constant (shown in the leftmost lane of Input). After incubation, the mixture was subjected to immunoprecipitation with anti-peptide antibodies to the unvaried counterpart: topo IIβ (upper panel) and SP120 (lower panel). The proteins on beads (Bound) and in solution (Unbound) were separated by SDS-PAGE and visualized by SYPRO Ruby staining. B, band densities for topo IIβ and SP120 in the complex were quantified by densitometry and plotted along the ordinate as molar ratios of the varied protein to the fixed protein on beads, assuming topo IIβ as monomeric form. Values shown are average and standard deviation from three independent experiments. C, RNase resistance assay. Purified FLAG-topo IIβ was mixed with equimolar amounts of tag-purified SP120 (FLAG-SP120-RNA). The mixture was treated (+) or untreated (−) with RNase and then immunoprecipitated with anti-topo IIβ antibody. As a control, SP120 that had been pretreated with RNase in high-salt extract was used (FLAG-SP120). Protein bands were visualized by SYPRO Ruby (top panel) and quantified by densitometry (middle panel). Note that the difference between RNase (+) and control is statistically significant by Student's t test (p < 0.01). RNA was purified from the complex on beads and quantified by using RiboGreen. RNA per 100 ng of topo IIβ was graphed (bottom panel). Values shown in graphs are average and standard deviation from three independent experiments.
As a high molecular weight poly(A) RNA is an essential component of the complex, topo IIβ and SP120 may not interact directly but are held together through RNA. If this were the case, however, the one-to-one stoichiometry between the proteins would never be observed. To confirm this notion, we looked at the stability of the ternary complex against RNase treatment (Fig. 5C). In the absence of physical contact between the proteins, their association should be dissolved completely upon disruption of the RNA bridge between them, which is sensitive to RNase. As shown in the middle panel, about half of SP120 associated with topo IIβ on beads was resistant to RNase with a statistical significance, suggesting that these proteins do form a direct complex. When RNA content of the RNase-treated complex was measured, it was comparable to that in the control, being indicative of the completion of treatment (bottom panel). The result also indicates that little RNA remains in the complex (however, see “Discussion”).
To further corroborate the importance of the C-terminal region of SP120 in the complex formation, we used Myc-C187 fixed on beads for the interaction with FLAG-topo IIβ in solution (supplemental Fig. S3). The molar ratio of topo IIβ against SP120 at saturation estimated from this experiment was 0.34, being significantly lower than unity. It appears, therefore, that C187 fixed on beads is under a steric hindrance resulting from immobilization. The RNase resistance assay shown in supplemental Fig. S3C gave similar results as in Fig. 5C, being consistent with the notion that C187 can mimic the full-length protein in the binding of both RNA and topo IIβ.
Effects of Complex Formation on the Catalytic Mode of Topo IIβ
A recent report has shown that human topo IIα interacts with RNA and as a consequence its activity (decatenation and relaxation) is inhibited (18). We have also observed an inhibition of topo IIβ relaxation activity with synthetic homoribopolymers and cellular RNA (results not shown). This fact raises a serious problem how the enzyme can operate in the nuclear environment that is filled with RNA molecules. The RNA isolated from immunopurified SP120 indeed inhibited the topo IIβ-catalyzed relaxation of supercoiled DNA (Fig. 6A). In contrast, no inhibition was observed when the same RNA was associated with SP120. It appears, therefore, that the formation of ternary complex with SP120 is an escape mechanism for topo IIβ from the inhibition by endogenous RNA.
FIGURE 6.
Modulation of topo IIβ reaction by SP120-RNA: Time course. A, tag-purified FLAG-topo IIβ (0.04 pmol) was preincubated with increasing amounts of FLAG-SP120 that contained endogenous RNA (SP120-RNA) or corresponding amounts of RNA purified from FLAG-SP120 (0.02, 0.04, 0.08, 0.16, and 0.32 pmol in lanes 1–5, respectively). Relaxation was started by addition of supercoiled DNA (pUC18) and the reaction products were separated by 1% agarose gel electrophoresis. S, substrate; E, enzyme alone. The lagging ladder bands are relaxed topoisomers. The reaction is totally dependent on the presence of ATP (not shown). B, two hypothetical modes of relaxation time course under restricted dose of enzyme. Note that a portion of substrate molecules remains unrelaxed when the reaction is completely processive, whereas all the substrate is converted eventually to relaxed forms in the distributive mode. C, relaxation of pUC18 by topo IIβ alone (0.01 pmol) or by topo IIβ-SP120-RNA equimolar complex (0.01 pmol each) was stopped at the times indicated. I, supercoiled DNA. D, unrelaxed DNA bands in C were quantified by densitometry and plotted in values relative to time 0.
After each catalytic cycle, type II topoisomerases can either dissociate from DNA or stay at the same site to enter the next cycle. In the former case, the enzyme redistributes to distant sites or different molecules before entering the next cycle. Thus, the mode is called distributive. In the latter that is called processive mode, the enzyme proceeds to the next cycle without releasing the G-segment. Which mode is preferred depends on conditions like ionic strength, being more processive in low salt. In theory, when topo II is associated with a DNA-binding protein, processive mode should be favored. These reaction modes can be discriminated by measuring the time course of relaxation at a low enzyme-to-substrate ratio. As shown schematically in Fig. 6B, the reaction never reaches completion in the processive mode. Under standard conditions, tag-purified topo IIβ relaxed supercoiled DNA distributively (Fig. 6C, left panel). However, when the enzyme turned into the ternary complex with SP120 and RNA, it appears to operate in a processive mode (Fig. 6C, right panel). This is more clearly demonstrated by the graph plotted with quantities of unrelaxed substrate (Fig. 6D).
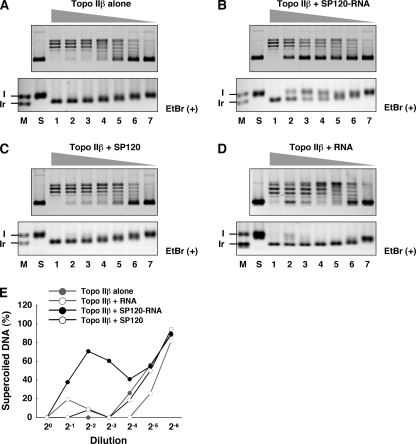
Processivity of the reaction can be demonstrated otherwise by varying the enzyme-to-substrate ratio (Fig. 7). The highest amount of topo IIβ used in this assay was sufficient to completely relax the supercoiled substrate. The enzyme was first mixed with equimolar amounts of SP120-RNA or RNase-treated SP120, and then the mixtures were diluted serially before the addition of substrate. In the reaction with free enzyme, average superhelicity of the product at the end of reaction increased gradually along with dilution, which is consistent with the distributive mechanism. This is reflected by the appearance of new topoisomer ladders in the intermediate dilutions (lanes 4–6 in Fig. 7A, upper panel). Electrophoresis in the presence of ethidium bromide also showed that the product band gradually shifts in mobility from fully relaxed (Ir) to supercoiled (I) forms.
FIGURE 7.
Modulation of topo IIβ reaction by SP120-RNA: Varying enzyme-substrate ratio. After incubating 0.08 pmol each of tag-purified topo IIβ and SP120 (with or without bound RNA), the mixture was 2-fold diluted serially, and then pUC18 was added to initiate the reaction. A, topo IIβ alone; B, topo IIβ and SP120-RNA; C, topo IIβ and SP120 treated with RNase; D, topo IIβ and RNA isolated from SP120. Reaction products were separated by 1% agarose gel electrophoresis in the absence (upper panels) or presence (lower panels) of ethidium bromide (EtBr). M, marker lane (I: supercoiled, Ir: relaxed); S, substrate DNA. Lanes 1–7 correspond to 20–2−6 dilutions. E, unrelaxed DNA bands in upper panels were quantified by densitometry and plotted as relative values to added substrate (lane S).
When topo IIβ was complexed with SP120 in the presence of RNA, the product topoisomer pattern was clearly different. In intermediate dilutions, unrelaxed substrate appeared abruptly without accompanying intermediate topoisomers (Fig. 7B). This is more clearly demonstrated by the ethidium bromide-containing gel shown in the lower panel (lanes 2–4). The data indicate that the enzyme operates almost processively in the form of ternary complex. However, at higher dilutions the enzyme appears to act distributively (lanes 5–7). The biphasic nature is evident in the densitometric readings shown in Fig. 7E. The result may imply that the complex dissociates in dilute conditions. In contrast, in the presence of SP120 without bound RNA, which is unable to form a complex with topo IIβ, the reaction proceeds distributively at all dilutions (Fig. 7C). The result also indicates that SP120 free from topo IIβ does not affect the reaction, although it does bind to substrate DNA (data not shown). No effect was observed with RNA alone, suggesting that the switch in the reaction mode is not caused by an inhibitory action of RNA (Fig. 7D).
SP120 Is Required for Stable Expression of Topo IIβ
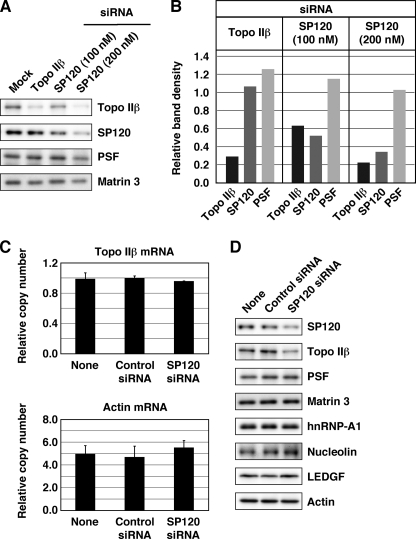
Topo IIβ in the developing cerebellar granule neurons is involved in the activation of a subset of neuronal genes (group A1) that are repressed in undifferentiated state (10). The targeted action of topo IIβ might depend on SP120 associated with topo IIβ. To test this, we knocked down SP120 by RNAi in differentiating granule cell culture to see whether it interferes with the A1 gene induction. This attempt was unsuccessful, however, because siRNA against SP120 also down-regulated topo IIβ expression in a dose-dependent manner (Fig. 8, A and B). Because the levels of topo IIβ mRNA remained constant (Fig. 8C), the result is not due to an off-target effect of SP120 siRNA. In contrast, the level of SP120 was not affected by topo IIβ siRNA, which decreased the enzyme to about 30% of control (mock transfection). Neither siRNAs caused significant decrease in the levels of PSF although the protein is one of the candidate proteins that interact with topo IIβ (Table 1).
FIGURE 8.
Topo IIβ is down-regulated in concert with SP120. A, cerebellar granule cells treated with siRNAs for topo IIβ, SP120, or control (mock) at day 0 were cultured for 2 days. Levels of proteins designated were estimated by Western blotting. A nuclear matrix protein, matrin 3, was detected with a rabbit polyclonal antibody and used as a loading control. B, band densities on immunoblots were measured and plotted in the graph after correction of loading errors and normalization by the mock. C, cerebellar granule cells treated with siRNA at day 0 were cultured for 5 days. RNA samples were prepared, and mRNA levels for topo IIβ and β-actin were determined by RT-qPCR. Copy numbers of mRNA in 20 ng of total RNA were calculated and plotted in the graph. The data represent average ± S.D. of triplicate analysis. D, protein samples from the same set of cells used in C were analyzed by Western blotting.
As SP120 has been shown to stabilize certain species of mRNA (19), there might be other proteins down-regulated in concert with SP120. We repeated the siRNA experiment in slightly different conditions to confirm the results shown above and to look at additional nuclear proteins (Fig. 8D). Among these, topo IIβ was the only protein down-regulated by SP120 siRNA. Similarly, expression of HP1α that was shown recently to interact directly with hnRNP U remains constant when hnRNP U was down-regulated (21). These results may suggest that topo IIβ is a unique protein that is unstable and degraded unless it is bound to SP120.
To see whether the SP120-dependent stabilization of topo IIβ is a general phenomenon, we performed a time course study using Neuro-2a cells, a mouse neuroblastoma cell line (supplemental Fig. S4). As shown in supplemental Fig. S4B, topo IIβ decreased much later (half-life∼72 h) after SP120 was practically knocked down (half-life∼15 h). The topo IIβ decrease appears to occur faster in granule neurons (Fig. 8B). The immunostaining patterns at 72 h (supplemental Fig. S4C) were consistent with the Western blots. As an ubiquitous protein, SP120 may be an indispensable partner of topo IIβ for its survival.
DISCUSSION
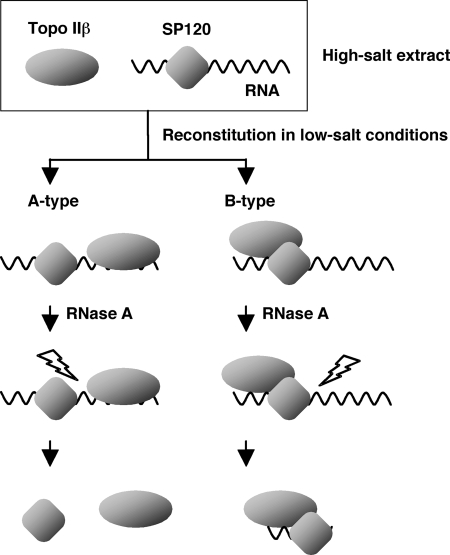
Using in vitro reconstitution experiments, we have demonstrated in the present study that topo IIβ forms a stoichiometric complex with SP120 if the protein is loaded with endogenous RNA. The RNA that is essential for the complex formation is susceptible to RNase A in the absence of bound topo IIβ in high-salt conditions. Possible interaction between these components in the ternary complex may be discriminated into two major types (Fig. 9). As both proteins bind RNA independently, they can be held together by a bridging RNA molecule (A type). Otherwise, these proteins may interact directly through the C-terminal domain of SP120, which is also involved in the binding of RNA (B type). Although these types are not mutually exclusive, the observed stoichiometry and the resistance of the complex to RNase indicate that the B-type interaction is dominant (see Fig. 5). While little RNA, compared with control, was detected in the ternary complex after RNase treatment, we speculate that a short stretch of poly(A) RNA protected from digestion is still bound to the complex, which is sufficient to keep the complex intact. The protection may be incomplete, however, because the complex was partially disrupted by RNase. Otherwise if no RNA survives after RNase treatment, RNA should be required only before topo IIβ comes in, and becomes dispensable after the interaction between the proteins is established.
FIGURE 9.
Possible interaction modes among the components of topo IIβ-SP120-RNA ternary complex reconstituted in vitro. In the A-type complex, both proteins are tethered by intervening RNA, which is susceptible to RNase. The molar ratio between the proteins varies in this type complex. However, it approximates unity in the B-type complex in that proteins are in close contact. Around the point of contact, a small fragment of RNA may escape from RNase digestion.
The C-terminal domain of SP120 (C187) contains an arginine-glycine-rich repetitive motif called RGG box that is involved in RNA binding (12). In the present study, the tag-purified C187 indeed contained tightly bound endogenous RNA, which is essential for the interaction with topo IIβ. When the primary sequence of SP120 was subjected to structural prediction such as the web-accessed software POODLE, the C-terminal region was largely disordered or unstructured. Importance of disordered protein structures has been recognized recently (20). In some proteins, these poorly structured regions undergo conformational change upon binding to RNA by an induced fit mechanism, which is required in many biological activities. As for topo IIβ, however, neither conventional RNA binding motifs like RNA recognition motif (RRM) nor biochemically demonstrated high affinity RNA binding sites were identified in the sequence. Therefore, the specific interaction between SP120 and topo IIβ is most likely to be based on some structural change in the C-terminal domain of SP120 that is induced by bound RNA, not on a simple RNA-mediated bridging mechanism.
A recent report has shown that human topo IIα interacts with RNA and its activity (decatenation and relaxation) is inhibited as a consequence (18). The association of RNA with topo IIα is unstable in high ionic strengths, which is different from the SP120 situation. We have also demonstrated in the present study that topo IIβ is inhibited by cellular RNA and the inhibition is canceled by formation of ternary complex with SP120. In a sense therefore, the protein can be regarded as an activator of the enzyme. Although the mechanism is different, a nuclear protein HMGB1 was shown to activate the relaxation activity of topo IIα through direct interaction (22).
Perhaps as a binder of both DNA and RNA, SP120 plays multiple roles in transcriptional regulation, both in repression and activation (23–26). The protein appears to be a pleiotropic regulator of gene expression and is very likely to take an important part in additional systems, as well. Recent study suggested that SP120 regulates the sonic hedgehog gene (Shh) expression by mediating a long-range interaction between distant chromatin sites (27), which resembles the action mode of topo IIβ on c2 sites (10). The result does not contradict the idea that topo IIβ cooperates with SP120 by direct interaction to catalyze a strand-passage event between distant genomic sites.
We have shown that topo IIβ protein is degraded when the cellular level of SP120 is lowered by RNAi. In other words, survival of topo IIβ in the cell nucleus depends on SP120. This is consistent with the observation that topo IIβ is particularly prone to proteolytic degradation during purification when its interaction with SP120 is disrupted. In addition, we have repeatedly observed a collateral expression of SP120 in differentiating cells and tissues in which topo IIβ expression becomes elevated (results not shown). As mentioned earlier, the degradation of topo IIβ in absence of SP120 becomes an obstacle when biological significance of the topo IIβ-SP120 complex is to be investigated by knocking down SP120 expression. Similar difficulty in this line of experimental scheme is represented by the embryonic lethality of hnRNP U hypomorphic mutants (28).
In summary, the present study proposes a novel regulation mechanism for type II DNA topoisomerases by identifying SP120 as the first protein factor that rescues topo IIβ from inhibition by RNA, protects the enzyme from degradation, and modulates its catalytic properties.
Supplementary Material
Acknowledgments
We thank Dr. Kunio Takeyasu (Kyoto University) for hybridoma cells producing monoclonal antibody 1-67D and Dr. Shinichi Nakagawa (RIKEN Advanced Research Institute) for Neuro-2a cells and mouse SP120 siRNA. We are grateful to Kazuko Kiyama for technical assistance.
This work was supported by grant-in-aid for scientific research funding (to K. T.) from the MEXT, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental materials and Figs. S1–S4.
- topo II
- topoisomerase II
- PSF
- polypyrimidine tract-binding protein-associated splicing factor
- NonO
- non-POU-domain-containing octamer-binding
- hnRNP U
- heterogeneous nuclear ribonucleoprotein U
- SAF-A
- scaffold attachment factor A
- SP120
- scaffold protein 120
- LEDGF
- lens epithelium-derived growth factor
- SAR
- scaffold attached region
- MAR
- matrix attachment region.
REFERENCES
- 1.Roca J., Berger J. M., Harrison S. C., Wang J. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang J. C. (1996) Annu. Rev. Biochem. 65, 635–692 [DOI] [PubMed] [Google Scholar]
- 3.Chung T. D., Drake F. H., Tan K. B., Per S. R., Crooke S. T., Mirabelli C. K. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9431–9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsutsui K., Tsutsui K., Okada S., Watanabe M., Shohmori T., Seki S., Inoue Y. (1993) J. Biol. Chem. 268, 19076–19083 [PubMed] [Google Scholar]
- 5.Tsutsui K., Tsutsui K., Hosoya O., Sano K., Tokunaga A. (2001) J. Comp. Neurol. 431, 228–239 [DOI] [PubMed] [Google Scholar]
- 6.Tsutsui K., Tsutsui K., Sano K., Kikuchi A., Tokunaga A. (2001) J. Biol. Chem. 276, 5769–5778 [DOI] [PubMed] [Google Scholar]
- 7.Yang X., Li W., Prescott E. D., Burden S. J., Wang J. C. (2000) Science 287, 131–134 [DOI] [PubMed] [Google Scholar]
- 8.Lyu Y. L., Wang J. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7123–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyu Y. L., Lin C. P., Azarova A. M., Cai L., Wang J. C., Liu L. F. (2006) Mol. Cell Biol. 26, 7929–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sano K., Miyaji-Yamaguchi M., Tsutsui K. M., Tsutsui K. (2008) PLoS One 3, e4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreyfuss G., Choi Y. D., Adam S. A. (1984) Mol. Cell Biol. 4, 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiledjian M., Dreyfuss G. (1992) EMBO J. 11, 2655–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romig H., Fackelmayer F. O., Renz A., Ramsperger U., Richter A. (1992) EMBO J. 11, 3431–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kipp M., Göhring F., Ostendorp T., van Drunen C. M., van Driel R., Przybylski M., Fackelmayer F. O. (2000) Mol. Cell Biol. 20, 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsutsui K., Tsutsui K., Okada S., Watarai S., Seki S., Yasuda T., Shohmori T. (1993) J. Biol. Chem. 268, 12886–12894 [PubMed] [Google Scholar]
- 16.Szemes M., Gyorgy A., Paweletz C., Dobi A., Agoston D. V. (2006) Neurochem. Res. 31, 237–246 [DOI] [PubMed] [Google Scholar]
- 17.Shav-Tal Y., Zipori D. (2002) FEBS Lett. 531, 109–114 [DOI] [PubMed] [Google Scholar]
- 18.Park S. W., Parrott A. M., Fritz D. T., Park Y., Mathews M. B., Lee C. G. (2008) Nucleic Acids Res. 36, 6080–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yugami M., Kabe Y., Yamaguchi Y., Wada T., Handa H. (2007) FEBS Lett. 581, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson J. R. (2000) Nat. Struct. Biol. 7, 834–837 [DOI] [PubMed] [Google Scholar]
- 21.Ameyar-Zazoua M., Souidi M., Fritsch L., Robin P., Thomas A., Hamiche A., Percipalle P., Ait-Si-Ali S., Harel-Bellan A. (2009) J. Biol. Chem. 284, 27974–27979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stros M., Baciková A., Polanská E., Stokrová J., Strauss F. (2007) Nucleic Acids Res. 35, 5001–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eggert H., Schulz M., Fackelmayer F. O., Renkawitz R., Eggert M. (2001) J. Steroid Biochem. Mol. Biol. 78, 59–65 [DOI] [PubMed] [Google Scholar]
- 24.Martens J. H., Verlaan M., Kalkhoven E., Dorsman J. C., Zantema A. (2002) Mol. Cell Biol. 22, 2598–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kukalev A., Nord Y., Palmberg C., Bergman T., Percipalle P. (2005) Nat. Struct. Mol. Biol. 12, 238–244 [DOI] [PubMed] [Google Scholar]
- 26.Onishi Y., Hanai S., Ohno T., Hara Y., Ishida N. (2008) Mol. Cell Biol. 28, 3477–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J., Ding J., Li Y., Ren K., Sha J., Zhu M., Gao X. (2009) Hum. Mol. Genet. 18, 3090–3097 [DOI] [PubMed] [Google Scholar]
- 28.Roshon M. J., Ruley H. E. (2005) Transgenic Res. 14, 179–192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.