Abstract
In mammals, the majority of DNA double-strand breaks are processed by the nonhomologous end-joining (NHEJ) pathway, composed of seven factors: Ku70, Ku80, DNA-PKcs, Artemis, Xrcc4 (X4), DNA-ligase IV (L4), and Cernunnos/XLF. Cernunnos is part of the ligation complex, constituted by X4 and L4. To improve our knowledge on the structure and function of Cernunnos, we performed a systematic mutagenesis study on positions selected from an analysis of the recent three-dimensional structures of this factor. Ten of 27 screened mutants were nonfunctional in several DNA repair assays. Outside amino acids critical for the expression and stability of Cernunnos, we identified three amino acids (Arg64, Leu65, and Leu115) essential for the interaction with X4 and the proper function of Cernunnos. Docking the crystal structures of the two factors further validated this probable interaction surface of Cernunnos with X4.
Keywords: DNA Repair, Protein Conformation, Protein Domains, Protein Folding, Protein Structure, Cernunnos, NHEJ, XLF, Xrcc4
Introduction
In higher eukaryotes, DNA double-strand breaks (dsb)4 are preferentially repaired by the nonhomologous end-joining (NHEJ) pathway composed of seven factors. The Ku70/Ku80 heterodimer recognizes the dsb and recruits the DNA-PK catalytic subunit (DNA-PKcs) forming the DNA-PK complex. DNA-ligase IV (L4) reseals the DNA break, in a complex including Xrcc4 (X4) and Cernunnos/XLF (Cernunnos), the most recently identified NHEJ factor (1–3). When necessary, the DNA ends are processed by the Artemis nuclease (4) prior to ligation. NHEJ defects in human and mice result in severe combined immune deficiency because of abortive V(D)J recombination, the diversification mechanism of T and B lymphocyte antigen receptor genes (5).
Co-immunoprecipitations (3) and yeast two-hybrid using X4 as a bait (1) demonstrated the physical interaction between Cernunnos and the X4-L4 complex. Cernunnos can also form homodimers (1, 3, 6, 7). Sensitive protein sequence analyses revealed similarities within the globular head domain, the central coiled-coil region, and the unstructured C-terminal domain of Cernunnos and X4 (1, 3), which were confirmed by the three-dimensional crystal structure of Cernunnos (6, 7). Cernunnos depends on L4 for its inclusion within the X4-L4 ligation complex, which itself depends on X4 for the stability of L4 (8–11). Although Cernunnos activity necessitates the entire coiled-coil region, the very C-terminal domain, which was not included in the three-dimensional structures, is dispensable (11) despite its in vitro DNA binding activity (8, 12, 13). Cernunnos does not have DNA-ligase activity on its own but stimulates the ligase activity of the X4-L4 complex (8, 9, 12, 14, 15). Accordingly, human patients with Cernunnos mutations present a major DNA repair defect leading to severe combined immunodeficiency and microcephaly (2), a phenotype recapitulated in Cernunnos knock-out mice to some extent (7).5 Cernunnos is therefore a “core” NHEJ component whose precise function during DNA repair still needs to be determined.
To define better the Cernunnos structure-function relationship, we performed site-directed mutagenesis on 27 residues, located in various regions of the protein or corresponding to positions mutated in Cernunnos-deficient patients. We initially screened these mutations for their impact on V(D)J recombination, a DNA recombination process that critically relies on effective NHEJ and selected 10 of them for additional analyses (expression, cellular localization, and dsb repair). We identified three amino acids (Arg64, Leu65, and Leu115), defining a probable interaction surface of Cernunnos with X4, which was further accredited using docking analysis. Altogether, these results propose a first mapping of Cernunnos/X4 interface and underline the major role of Cernunnos-X4 interaction on final steps of V(D)J recombination and on overall dsb repair process.
EXPERIMENTAL PROCEDURES
Cells
All cells were maintained in culture at 37 °C, 5% CO2 and 95% air. Cernunnos-deficient cells (Patient P2 described in Ref. 2) and OTel control cells are SV40-transformed and telomerase-immortalized skin fibroblasts (16).
DNA Cloning
The WT Cernunnos was PCR-amplified from the cDNA library and cloned in fusion with a V5 tag into pcDNA5.1 vector (Invitrogen). Mutagenesis was performed using Pfu Turbo Taq polymerase (Stratagene) according to the manufacturer's recommendations. All constructs were subcloned into the pMND-myc-ires-EGFP retroviral vector, using BD In-Fusion (Clontech). High titer virus supernatants and transduction were performed as described (2).
V(D)J Recombination Assays
In-chromosome V(D)J recombination assays were performed, with or without co-transfection of Cernunnos-expressing plasmids (2.5 μg), as described (2, 16).
Western Blotting (WB) and Immunoprecipitation (IP) Experiments
Cernunnos, WT, and point mutants expressing constructs were transfected in 293T cells. IPs were performed on precleared lysates using rabbit polyclonal anti-V5 (Abcam) and rabbit polyclonal anti-IgG (Santa Cruz Biotechnology) as described (3) and revealed by WB using mouse monoclonal anti-V5 (Invitrogen) or rabbit polyclonal anti-L4 (Acris), anti-X4 (Serotec), or anti-Cernunnos (Bethyl) antibodies. Protein loading was verified using mouse monoclonal anti-Ku70 and anti-myc (Santa Cruz Biotechnology) antibodies.
Immunofluorescence Detection
Patient P2's cells, transfected with the Cernunnos-V5 (WT, R57G, C123R, F117D, W119A, K26A, L115D, R64E, L65D, R178A, and L24A) cDNA constructs, were seeded on coverslides. 24 h later, cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 15 min. After each step, coverslides were rinsed three times with PBS. Cells were incubated for 20 min with PBS containing 0.1 m glycine, permeabilized with 0.5% Triton X-100 for 15 min, incubated for 30 min with PBS-BSA solution, and finally labeled with the mouse monoclonal anti-V5 (Invitrogen) antibody, followed by washes with PBS-BSA solution and incubation with Alexa Fluor 546 goat-F(Ab′) secondary anti-mouse IgG antibody (Molecular Probes). Slides were counterstained with 0.1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) and mounted in Fluorsave (Calbiochem). Slides were viewed by epifluorescence microscopy (Axioplan; Zeiss). Images were taken by a cooled charge-coupled device camera and processed using Adobe Photoshop 9.0.
Immunofluorescence Detection of Ionizing Radiation-induced Foci (IRIFs)
Cernunnos-deficient fibroblasts transduced with the empty MND-myc-ires-EGFP retrovirus or containing Cernunnos (WT, R57G, C123R, F117D, W119A, K26A, L115D, R64E, L65D, R178A, and L24A) were seeded on coverslides and γ-irradiated (2 Gray). DNA repair foci were labeled with mouse monoclonal anti-γH2AX (Millipore) antibody, revealed by the Alexa Fluor 546 goat-F(Ab′) secondary anti-mouse IgG antibody (Molecular Probes), 2 h and 24 h after IR as described (2).
Bleomycin Sensitivity Assay
Cernunnos-deficient cells were transduced with MND-myc-ires-EGFP virus expressing Cernunnos (mock, WT, R57G, C123R, F117D, W119A, K26A, L115D, R64E, L65D, R178A, and L24A) as above to generate mixed populations of untransduced cells and transduced cells expressing Cernunnos-myc and EGFP. Established cell lines were treated with increasing doses of bleomycin for 1 h. The frequency of GFP-expressing cells was analyzed by FACS over 14 days (Becton Dickinson FACScalibur flow cytometer). The selective advantage conferred by Cernunnos-myc was determined by calculating the ratio of GFP+ (transduced) to GFP− (untransduced) cells, set to 1 prior to bleomycin treatment.
Purification of Cernunnos (WT and Mutants) and X4 Proteins
Cernunnos 1–224 was subcloned into an expression vector derived from pETM13 (G. Stier, EMBL, Heidelberg) with a C-terminal His6 tag. Cernunnos mutants (L65D, R64E, and L115D) were generated using the QuikChange kit (Stratagene). Cernunnos 1–224 (WT and mutants) expressed in BL21(DE3) cells were grown at 37 °C and induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h. Cells were resuspended in buffer T (20 mm Tris, pH 8.0, 10 mm β-mercaptoethanol, 10% glycerol, 100 mm KCl). After lysis and clarification, lysate was brought to 1.5 m KCl. Sample was applied to a nickel-nitrilotriacetic acid-agarose (Qiagen). Bound Cernunnos 1–224 was washed with 150 ml of buffer T + 25 mm imidazole and step-eluted with buffer T + 220 mm imidazole. Pooled fractions were loaded onto a Resource Q column (GE Healthcare) equilibrated with 10 mm sodium phosphate, pH 8.0, 10 mm β-mercaptoethanol. Proteins were eluted with a linear gradient from 0 to 400 mm NaCl. Purified Cernunnos 1–224 was dialyzed in buffer P (25 mm sodium phosphate, pH 8.0, 150 mm NaCl, 10 mm β-mercaptoethanol) and 25 units of benzonase nuclease (Novagen). Human X4 1–203 was subcloned into the pExp vector with a His6 tag in N-terminal (17). X4 1–203 was expressed in Rosetta (DE3) cells (Novagen) and induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 16 h at 20 °C. Cell pellets were resuspended in 50 mm Tris, pH 8.0, 1 m KCl, 2 mm EDTA, 2 mm DTT, 1 mm PMSF, 10% glycerol. After lysis and clarification, X4 was purified by nickel affinity chromatography (GE Healthcare). The column was washed, and the sample was eluted with a gradient from 40 to 300 mm imidazole. X4 was further purified on a Hiprep 26/60 Sephacryl S-200 column (GE Healthcare) equilibrated with buffer P (25 mm sodium phosphate, pH 8.0, 150 mm NaCl, 10 mm β-mercaptoethanol).
Isothermal Titration Calorimetry (ITC)
The calorimetric titration experiments were performed using a VP-ITC (MicroCal). All solutions were degassed under vacuum prior to measurements. X4 at 20 μm in the calorimeter cell was titrated by WT or mutant Cernunnos at 200 μm at 10 °C by automatic injections of 10 μl. The first injection of 2 μl was ignored in the final data analysis. Integration of the peaks corresponding to each injection and correction for the base line were done using Origin-based software provided by the manufacturer. The fit of the data to a single-site interaction model results in the stoichiometry (n), equilibrium binding constant (Ka), and enthalpy of complex formation (ΔH). Control experiments were performed with WT or mutant Cernunnos injected into the buffer to evaluate the heat of dilution.
Docking of Cernunnos and X4 Crystal Structures
The coordinates of the X4 and Cernunnos proteins (Protein Data Bank codes 1FU1 and 2QM4) were used as starting structures for docking experiments. Regions exhibiting sequence conservation among different species and located at the solvent-exposed surface of both Cernunnos and X4 were analyzed using the Rate4Site algorithm (18). The docking procedure was performed following the guidelines published in Ref. 19. First, a set of structural models satisfying the constraints from conservation analyses were generated, clustered, and filtered using the HADDOCK software (20, 21). A refinement procedure was then performed on the most promising structural models using the RosettaDock software and in the absence of any constraints (22, 23). Extensive structural perturbations with RosettaDock calculated from the initial best HADDOCK model led to an ensemble of low energy conformations that clustered around a specific solution. Interestingly, this assembly can be reached with almost no modification of the unbound structures (root mean square deviation to the monomeric proteins is 1.0 Å and 1.13 Å for X4 and Cernunnos, respectively).
RESULTS
Rationale for Site-directed Mutagenesis
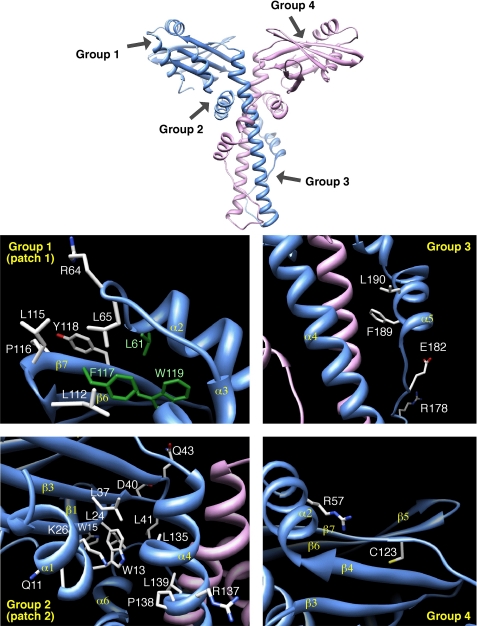
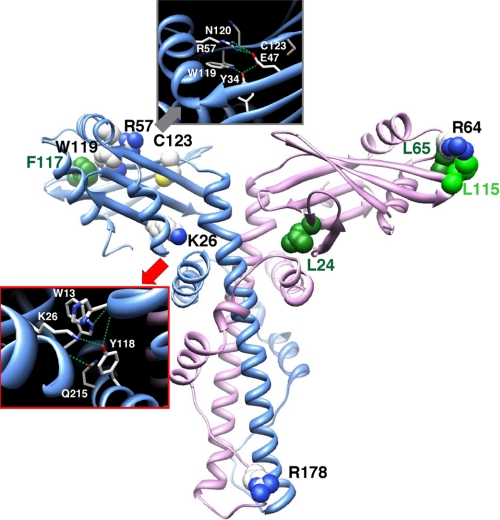
We defined four groups of Cernunnos residues to mutate based on our initial in silico protein sequence analysis (3) and published three-dimensional structures (6, 7). Group 1 consists of 9 amino acids (Leu61, Arg64, Leu65, Leu112, Leu115, Pro116, Phe117, Tyr118, and Trp119) located in the globular head of Cernunnos and highly conserved among Cernunnos orthologs (3). These positions are located at the end of helix α2 and in the β6–β7 β-hairpin at the distal end of the head domain. Leu61, Phe117, and Trp119 are buried into the hydrophobic core of the head domain (green in Fig. 1), whereas the other six positions define a contiguous patch exposed to the solvent (Fig. 1). The second group contains 13 residues located at the interface between the globular head domain and the beginning of the long helix forming the coiled-coil structure. They include Gln11, Trp13, and Trp15 in β1 strand; Leu24 and Lys26 in β2 strand; Leu37, Asp40, Leu41, and Gln43 in β3 strand and β3–β4 loop; and Leu135, Arg137, Pro138, and Leu139 in the beginning of the coiled-coil region. Importantly, Lys26 and Asp40 are very well conserved among Cernunnos orthologs. Amino acids with shared sequence similarities between X4 and Cernunnos and corresponding to the L4 binding region of X4 constitute group 3. This region is of particular interest as it is the site of a major difference in the three-dimensional structure of Cernunnos and X4 (6, 7). It is part of the hinge, which allows the folding back of helix α5 (or αE) onto helix α4 (or αD) and in the helix α5 (or αE) in Cernunnos, whereas it is linear in X4. The structure of this region is therefore more compact in Cernunnos, occluding a potential L4 binding site. Li et al. proposed that this region of Cernunnos could rearrange to adopt an X4-like extended coiled-coil structure (7). We designed two simple mutants affecting Glu182 and Arg178 in this region. The X4 F180IXXL cluster in X4 is critical for the interaction with L4 (24). Because these residues are conserved in Cernunnos (F189LXXF), we designed a double mutant (F189D/L190D) based on the one tested for X4 by Modesti et al (24). The last group (group 4) comprises two amino acids, Arg57 and Cys123, whose mutations were identified in human Cernunnos-deficient patients (Fig. 1; see “Discussion”) (2).
FIGURE 1.
Detailed views of the experimental three-dimensional structure of human Cernunnos (Protein Data Bank 2R9A (6)). This figure shows the positions of the amino acids of four groups, as selected for this point mutational analysis. Group 1 corresponds to two sets of conserved amino acids, which were predicted to cluster at the surface of the globular head domain (with Leu61, Phe117, and Trp119 (green) actually buried in the experimental structure). Group 2 corresponds to a set of amino acids predicted to be included in the interface between the head and stalk domains. Group 3 includes conserved amino acids in the stalk domain (L4 binding region of X4), whereas group 4 is formed by amino acids, which are associated with naturally occurring mutations.
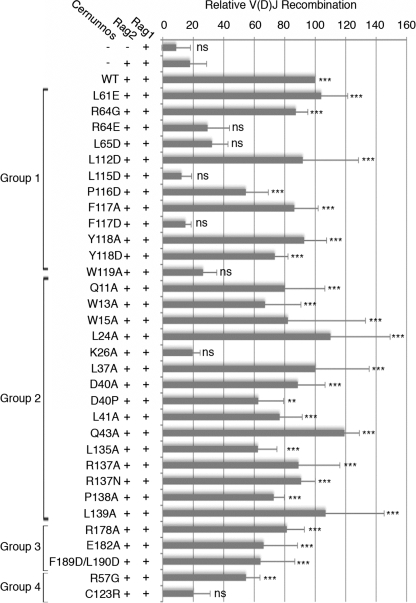
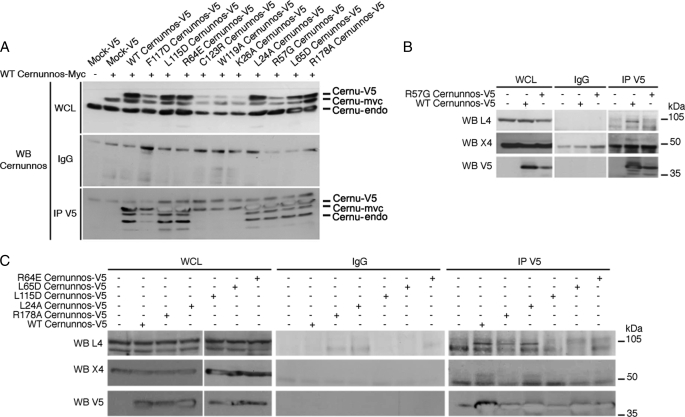
Each mutant was analyzed for its capacity to complement the intrachromosomal V(D)J recombination assay (2) in Cernunnos-defective fibroblasts (Fig. 2 and supplemental Fig. S1). Seven of the 27 tested mutants failed to complement in vivo V(D)J recombination (K26A, R64E, L65D, L115D, F117D, W119A, and C123R), and one (R57G) retained partial activity. Although the R178A mutant retained full activity in our initial in vivo screen, it was analyzed further because of its previously described low activity using an in vitro assay of mismatched DNA ends (6). L24A was kept as a control in subsequent experiments. None of these 10 selected mutants exerted a dominant negative effect on V(D)J recombination (supplemental Fig. S1).
FIGURE 2.
Point mutants of Cernunnos in V(D)J recombination. Mean results of three V(D)J recombination assays on chromosomal substrates, stably integrated in the genome of Cernunnos-deficient fibroblasts (Cer-RSS) are shown. Relative V(D)J recombination is calculated based on the recombination frequency obtained with transfection of Rag1, Rag2, and WT Cernunnos. ns, nonsignificant difference (p > 0.005); **, statistically significant (p < 0.005); ***, highly statistically significant (p < 0.001).
Cellular Phenotype of the Selected Mutant
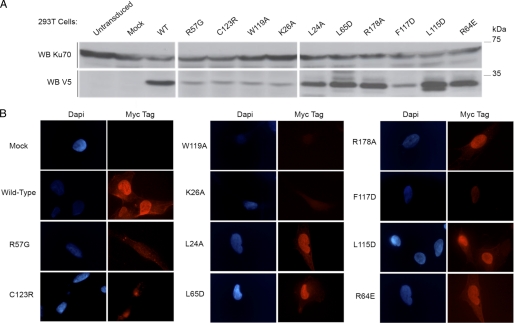
Protein expression level of the V5-tagged mutants was verified by WB analysis (Fig. 3A) on whole cell extracts from 293T cells. L24A, R64E, L65D, L115D, and R178A had an expression similar to WT. In contrast, K26A, R57G, F117D, W119A, and C123R mutations resulted in a weaker protein level most likely reflecting a decreased stability of these mutant proteins. We derived Cernunnos-defective fibroblasts stably expressing myc-Cernunnos mutant (WT or mutant) and co-expressing GFP, to verify proper cellular localization by immunofluorescence (Fig. 3B). Although the weak expression of R57G, C123R, W119A, K26A, and F117D makes it difficult to conclude with certainty on their proper nuclear localization, it appears that the L24A, R64E, L65D, L115D, and R178A mutants are correctly localized in the nucleus.
FIGURE 3.
Protein expression and localization of the selected point mutants of Cernunnos. A, 100 μg of total proteins obtained from 293T cells, transfected with V5-tagged, WT or mutant Cernunnos expression constructs or not, were analyzed by WB with the indicated antibodies. B, immunofluorescence experiments were performed on glass slide-immobilized Cernunnos-defective cells, which have been transduced with the pMND-myc-ires-GFP retrovirus containing Cernunnos (WT or the indicated mutant). Nuclei were stained with DAPI. The secondary anti-mouse Alexa Fluor 546 antibodies were exposed myc-tagged proteins.
DNA Repair Functions of Cernunnos Mutants
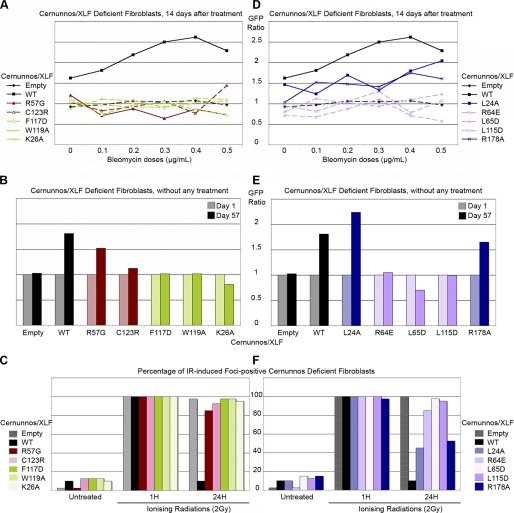
We next analyzed the capacity of each mutant to confer a selective advantage to Cernunnos-deficient cells upon treatment with increasing doses of the radiomimetic drug bleomycin (Fig. 4, A and B). A selective advantage, as judged by an increase in the frequency of GFP+-transduced cells, was obtained with Cernunnos-WT, but not with the mutants K26A, R57G, F117D, W119A, and C123R, whose expression was reduced compared with WT and which we consider as unstable (Fig. 4A). In addition, we noticed a persistence of γH2AX IRIFs (a hallmark of Cernunnos deficiency (2)) 24 h after IR in cells transduced with these mutants (Fig. 4C). Strikingly, a slight spontaneous selective advantage (in the absence of bleomycin treatment), comparable with that of WT, was noticed with the R57G mutant (Fig. 4B), suggesting a very weak DNA repair efficiency sufficient to cope with the few DNA lesions introduced during V(D)J recombination (Fig. 2) but not with a larger amount of dsb provoked by IR or bleomycin (Fig. 4, A–C). Using myc-tagged WT-Cernunnos and V5-tagged WT or mutants Cernunnos, we found that the R57G and F117D mutants retained the capacity to dimerize with WT Cernunnos. In case of K26A, W119A, and C123R, this analysis was hampered by the weak expression of these mutants. Moreover, in our experimental conditions, the R57G mutant has lost the interaction with X4 (Fig. 4B) (8). Phe117, Trp119, and Cys123, which are buried within the hydrophobic core, are likely to be involved in the correct conformation of the head (see Fig. 6, left panel). Lys26 and Arg57, which are not present in the hydrophobic core, may participate in the overall stabilization of the head domain through bonding with other critical amino acids (see Fig. 6, left panel, gray and red boxes; see “Discussion”). R57G can be considered as a hypomorphic mutation in human patients (2) given its general weak activity, whereas the other patient-driven mutation, C123R, can be considered as a complete loss of function (Figs. 2, 4, 5 and supplemental Fig. S1).
FIGURE 4.
DNA repair efficiency of Cernunnos mutants. A and D, survival of Cernunnos-deficient cells, transduced with the pMND vector containing WT or the indicated Cernunnos mutants, 14 days after bleomycin treatment is shown. B and E, survival of empty MND vector, WT or the indicated mutant Cernunnos, complemented Cernunnos-deficient cells, without any treatment. Cells were followed for 57 days. C and F, percentage of IRIF-positive cells is shown. 40 transduced cells were scored for DAPI and γH2AX foci and IR-untreated, 2 and 24 h after IR (2 Gray). A cell was considered IRIF-positive if it had >10 IRIFs.
FIGURE 6.
Structuring and interacting residues of human Cernunnos. Ribbon representation of the three-dimensional structure of human Cernunnos (Protein Data Bank code 2R9A (6)) highlights residues analyzed in this study and playing important role for the Cernunnos structure (“structuring” residues, left) and function (“interacting” residues, right). At the left are two boxes, with emphasis on the neighborhood of residues participating in the hydrophobic core of the head domain (gray box: Phe117, Trp119, Cys123) or in the stabilization of the head domain structure (gray box: Arg57; red box: Trp13, Lys26).
FIGURE 5.
Interactions between mutants of Cernunnos and the X4-L4 complex. A, 293T cells were co-transfected with myc-tagged WT Cernunnos and the V5-tagged Cernunnos (WT or the indicated mutant). The obtained whole cell lysates (WCL) were immunoprecipitated (IP) with rabbit irrelevant (IgG) and anti-V5 antibodies and were analyzed by WB with the indicated antibodies. B, 293T cells were transfected with V5-tagged WT or R57G Cernunnos. Whole cell lysates were immunoprecipitated with rabbit irrelevant (IgG) and anti-V5 antibodies and revealed as indicated. C, 293T cells were transfected with V5-tagged WT or mutant Cernunnos. Whole cell lysates were immunoprecipitated and treated as Fig. 4C.
A second group of mutants selected on their WT expression level (R64E, L65D, L115D, R178A, and L24A) was analyzed using the same procedures. L24A, which was initially considered as a control based on its full V(D)J recombination complementation, turned out to only partly complement the general dsb repair defect as judged by the intermediate selective advantage following bleomycin (Fig. 4D) and the incomplete disappearance of γH2AX foci 24 h after IR (Fig. 4F and supplemental Fig. S2). Consistent with this slightly reduced activity, L24A still provided a spontaneous selective advantage to Cernunnos-deficient cells as noted above with R57G (Fig. 4E). The same overall cellular phenotype was observed with R178A, whose effect on a V(D)J recombination was only minimal. These two mutants can thus be considered as hypomorphic. R64E, L65D, and L115D mutants, whose expression level and cellular localization were unperturbed, were unable to complement either the spontaneous or bleomycin-induced decrease in cell survival (Fig. 4, D–F) or the γH2AX foci kinetics following IR (Fig. 4F), arguing for a critical function of these residues. Interestingly, these three amino acids are exposed to the solvent and thus may participate in the interface with a Cernunnos partner (Fig. 6, right panel, see below and “Discussion”).
Cernunnos Interacts Directly and Specifically with X4 through a Conserved Site in the N-terminal Head
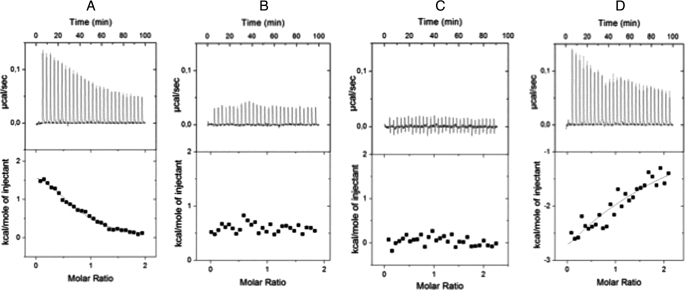
To clarify the loss of function of the mutants R64E, L65D, and L115D, we analyzed their interaction with the Cernunnos cognate partner X4 by co-immunoprecipitation experiments from 293T-transfected cells (Fig. 5). Although they were able to interact with WT full-length Cernunnos (Fig. 5A), they did not associate with the X4-L4 complex (Fig. 5C). To quantify further the strength of the interactions between the various Cernunnos mutants and X4, we purified X4 (amino acids 1–203) and Cernunnos (amino acids 1–224) fragments as well as the three Cernunnos mutants to homogeneity from Escherichia coli (supplemental Fig. S3) and verified their secondary structure content and Stokes radius by circular dichroism and analytical size exclusion chromatography, respectively (supplemental Figs. S4 and S5 and supplemental Materials and Methods). The WT and mutant Cernunnos fragments presented similar spectra with two minima at 208 and 222 nm that are characteristic of α+β proteins in agreement with the x-ray structure of Cernunnos (6, 7, 25, 26). These proteins presented a similar size exclusion chromatography profile in agreement with homodimerization as observed by crystallography. Altogether, these results attest for the correct folding of the in vitro produced various Cernunnos proteins. We performed calorimetry studies to measure the dissociation constant of the Cernunnos-X4 interaction in solution (Fig. 7). The binding reaction (ITC performed at 10 °C) between the WT Cernunnos and X4 gave an endothermic heat exchange (positive enthalpy) (Fig. 7A). Integrated thermograms were fitted to a one-site binding model and gave a dissociation constant of 4.2 μm in agreement with the previously 7.8 μm value determined by BiAcore (7). The isotherms obtained with R64E and L115D Cernunnos mutants confirmed their lack of interaction with X4 in the same experimental conditions (Fig. 7, B and C). The L65D mutant led to a 20-fold decrease (Kd > 80 μm) of affinity for X4 compared with the WT protein (Fig. 7D).
FIGURE 7.
Physical interactions between WT or mutant Cernunnos/XLF and Xrcc4 proteins measured by calorimetry. All experiments were performed at 10 °C. Upper panels show the binding isotherms. Lower panels show integrated heats, after subtraction of heats from control experiments. The black lines represent least-square fits of data. A, ITC binding isotherm obtained when WT Cernunnos was injected into the X4-containing cell. B, ITC binding isotherm obtained for Cernunnos L115D injected into the X4-containing cell. C, interaction between Cernunnos R64E and X4. D, interaction between Cernunnos L65D and X4.
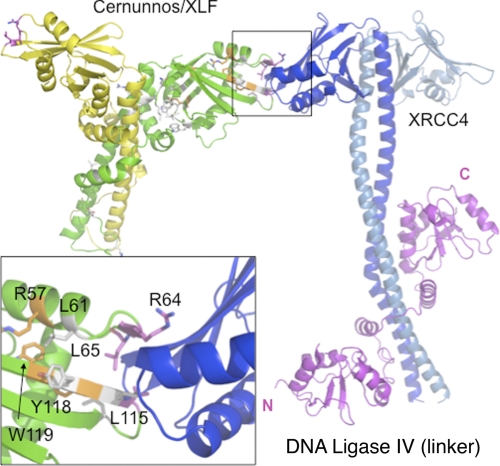
To explore further the sites involved in Cernunnos-X4 interaction, a molecular modeling of the complex was realized by docking the crystal structures of Cernunnos and X4 (see “Experimental Procedures,” supplemental Figs. S6 and S7, and supplemental Tables 1, 2, and 3) Based on these observations, we propose that Cernunnos and X4 interact through the distal part of their respective head domains (Fig. 8). The co-crystal structure of the two proteins should help clarify this issue in the future.
FIGURE 8.
Docking of Cernunnos and X4 crystal structures. Both dimeric proteins are represented as schematics with Cernunnos monomers in green and yellow and X4 monomers in blue and light blue. The model was extracted from a set of structural models satisfying the constraints from conservation. Interaction is mediated by the distal end of the head of Cernunnos and X4 and is remote from L4 (magenta) binding site that is located on the X4 coiled-coil. Positions of the variants analyzed in this study are represented in stick in gray for VDJ neutral position, in orange for unstable variant, and in magenta for interacting residues. The inset presents a focus of the interface proposed with the residues identified in this study as essential for the interaction.
DISCUSSION
Human Mutations of Cernunnos
Two naturally occurring missense mutations (R57G and C123R) have been described in humans (2). Our results confirmed that R57G results in a cytoplasmic mislocalization, as previously described (7, 8). According to the three-dimensional structure of human Cernunnos (6, 7), R57G probably participates in the stabilization of the head conformation through hydrogen bonding via its guanidinium group with the side chain of Glu47 and the main chain of Asn120 (Fig. 6, left panel, gray box). All of our functional assays are consistent with R57G being hypomorphic. The residue Cys123 is located in the hydrophobic core of the head domain (Fig. 6, left panel). The mutation C123R has been predicted to highly destabilize the head domain with consequences on the whole protein (7). We confirmed that prediction by visualizing an abnormal and granular cellular localization of this mutant.
Structure of the Head Domain
The three-dimensional experimental structures highlighted several very conserved Cernunnos residues, critical for the maintenance of the hydrophobic core such as Trp45 and Trp119 (6, 7). In this study, we noticed that W119A leads to the destabilization of the protein (Fig. 6, left panel, upper box), arguing for a structural rather than a “functional” role for this residue, as first noticed by the initial sequence alignments between Cernunnos and X4 (3). Trp119 probably contributes to the head stability through several contacts with hydrophobic residues such as Leu26, Phe121, and Val54 but also as it participates in hydrogen bond networks involving Tyr34 and Glu47. One particular feature of Cernunnos highlighted by the three-dimensional structures is the unexpected presence of a kink in the stalk domain, which reverses the direction of the polypeptidic chain and allows additional interactions between the head domain and the last helix (helix α6 or αF) of the C-terminal region (6, 7). Helix α6 indeed wedges itself between the head domain and helix α4 (or αD) with, as a consequence, a greater angle between the head and stalk domains than in X4. According to these structures, Trp13 and Lys26, which were selected in our mutational studies, participate in an intricate network of hydrogen bonds, involving Gln215 and Tyr218 in the C-terminal helix α6, as well as His134 in helix α4 (Fig. 6, left panel, red box). In our analysis, the mutation of Trp13 in alanine, however, did not have any impact on the V(D)J recombination efficiency and was not included in the subsequent experiments (Fig. 2). On the contrary, K26A led to the destabilization of the protein and to the loss of the DNA repair function (Fig. 6). These results demonstrated that the fold of the Cernunnos head domain, as well as the stabilization of this fold through intramolecular interactions, are critical for the function.
Mapping the Interaction with X4
The main conclusion driven by our mutational study resides in the better definition of the interacting surface of Cernunnos with X4. Leu115 had previously been shown to be critical for this interaction using native gels (6). We extended this observation by demonstrating the critical role of Arg64, Leu65, and Leu115 in this interaction and the functional consequences of its loss on Cernunnos function. Arg64, Leu65, and Leu115 belong to two patches of conserved amino acids (57–65 and 108–123 patches) (6). Their respective mutants preserve protein stability, cellular localization, and secondary structure content but abrogate the interaction with X4. The crystal structures of Cernunnos indeed established that these three amino acids are clustered at the surface of the head domain as we had proposed based on homology modeling, revealing the probable surface of interaction of Cernunnos with X4 (Fig. 6, right panel). Our results are in good agreement with two-hybrid and pulldown results that identify an interaction between N-terminal region (1–128) of Cernunnos with X4 (27). Two preceding studies report on the quantitative analysis of the Cernunnos/X4 interaction either through measurement on native gel (6) or on immobilized surface (7). We used microcalorimetry to quantify this interaction in solution. The isotherm analysis revealed an interaction between Cernunnos and X4 driven by entropy term with a weak unfavorable enthalpy term. Such behavior has already been observed in interaction between HIV proteases and its inhibitors, for example (28) and suggests an interface with reduced area (weak enthalpy term) and significant hydrophobic contribution (entropy-driven). Altogether, our thermodynamic data are in agreement with the rather small patch identified by molecular docking between Cernunnos and X4 and centered on residues Arg64, Leu65, and Leu115 for Cernunnos. Interestingly, the triad Arg64, Leu65, and Leu115 coincides perfectly with one of the most conserved amino acid patches in Cernunnos (supplemental Fig. S6) further arguing for the functional relevance of this X4 interacting region. The interaction site proposed for X4 is also in agreement with mutational analyses reported by Andres et al. (6) because this interface is close to positions Lys63, Lys65, and Lys99 on X4, whose substitutions disrupt the interactions with XLF. Andres et al. proposed a docking between X4 and Cernunnos/XLF different from our model because they predict a role for the X4 coiled-coil region. Further experimental structural data of the complex will help to precise the arrangement between X4 and Cernunnos. The arrangement of the two dimeric proteins in our docking model (supplemental Fig. S8) suggests that additional interactions may occur between the Cernunnos-X4 complex and additional Cernunnos or X4 monomers. The crystal structure of the combined Cernunnos and X4 should help clarify this issue.
Cernunnos-X4 interaction in vivo relies on the presence of L4 that makes an almost constitutive complex with X4. How may the presence of L4 affect the Cernunnos/X4 interactions described here is of particular interest. Additional studies in vivo and in vitro are needed to be definitive on this point. However, the low resolution structure of the X4-L4 complex recently reported by electronic microscopy (29) suggests that the large catalytic domain of L4 would occupy a position to one of the two X4 dimer heads. These data and our study thus suggest a stoichiometry of Cernunnos/X4/L4 that can be 1/2/2 with Cernunnos facilitating the load of two X4-L4 complexes close to the DNA dsb (supplemental Fig. S8).
Supplementary Material
Acknowledgment
We thank C. Tellier for vectors design used for X4 and Cernunnos purifications.
This work was supported by institutional grants from INSERM as well as grants from the Ligue National contre le Cancer (Equipe labellisée LA LIGUE), the Institut National du Cancer a/Cancéropôle Ile-de-France, Agence Nationale pour la Recherche (ANR'06), and Commissariat à l'Energie Atomique (LRC-CEA 40V).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. 1–8, Tables 1–3, and additional references.
P. Rivera-Muñoz, L. Malivert, P. Revy, and J.-P. de Villartay, unpublished observations.
- dsb
- double-strand break
- EGFP
- enhanced green fluorescent protein
- IP
- immunoprecipitation
- IRIF
- ionizing radiation-induced foci
- ITC
- isothermal titration calorimetry
- L4
- DNA·ligase 4
- NHEJ
- nonhomologous end-joining
- WB
- Western blotting
- X4
- Xrcc4.
REFERENCES
- 1.Ahnesorg P., Smith P., Jackson S. P. (2006) Cell 124, 301–313 [DOI] [PubMed] [Google Scholar]
- 2.Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M. C., Sanal O., Plebani A., Stéphan J. L., Hufnagel M., le Deist F., Fischer A., Durandy A., de Villartay J. P., Revy P. (2006) Cell 124, 287–299 [DOI] [PubMed] [Google Scholar]
- 3.Callebaut I., Malivert L., Fischer A., Mornon J. P., Revy P., de Villartay J. P. (2006) J. Biol. Chem. 281, 13857–13860 [DOI] [PubMed] [Google Scholar]
- 4.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N., Fischer A., de Villartay J. P. (2001) Cell 105, 177–186 [DOI] [PubMed] [Google Scholar]
- 5.de Villartay J. P., Fischer A., Durandy A. (2003) Nat. Rev. Immunol. 3, 962–972 [DOI] [PubMed] [Google Scholar]
- 6.Andres S. N., Modesti M., Tsai C. J., Chu G., Junop M. S. (2007) Mol. Cell 28, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Chirgadze D. Y., Bolanos-Garcia V. M., Sibanda B. L., Davies O. R., Ahnesorg P., Jackson S. P., Blundell T. L. (2008) EMBO J. 27, 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu H., Pannicke U., Schwarz K., Lieber M. R. (2007) J. Biol. Chem. 282, 11155–11162 [DOI] [PubMed] [Google Scholar]
- 9.Riballo E., Woodbine L., Stiff T., Walker S. A., Goodarzi A. A., Jeggo P. A. (2009) Nucleic Acids Res. 37, 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryans M., Valenzano M. C., Stamato T. D. (1999) Mutat. Res. 433, 53–58 [DOI] [PubMed] [Google Scholar]
- 11.Malivert L., Callebaut I., Rivera-Munoz P., Fischer A., Mornon J. P., Revy P., de Villartay J. P. (2009) Mol. Cell. Biol. 29, 1116–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hentges P., Ahnesorg P., Pitcher R. S., Bruce C. K., Kysela B., Green A. J., Bianchi J., Wilson T. E., Jackson S. P., Doherty A. J. (2006) J. Biol. Chem. 281, 37517–37526 [DOI] [PubMed] [Google Scholar]
- 13.Yano K., Morotomi-Yano K., Wang S. Y., Uematsu N., Lee K. J., Asaithamby A., Weterings E., Chen D. J. (2008) EMBO Rep. 9, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu P. Y., Frit P., Meesala S., Dauvillier S., Modesti M., Andres S. N., Huang Y., Sekiguchi J., Calsou P., Salles B., Junop M. S. (2009) Mol. Cell. Biol. 29, 3163–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai C. J., Kim S. A., Chu G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poinsignon C., Moshous D., Callebaut I., de Chasseval R., Villey I., de Villartay J. P. (2004) J. Exp. Med. 199, 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braud S., Moutiez M., Belin P., Abello N., Drevet P., Zinn-Justin S., Courçon M., Masson C., Dassa J., Charbonnier J. B., Boulain J. C., Ménez A., Genet R., Gondry M. (2005) J. Proteome Res. 4, 2137–2147 [DOI] [PubMed] [Google Scholar]
- 18.Pupko T., Bell R. E., Mayrose I., Glaser F., Ben-Tal N. (2002) Bioinformatics 18, Suppl. 1, S71–S77 [DOI] [PubMed] [Google Scholar]
- 19.Vajda S., Kozakov D. (2009) Curr. Opin. Struct. Biol. 19, 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Vries S. J., van Dijk A. D., Krzeminski M., van Dijk M., Thureau A., Hsu V., Wassenaar T., Bonvin A. M. (2007) Proteins 69, 726–733 [DOI] [PubMed] [Google Scholar]
- 21.Dominguez C., Boelens R., Bonvin A. M. (2003) J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 22.Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 23.Wang C., Bradley P., Baker D. (2007) J. Mol. Biol. 373, 503–519 [DOI] [PubMed] [Google Scholar]
- 24.Modesti M., Junop M. S., Ghirlando R., van de Rakt M., Gellert M., Yang W., Kanaar R. (2003) J. Mol. Biol. 334, 215–228 [DOI] [PubMed] [Google Scholar]
- 25.Junop M. S., Modesti M., Guarné A., Ghirlando R., Gellert M., Yang W. (2000) EMBO J. 19, 5962–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sibanda B. L., Critchlow S. E., Begun J., Pei X. Y., Jackson S. P., Blundell T. L., Pellegrini L. (2001) Nat. Struct. Biol. 8, 1015–1019 [DOI] [PubMed] [Google Scholar]
- 27.Deshpande R. A., Wilson T. E. (2007) DNA Repair 6, 1507–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd M. J., Luque I., Velázquez-Campoy A., Freire E. (2000) Biochemistry 39, 11876–11883 [DOI] [PubMed] [Google Scholar]
- 29.Recuero-Checa M. A., Doré A. S., Arias-Palomo E., Rivera-Calzada A., Scheres S. H., Maman J. D., Pearl L. H., Llorca O. (2009) DNA Repair 8, 1380–1389 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.