Abstract
A missense C1858T single nucleotide polymorphism in the PTPN22 gene recently emerged as a major risk factor for human autoimmunity. PTPN22 encodes the lymphoid tyrosine phosphatase (LYP), which forms a complex with the kinase Csk and is a critical negative regulator of signaling through the T cell receptor. The C1858T single nucleotide polymorphism results in the LYP-R620W variation within the LYP-Csk interaction motif. LYP-W620 exhibits a greatly reduced interaction with Csk and is a gain-of-function inhibitor of signaling. Here we show that LYP constitutively interacts with its substrate Lck in a Csk-dependent manner. T cell receptor-induced phosphorylation of LYP by Lck on an inhibitory tyrosine residue releases tonic inhibition of signaling by LYP. The R620W variation disrupts the interaction between Lck and LYP, leading to reduced phosphorylation of LYP, which ultimately contributes to gain-of-function inhibition of T cell signaling.
Keywords: Immunology, Protein Phosphorylation, Signal Transduction, T cell Receptor, Tyrosine-protein Phosphatase (Tyrosine Phosphatase), PEP, PTPN22, Autoimmunity
Introduction
A C1858T single nucleotide polymorphism in PTPN22 was first reported to be associated with type 1 diabetes (1) and rheumatoid arthritis (2) and was subsequently found to predispose humans to a wide range of autoimmune diseases (reviewed in Refs. 3 and 4). In Caucasian populations PTPN22 currently ranks in third and in second place in terms of single-gene contribution to the etiology of type 1 diabetes and rheumatoid arthritis, respectively (5). PTPN22 *T1858 acts as a dominant allele and confers significant predisposition to autoimmunity even when present as a single copy (3, 4).
PTPN22 encodes the lymphoid tyrosine phosphatase LYP,5 which acts as a critical negative regulator of T cell receptor (TCR) signaling (2, 6–9) through dephosphorylation of several key substrates, including the Src family kinases Lck and Fyn, ZAP70, and TCRζ (8, 10).
LYP and its mouse homolog PEST-enriched phosphatase (PEP) are ∼105-kDa proteins characterized by a ∼300-aa N-terminal protein-tyrosine phosphatase (PTP) domain and a ∼200-aa C-terminal domain that includes four putative polyproline motifs (termed P1–P4). The catalytic domain and the C-terminal domain are separated by a ∼300-aa region called the “interdomain.” A second shorter isoform of LYP called LYP2 has been identified in resting T cells (11). The most N-terminal P1 motif of LYP mediates the interaction of PEP/LYP with the SH3 domain of the protein-tyrosine kinase (PTK) Csk, also a negative regulator of TCR signaling (8, 12). The PTPN22 C1858T polymorphism causes an R620W substitution within the P1 motif of the protein. The pathogenic LYP-W620 variant exhibits reduced interaction with Csk (1, 2), shows increased phosphatase activity, and is a gain-of-function inhibitor of signaling in T cells (6, 13). T cells from type 1 diabetes and healthy subjects carrying the LYP-W620 variant show reduced production of interleukin-2 and other cytokines following TCR stimulation (6, 13, 14). Reduced TCR signaling has recently been recognized as a major risk factor for autoimmunity, and it affects tolerance through multiple mechanisms (15, 16). Here we sought to identify molecular mechanisms that contribute to the gain-of-function phenotype of LYP-W620 in T cells.
EXPERIMENTAL PROCEDURES
Plasmids
Full-length LYP-R620, LYP-W620, LYP2-R620, and their C227S mutants were cloned in the BamHI site of the plasmid pEF5-HA (17), whereas full-length PEP-R619 and PEP-W619 and their C227S mutants were cloned in the pEFHA vector (18). Point mutagenesis of LYP constructs was performed by PCR using primers containing the desired mutation. FLAG-tagged LYP-R620 C227S and N-terminal truncation mutants of LYP were performed by PCR using LYP-R620 or LYP-W620 in pEF5-HA (Δ288LYP) or in pEFHA (Δ399LYP and Δ517LYP) vector as templates. The primers were designed to anneal around the truncated regions of the gene and replace the HA tag with a FLAG tag. An S-tag (15 aa; see Ref. 19) was cloned for 3′ and was in-frame with the HA tag in the pEF5 vector, thus generating the pEF5HA-S vector. LYP mutants were then subcloned into the BamHI site.
Antibodies and Other Reagents
The anti-HA monoclonal Ab (clone 16B12) was from Covance (Berkeley, CA). The anti-Tyr(P) Ab (clone 4G10) was from Chemicon International (Temecula, CA). The anti-LYP polyclonal Ab was from R & D Systems (Minneapolis, MN). The anti-PEP polyclonal Ab has been previously described (20). The monoclonal anti-Lck, the polyclonal anti-Csk, and the polyclonal anti-Fyn were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-Lck polyclonal Ab, the monoclonal anti-Fyn, the monoclonal anti-Csk, the anti-huCD4, anti-moCD4, and anti-moCD28 were from BD Biosciences (Carlsbad, CA). The anti-ZAP70 Ab was from Invitrogen, whereas the anti-Itk Ab was from Cell Signaling Technology (Boston, MA). OKT3 (21) was purified from hybridoma supernatants. F(ab′)2 Ab and anti-mouse IgG used for cross-linking were purchased from Jackson Immunoresearch (West Grove, PA) and Upstate/Millipore (Billerica, MA), respectively. Agarose-conjugated M2-FLAG and PT66 Abs were from Sigma. The normal rabbit serum used for control precipitations was purchased from Thermo Fisher Scientific (Rockford, IL). The control goat IgG was purchased from Sigma. 4-Amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4,d]pyrimidine was from EMD Calbiochem (Gibbstown, NJ). The anti-Fyn small interfering RNA was a commercially available oligonucleotide from Santa Cruz, and the anti-Csk and anti-Lck small interfering RNAs were custom ordered from Dharmacon (Lafayette, CO) (22, 23). PfuUltra polymerase was from Stratagene (La Jolla, CA), and Taq polymerase was from Invitrogen. The AlexaFluor-conjugated anti-HA antibody was from Cell Signaling Technology (Boston, MA). The phycoerythrin-conjugated anti-phospho-ZAP70(Y319) antibody was from BD Biosciences. The allophycocyanin-conjugated anti-human CD69 was purchased from BioLegend (San Diego, CA).
Purification of Recombinant Full-length LYP
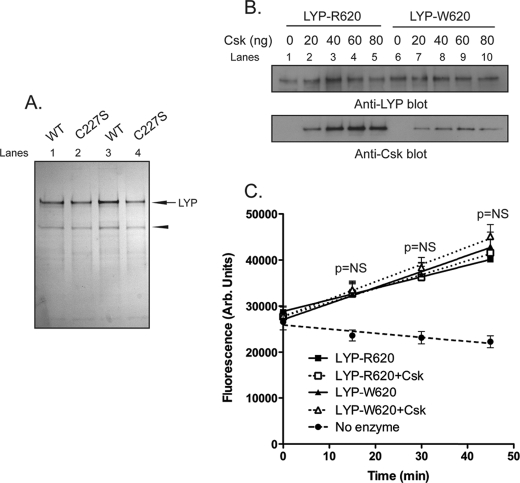
Full-length LYP-R620 and -W620 and their C227S mutants were cloned in the BamHI site of the pFastBac-HTa (Invitrogen) in-frame with a FLAG tag, and recombinant bacmids and baculoviruses were produced using the Bac-to-Bac® method (Invitrogen). Virus titers/times of incubation were optimized to obtain high expression of full-length recombinant proteins in Spodoptera fungiperda 9 cells. The protein was purified from lysates of insect cells using single-step affinity chromatography on FLAG-M2 beads and eluting with a combination of FLAG peptide and high concentrations of DTT. The final buffer was 50 mm Tris/HCl, pH 8.0, 0.5 mm EDTA, and 1 mm DTT. The purity of the recombinant proteins was more than 80% as assessed by silver staining of polyacrylamide gels (see Fig. 1). The yield of the isolation was ∼1 μg of full-length protein/150-mm plate of infected Spodoptera fungiperda 9 cells.
FIGURE 1.
Binding of Csk to LYP does not directly affect the phosphatase activity. A and B, in vitro reconstitution of the LYP·Csk complex. A, isolation of recombinant full-length LYP-R620 and LYP-W620. The figure shows a silver-stained polyacrylamide gel with 300 ng of purified LYP-R620 (lane 1), LYP-R620/S227 (lane 2), LYP-W620 (lane 3), or LYP-W620/S227 (lane 4). The arrow indicates LYP, and the arrowhead indicates a nonspecific protein that co-purifies with LYP in our preparation. B, LYP-R620 binds Csk more efficiently than LYP-W620. 25 ng of recombinant LYP-R620 (lanes 1–5) or LYP-W620 (lanes 6–10) bound to M2-FLAG beads were incubated with increasing amounts of recombinant His-Csk in 20 mm Tris/HCl, pH 7.4, 150 mm NaCl, and 1 mm EDTA overnight at 4 °C. The complex was washed twice with the same buffer and run on a polyacrylamide gel. C, binding to Csk does not directly affect the activity of LYP. The activity of 2 ng of recombinant LYP-R620 (squares) or LYP-W620 (triangles) was assayed in the presence (open symbols, dotted lines) or absence (filled symbols, continuous lines) of 60 ng of recombinant His-Csk using 0.4 mm 14LckpCAP394 peptide as substrate in 50 mm Bis-Tris, pH 6.0, 1 mm DTT. The diamond symbols and dashed line show fluorescence of control reaction carried out without adding any enzyme. The reaction was followed continuously to ensure initial rate conditions. The symbols show averages ± S.D. activity at various incubation time. Regression lines are shown. The significance of the differences has been calculated using analysis of variance. Identical results were obtained when the assays on LYP-R620 were repeated using a 50 mm Tris/HCl, 1 mm DTT, pH 7.4 buffer (data not shown). WT, wild type.
Cell Culture, Transfection, and Stimulation
Jurkat E6.1, JTAg (24), JCaM1, Hut78, and primary T cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mm l-glutamine, 1 mm sodium pyruvate, 10 mm HEPES, pH 7.3, 2.5 mg/ml d-glucose, 100 units/ml of penicillin, and 100 μg/ml streptomycin. COS-7 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum, 100 units/ml of penicillin, and 100 μg/ml streptomycin. Jurkat and JTAg transfections were performed as described (6). To generate JTAg cells stably expressing Δ288LYP, the cells were transfected with linearized plasmid and after 2 days were subjected to selection with 0.2 mg/ml Zeocin (Invitrogen). Stable transfectants were used for experiments at the polyclonal stage. COS cells were transfected using Lipofectamine and Plus reagent (Invitrogen). For pervanadate (PV) stimulation of JTAg, a PV solution was added to RPMI medium to achieve a 200 μm PV final concentration. C305 stimulation of JTAg and Hut78 cells was performed using C305 hybridoma (25) supernatant. For stimulation of primary human T cells, the cells were incubated in RPMI medium with OKT3 (1 μg/ml) and anti-huCD4 (1 μg/ml) or anti-huCD28 (1 μg/ml), followed by cross-linking with 10 μg/ml F(ab′)2 rabbit anti-mouse IgG for the time indicated in the figure. For stimulation of mouse thymocytes, the cells were incubated in RPMI medium with biotinylated anti-moCD3 (20 μg/ml) and biotinylated anti-moCD4 (20 μg/ml) or biotinylated anti-moCD28 (20 μg/ml) Ab, followed by cross-linking with streptavidin for the time indicated in the figure.
Mouse Models
129/Ola mice were purchased from Harlan (Indianapolis, IN). Fyn KO mice (B6;129S7-Fyntm1Sor/J) (26) were purchased from JAX® mice and services (Bar Harbor, ME; stock number 002385), whereas the conditional KO mice carrying deletion of Csk exclusively in CD4+ T cells have already been described (27). Thymi were isolated from 4–6-week-old mice, and thymocytes were purified using standard protocols. All of the procedures involving animals described in this manuscript were approved by the University of Southern California (protocol numbers 10714 and 10853) and LIAI (protocol number AP-NB1-0709) Institutional Animal Care and Use Committee.
Isolation of Primary Human T Cells and Genotyping
Anonymous buffy coats were purchased from Advanced Bioservices, LLC (Reseda, CA) or obtained from the blood bank of the Tor Vergata University Hospital in Rome, Italy. T cells were isolated by Lymphoprep (VWR) or Ficoll-Paque (GE Healthcare) gradient centrifugation followed by depletion of B cells and monocytes by anti-CD19 and anti-CD14 Dynabeads (Invitrogen). If necessary to induce expression of LYP, the cells were cultured in the presence of 10 ng/ml phorbol 12-myristate 13-acetate for 24 h. When needed, genomic DNA was extracted from 100 μl of peripheral blood using a genomic DNA extraction kit (Qiagen, Inc.), and the genotype at the LYP-R620W locus (single nucleotide polymorphism rs2476601) was determined by restriction fragment length polymorphism-PCR as described in Ref. 1. All of the procedures involving human subjects described in this manuscript were approved by the University of Southern California Institutional Review Board (exempt approval number 053029) or by the Ethical Committee of the Tor Vergata University Hospital.
Immunoprecipitations
For IPs, the cells were lysed in 20 mm Tris/HCl, pH 7.4, 150 mm NaCl, 5 mm EDTA with 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin/leupeptin, and 10 μg/ml soybean trypsin inhibitor. The lysis buffer also contained either 5 mm iodoacetamide or Na3VO4 in concentrations variable between 1 and 10 mm. 10 mm Na3VO4 was added when it was necessary to preserve the phosphorylation of active LYP/PEP in TCR-stimulated cells. The IP of LYP by PT66 or 4G10 Ab was not affected by the presence of Na3VO4 in the lysis buffer up to 5 or 10 mm, respectively (data not shown). Denaturing IPs were carried out by adding 1% SDS to the lysate. The lysates were diluted to reduce the final concentration of SDS to 0.1% immediately prior to the addition of the Ab.
Phospho-mass Spectrometry, In-gel Digestion, and Phosphopeptide Enrichment
Coomassie gel bands of interest were excised and chopped into small pieces and digested by trypsin as described elsewhere (28). In brief, the proteins were reduced in 10 mm DTT for 30 min at room temperature, alkylated in 55 mm iodoacetamide for 30 min at room temperature in the dark, and digested overnight at room temperature with 12.5 ng/μl trypsin (proteomics grade; Sigma). The digestion medium was then acidified to 2% trifluoroacetic acid. The supernatant was loaded onto a self-packed 200-μl pipette tip plugged with C18 material (3M EmporeTM C18 disk; 3M Bioanalytical Technologies, St. Paul, MN) filled first with 2 mm of TiO2 beads (GL Sciences Inc. Tokyo, Japan) and then another C18 disk. Bound peptides to the first upper C18 disk were desalted by a wash with 0.1% trifluoroacetic acid and then eluted onto underneath TiO2 beads with 30 mg/ml 2,5-dihydroxybenzoic acid in 80% acetonitrile and 0.1% trifluoroacetic acid. The bound peptides to TiO2 were washed once with the previous buffer and then once with a similar buffer without 2,5-dihydroxybenzoic acid. The peptides were eluted using 50 μl of 20% NH4OH in 40% acetonitrile in water, pH ≥ 10.5. Phosphopeptide mixture was almost dried using a SpeedVac concentrator (Concentrator 5301; Eppendorf AG, Hamburg Germany) and then resuspended in 0.1% of trifluoroacetic acid for LC-MS/MS analysis.
Mass Spectrometry Analysis
An LTQ-Orbitrap mass spectrometer (ThermoElectron, Bremen, Germany) coupled on-line to nano-LC (Ultimate, Dionex) was used. To prepare an analytical column C18 material (ReproSil-Pur C18-AQ 3 μm; Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) was packed into a spray emitter (75-μm inner diameter, 8-μm opening, 70-mm length; New Objectives) using a high pressure packing device (NanobaumeTM; Western Fluids Engineering). Mobile phase A consisted of water, 5% acetonitrile, and 0.5% acetic acid, and mobile phase B consisted of acetonitrile and 0.5% acetic acid. The five most intense peaks of the MS scan were selected in the ion trap for MS2 (normal scan, filling 5 × 105 ions, 500-ms maximum fill time for MS scan, 2 × 105 ions for MS2, multistage activation enabled, 200-ms maximum fill time, dynamic exclusion for 60 s). The raw files were processed using DTAsupercharge v.1.18. The generated peak lists were searched against the IPI human data base using Mascot 2.2 with the parameters: monoisotopic masses, 10 ppm on MS, 0.5 Da on MS/MS, electrospray ionization trap parameters, full tryptic specificity, cysteine carbamidomethylated as fixed modification, oxidation on methionine, phosphorylation on serine, threonine, or tyrosine, protein N-acetylation, and deamidation on glutamine and asparagine as variable modifications, with three missed cleavage sites allowed. The results were parsed through MSQuant 1.4.3a74. The identified phosphopeptide was manually validated.
Luciferase Assays
Luciferase assays were performed as described in Ref. 6. The difference in the ratio between firefly and Renilla luciferase activity in stimulated versus unstimulated cells (TCR-induced increased activation of reporter) was then plotted against the expression of LYP assessed by densitometric scanning of anti-HA blots of total lysates.
Flow Cytometry Assays
All of the samples were acquired on a FACSCanto II (BD Biosciences). The data were analyzed using FlowJo software (TreeStar, Ashland, OR). For induction of CD69, the cells were stimulated with 5 μg/ml OKT3 for 4 h at 37 °C. The cells were then washed, fixed, permeabilized, blocked with 10% mouse serum, and co-stained with AlexaFluor488-conjugated anti-HA antibody (Cell Signaling Technology) and allophycocyanin-conjugated anti-CD69 antibody. The cells overexpressing LYP were gated by comparing AlexaFluor488 fluorescence of cells transfected with HA-LYP versus cells transfected with vector alone.
Phosphatase Assays Using a Novel Peptidic Fluorogenic Substrate
Autodephosphorylation of PTPs is a well known phenomenon that complicates the assessment of the effect of phosphorylation on the phosphatase activity of PTPs. We observed initial autodephosphorylation of immunoprecipitated LYP already after 5 min of incubation in phosphatase buffer (Bis-Tris, pH 6.0, 5 mm DTT) (data not shown). To study the activity of phospho-LYP in conditions of fast enzyme autodephosphorylation, we used a fluorogenic peptide based on a Tyr(P) mimicking coumarin amino acid, which has recently been developed by the Barrios group (29). The enantiomerically pure, appropriately protected, phosphorylated coumaryl amino propionic acid (pCAP) can be incorporated into peptide substrates using standard N-(9-fluorenyl)methoxycarbonyl (Fmoc)-based solid phase peptide synthesis methodologies and undergoes enzymatic dephosphorylation by PTPs to CAP (29). Upon excitation ∼340 nm, CAP-containing peptides are over 104 times more fluorescent than pCAP-containing peptides (λem = 460 nm). Thus, PTP-catalyzed hydrolysis of pCAP-containing peptides results in a fluorogenic, continuous, and direct assay for PTP activity. The assay is extremely sensitive compared with the standard Tyr(P) peptide assay, and minimal spontaneous hydrolysis of the fluorogenic peptide ensures that the signal/background ratio of this assay is optimal even for short assay times (29, 30). Also because the assay is direct, continuous monitoring of linearity of the reaction is possible. We synthesized and purified the pCAP-containing peptide substrate ARLIEDNE(pCAP)TAREG (peptide 14LckpCAP394), a sequence based on residues around the Lck Tyr394 autophosphorylation site, which is a physiological substrate of LYP. A shorter version of this peptide has been reported to be an excellent LYP substrate (30), and we found that the recombinant catalytic domain of LYP dephosphorylates the 14LckpCAP394 peptide following Michaelis-Menten kinetics, with Km and kcat values equal or better than the corresponding 14-aa Tyr(P) peptide (kcat was 11.7 and 4.4 s−1, and Km was 81 and 71 μm for the pCAP and Tyr(P) peptides, respectively).6 For detection of phosphatase activity of LYP IPed from transfected cells, the IPs were washed in Bis-Tris, pH 6.0, and then resuspended in phosphatase buffer (Bis-Tris, pH 6.0, 5 mm DTT). After the addition of 0.4 mm peptide, the reaction was monitored continuously by measuring the increase in fluorescence (λex = 340 nm and λem = 460 nm) at 60-s intervals for 30 min. The activity measured in triplicate was corrected for the nonspecific signal of identical reactions performed also in triplicate without the addition of enzyme. The activity corrected for background fluorescence of substrate alone was then normalized for LYP expression as assessed by anti-HA Western blotting of fractions of IPs taken after resuspension in the final phosphatase buffer.
Graphs and Statistics
Graphs, curve fittings, and kinetic parameter calculations were performed using the Graphpad Prism software package (Graphpad, San Diego, CA). All of the S.D. values of the differences and ratios were calculated according to the error propagation rules described by Taylor (31).
RESULTS
Binding of Csk to LYP Does Not Directly Affect the Phosphatase Activity
To explain the gain-of-function phenotype of LYP-W620, we first assessed whether (i) the polymorphism directly affects the protein to induce increased enzymatic activity or (ii) simple binding of LYP to Csk is sufficient to inhibit the activity of the phosphatase. We purified recombinant full-length LYP-R620 and LYP-W620 and their inactive C227S mutants from lysates of insect cells (Fig. 1A). Because insect Csk lacks a functional SH3 domain (32), we obtained Csk-free LYP, which could be used to in vitro reconstitute the complex between LYP and Csk (Fig. 1B). When we measured the activity of recombinant LYP using an Lck-derived peptide as a substrate, we observed no significant difference between the activity of LYP-R620 and LYP-W620 (Fig. 1C). In control assays both LYP-C227S variants did not show any activity (data not shown, and see also Ref. 33). The addition of Csk to the reaction did not affect the activity of LYP-R620 or LYP-W620 (Fig. 1C). Despite the limitations imposed by our in vitro system (for example, differences in post-translational modifications of LYP between mammalian and insect cells), these data suggest that the polymorphism does not induce gain-of-function through a direct effect on the protein and that binding of Csk to LYP is not sufficient per se to induce changes in the enzymatic activity.
TCR Stimulation Induces Tyrosine Phosphorylation of LYP
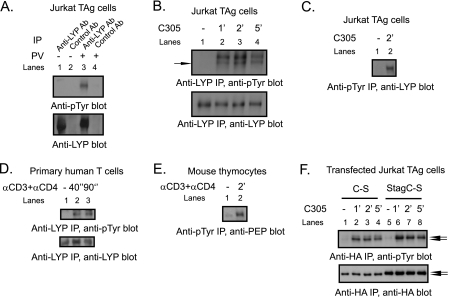
We next hypothesized that differences in post-translational modifications between LYP-R620 and LYP-W620 contribute to the gain-of-function phenotype of LYP-W620. Given that LYP forms a complex with Csk, a PTK, and the polymorphism affects the PTP/PTK interaction, we considered the possibility that the phosphatase could be phosphorylated on tyrosine. Several PTPs are regulated by tyrosine phosphorylation (34, 35), including PTP20 (36), which is homolog to LYP. We found that treatment of human JTAg cells (24) with pervanadate, a powerful PTP inhibitor, induced phosphorylation of LYP on tyrosine (Fig. 2A). TCR engagement also caused phosphorylation of LYP in the Jurkat E6.1, JTAg, and Hut78 T cell lines (Fig. 2, B and C, and data not shown) and in primary human T cells (Fig. 2D). The kinetics of LYP phosphorylation were fast (in JTAg cells it was detected after 15 s of stimulation; data not shown) and peaked between 1 and 2 min (Fig. 2, B–D). Endogenous PEP was also tyrosine-phosphorylated after TCR stimulation of mouse thymocytes (Fig. 2E). Transfected HA-tagged LYP and PEP were similarly phosphorylated after TCR engagement in JTAg cells (Fig. 2F and data not shown). Inactive PTP mutants are often used to study their regulation by phosphorylation on tyrosine residues, because PTPs often show a strong tendency toward auto-dephosphorylation (34, 36). Indeed, we observed that TCR-induced phosphorylation of transfected inactive C227S mutants of LYP and PEP was severalfold more prominent than that of the wild type phosphatases (data not shown). The identity of anti-Tyr(P)-reactive LYP and PEP immunoprecipitated (IPed) from lysates of transfected T cells was confirmed by using an S-tagged mutant of LYP and PEP (Fig. 2F and data not shown).
FIGURE 2.
TCR-induced tyrosine phosphorylation of LYP. A, PV treatment of JTAg cells induces LYP phosphorylation. LYP was IPed from lysates of JTAg cells left unstimulated (lanes 1 and 2) or treated with 200 μm PV for 15 min (lanes 3 and 4). The cell lysates were subjected to IP using an anti-LYP Ab (lanes 1 and 3) or to control precipitation using a control goat IgG (lanes 2 and 4). B–D, TCR stimulation induces LYP phosphorylation. B, LYP was IPed from lysates of JTAg cells left unstimulated (lane 1) or stimulated with C305 for 1, 2, or 5 min (lanes 2–4). C, anti-Tyr(P) IPs were performed from lysates of JTAg cells left unstimulated (lane 1) or stimulated with C305 for 2 min (lane 2). D, IP of LYP from lysates of primary human T cells left unstimulated (lane 1) or stimulated with anti-CD3 + anti-CD4 for 40 s (lane 2) or 90 s (lane 3). E, TCR-induced Tyr phosphorylation of PEP. Anti-Tyr(P) IPs were performed from lysates of primary mouse thymocytes left unstimulated (lane 1) or stimulated with anti-CD3 + anti-CD4 for 2 min (lane 2). Similar results were obtained in additional sets of experiments that were performed by immunoprecipitating PEP and blotting IPs with anti-Tyr(P) Ab and stimulating cells with anti-CD3 + anti-CD4 or anti-CD3 + anti-CD28 (data not shown). F, TCR stimulation of JTAg cells induces phosphorylation of transfected LYP. JTAg cells were transfected with the inactive C227S mutant of LYP-R620 (lanes 1–4) or with a construct expressing the same mutant in fusion with an additional N-terminal 15-aa S tag (lanes 5–8, the S tag slows the migration of HA-LYP on polyacrylamide gels). The cells were left unstimulated (lanes 1 and 5) or stimulated with C305 for 1 min (lanes 2 and 6), 2 min (lanes 3 and 7), or 5 min (lanes 4 and 8). The black arrows indicate the positions of HA-LYP and HA-S-LYP. Similar results were obtained in additional experiments where JTAg cells were transfected with an inactive C227S mutant of PEP in fusion or not with an N-terminal 15 aa S-tag (data not shown).
Autoimmune-associated LYP-R620W Polymorphism Affects TCR-induced Phosphorylation of LYP
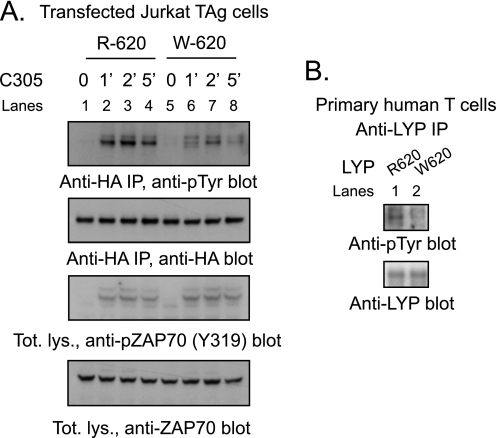
Next, we assessed whether LYP-R620 and LYP-W620 show any difference in TCR-induced phosphorylation. TCR stimulation induced much higher phosphorylation of LYP-R620 than of LYP-W620 in JTAg and Jurkat E6.1 cells (Fig. 3A and data not shown). Phosphorylation of LYP-R620 in resting cells was also higher than LYP-W620 (data not shown). The difference in phosphorylation between Arg620 and Trp620 was independent of the expression levels of the two LYP variants and was observed even at very low overexpression levels (data not shown). Similar results were obtained when the two homolog variants of PEP (PEP-R619 and PEP-W619) were transfected in JTAg cells (data not shown). Fig. 3B shows that LYP IPed from primary T cells isolated from healthy subjects of RR genotype have greater phosphorylation than LYP IPed from cells of RW genotype. We concluded that LYP and PEP are phosphorylated on tyrosine at levels that are detectable in resting T cells and are strongly induced in the early phase of TCR signaling. In transfected cells LYP-R620 also has greater phosphorylation on tyrosine than LYP-W620, in resting and stimulated T cells. We have preliminary evidence that this is true also in primary cells, although more experiments are warranted, especially comparing subjects of RR genotype with subjects of WW genotype.
FIGURE 3.
LYP-W620 is less phosphorylated in resting and TCR-stimulated T cells. A, IP from transfected JTAg cells. Anti-HA IPs were performed from lysates of JTAg cells transfected with LYP-R620 (lanes 1–4) or LYP-W620 (lanes 5–8). The cells were left unstimulated (lanes 1 and 5) or stimulated with C305 for 1 min (lanes 2 and 6), 2 min (lanes 3 and 7), or 5 min (lanes 4 and 8). The efficiency of TCR stimulation was similar in LYP-R620 and LYP-W620 transfected cells, as shown by anti-pZAP70(Y319) and anti-ZAP70 blots of total lysates. B, LYP-W620 is less phosphorylated than LYP-R620 in resting T cells. IPs of endogenous LYP from primary human T cells from healthy subjects of RR (lane 1) or RW (lane 2) genotype are shown. The observation was replicated on an additional couple of unrelated control subjects of RR and RW genotype.
Lck Phosphorylates LYP in T Cells
Next, we set out to assess which PTK is responsible for the phosphorylation of LYP in T cells. Incubation of JTAg cells with 10 μm of the Src family kinase inhibitor 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4,d]pyrimidine completely abolished the basal and TCR-induced phosphorylation of endogenous LYP (data not shown), suggesting that phosphorylation of LYP is dependent on the activity of Src family PTKs. When we co-transfected LYP with a set of candidate PTKs in COS cells, we observed that Lck, Fyn, and Csk could phosphorylate LYP, whereas ZAP70 and Itk could not (Fig. 4A). Lck was the most efficient LYP kinase in this assay and in additional in vitro kinase assays (Fig. 4B). RNA interference-mediated knockdown of Lck and Csk in T cells respectively abolished and reduced TCR-induced tyrosine phosphorylation of endogenous LYP (Fig. 4C and data not shown), whereas knockdown of Fyn did not seem to substantially affect LYP phosphorylation (Fig. 4D). The phosphorylation of PEP was conserved in Fyn−/− (26) thymocytes (Fig. 4E), further arguing against a role of Fyn as the major LYP kinase in T cells. On the other hand, Fig. 4F shows that the phosphorylation of PEP was reduced but not abolished in thymocytes isolated from mice carrying a conditional deletion of Csk in CD4+ cells. In these experiments all of the TCR-expressing thymocytes were virtually Csk KO. Assuming the absence of significant compensatory mechanisms in the cells analyzed, these data suggest that Lck and Csk are the major mediators of TCR-induced tyrosine phosphorylation of LYP in T cells, although we cannot formally exclude the role of other kinases.
FIGURE 4.
Phosphorylation of LYP in T cells depends on Csk and Lck. A, kinase screening in COS cells. COS cells were co-transfected with LYP-R620/S227 and Lck (lane 1), Fyn (lane 2), Csk (lane 3), ZAP70 (lane 4), Itk (lane 5), or empty vector (lane 6), and total lysates were subjected to denaturing anti-Tyr(P) IP. Similar results were obtained in separate experiments, performed by immunoprecipitating LYP and blotting with anti-Tyr(P) Ab (data not shown). B, in vitro phosphorylation of LYP by PTKs. LYP-R620/S227 was IPed from transfected COS cells and in vitro phosphorylated with recombinant Csk (lane 2), Fyn (lane 3), or Lck (lane 4). Lane 1 is a control reaction without PTK. C and D, phosphorylation of LYP is reduced by knockdown of Lck and Csk but is not affected by knockdown of Fyn. C, endogenous LYP was IPed from lysates of JTAg cells transfected with RNA interference oligonucleotides specific for Lck (lanes 1 and 2), Csk (lanes 5 and 6), or medium alone (lanes 3 and 4). The cells were left unstimulated (lanes 1, 3, and 5) or subjected to 2 min of stimulation with C305 (lanes 2, 4, and 6). Similar results were obtained by performing denaturing anti-Tyr(P) IPs followed by Western blotting with anti-LYP Ab (data not shown). D, endogenous LYP was IPed from lysates of JTAg cells transfected with a nontargeting oligonucleotide (lanes 1 and 2) or an oligonucleotide specific for Fyn (lanes 3 and 4). The cells were left unstimulated (lanes 1 and 3) or subjected to 2 min of stimulation with C305 (lanes 2 and 4). E, PEP phosphorylation is conserved in Fyn−/− thymocytes. Anti-PEP IPs were performed from lysates of thymocytes isolated from Fyn−/− mice (lanes 3 and 4) and from wild type littermates (lanes 1 and 2). The cells were left unstimulated (lanes 1 and 3) or stimulated with anti-CD3 + anti-CD4 for 1 min (lanes 2 and 4). Identical results were obtained by performing anti-PEP blot of anti-Tyr(P) IPs (data not shown). F, PEP phosphorylation is reduced in T cells from Csk−/− mice. Thymocytes were isolated from Csk conditional KO mice (lanes 1, 2, 5, and 6) and control littermates (lanes 3, 4, 7, and 8). The cells were left unstimulated (lanes 1–4) or stimulated with anti-CD3 + anti-CD4 for 1 min (lanes 5 and 6). The cell lysates were subjected to IP using an anti-Pep Ab (lanes 1, 3, 5, and 7) or to control precipitation using normal rabbit serum (lanes 2, 4, 6, and 8).
Lck Interacts with LYP in a Csk-dependent Manner
We next tested whether Lck co-precipitates with a deletion mutant of LYP, which is missing the catalytic domain (Δ288LYP, including aa 289–807). This mutant was used to exclude interactions caused by the substrate trapping activity of LYP (10). We found that Δ288LYP is able to form a complex with Csk as well as Lck but not with other PTKs in T cells. Both complexes were constitutive and apparently unaffected by TCR stimulation (Fig. 5A). The anti-Lck antibody could not precipitate LYP from lysates of the Lck-negative JCaM1 cells (37), further supporting the specificity of the interaction. Interestingly, the R620W mutation of Δ288LYP reduced the co-precipitation of Δ288LYP with Lck and Csk (Fig. 5B), although the effect of the mutation on the interaction with Lck was less dramatic than on the interaction with Csk. The co-precipitation between full-length LYP and Lck was decreased by the R620W mutation as well (Fig. 5C). Because the effect of the R620W mutation cannot be due to a direct interaction between the SH3 domain of Lck and the P1 domain of LYP (12),6 we assessed whether Csk affects the interaction between LYP and Lck. Fig. 5D shows that knockdown of Csk in T cells reduced the co-precipitation of LYP with Lck and abolished the difference between LYP-R620 and LYP-W620. Co-precipitation between Lck and PEP was also much decreased in thymocytes from Csk conditional KO mice (Fig. 5E). It is unlikely that the results shown in Fig. 5 (D and E) are due to decreased trapping of Lck by LYP, because knockdown/knockout of Csk rather leads to increased phosphorylation of Lck on Tyr394 (22, 27). The above-mentioned data support a model where recruitment of Csk to the P1 motif of LYP facilitates (i) the interaction between LYP and Lck and (ii) the phosphorylation of LYP on tyrosine residue(s). Reduced binding of Csk to LYP-W620 leads to reduced recruitment of Lck to the LYP protein complex and reduced phosphorylation of LYP-W620. In support of our model we also observed that (i) there was no significant in vitro co-precipitation between recombinant LYP and Lck purified from insect cell lysates (data not shown); (ii) a C-terminal truncation of LYP at aa 517 effectively abolished any interaction between LYP and Lck in T cells and in co-transfected COS cells (data not shown); and (iii) LYP-R620 and LYP-W620 purified from insect cell lysates showed identical low levels of phosphorylation on tyrosine, as assessed by Western blotting using an anti-Tyr(P) Ab (data not shown). The mechanism of Csk-mediated recruitment of Lck to LYP is unclear; however, as shown in Fig. 5F, overexpression of a kinase-dead mutant of Csk reduced co-precipitation between Lck and LYP, suggesting that recruitment of Lck depends at least in part on the kinase activity of Csk.
FIGURE 5.
Csk-dependent co-precipitation of Lck with LYP. A, Lck co-precipitates with Δ288LYP. JTAg cells were transfected with Δ288LYP, and IPs were performed from lysates of resting cells (lanes 1, 3, 5, 7, and 9) or cells stimulated for 2 min with C305 (lanes 2, 4, 6, 8, and 10) using an Ab against Lck (lanes 1 and 2), Fyn (lanes 3 and 4), Itk (lanes 5 and 6), Csk (lanes 7 and 8), or ZAP70 (lanes 9 and 10). B and C, more Lck co-precipitates with LYP-R620 than with LYP-W620. B, JTAg cells were transfected with Δ288LYP-R620 (lanes 1 and 4), Δ288LYP-W620 (lanes 2 and 5), or empty vector (lanes 3 and 6), and IPs were performed from lysates of resting cells using an Ab against Lck (lanes 1–3) or Csk (lanes 4–6). C, JTAg cells were transfected with LYP-R620 (lane 1), LYP-W620 (lane 2), or empty vector (lane 3), and IPs were performed from lysates of resting cells using an anti-Lck Ab. D–F, the co-precipitation of Lck with LYP is dependent upon Csk activity. D, JTAg cells were co-transfected with LYP-R620 (lanes 1–3 and 7–9) or LYP-W620 (lanes 4–6 and 10–12) and with RNA interference oligonucleotides specific for Csk (lanes 2, 5, 8, and 11) or nontargeting ones (lanes 1, 3, 4, 6, 7, 9, 10, and 12). The cells were left unstimulated (lanes 1–6) or stimulated for 2 min with C305 (lanes 7–12). The cell lysates were subjected to IP using an anti-Lck Ab. E, thymocytes were isolated from Csk conditional KO mice (lanes 2 and 4) and control littermates (lanes 1 and 3). The cells were left unstimulated (lanes 1 and 2) or stimulated with anti-CD3 + anti-CD4 for 1 min (lanes 3 and 4). The cell lysates were subjected to IP using an anti-PEP Ab. F, JTAg cells stably overexpressing Δ288LYP-R620 were transfected with HA-Csk (lane 1) or decreasing amounts (3, 2, or 1 μg of plasmid DNA) of the catalytically inactive mutant HA-Csk K222R (lanes 2–4) or vector alone (lane 5). The cells were stimulated for 2 min with C305, and IPs were performed from lysates using an anti-Lck Ab.
Tyr536 Is a Major Lck Phosphorylation Site of LYP
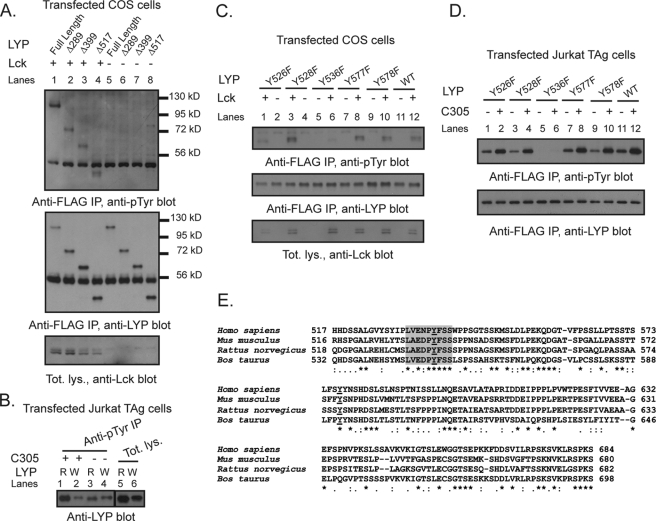
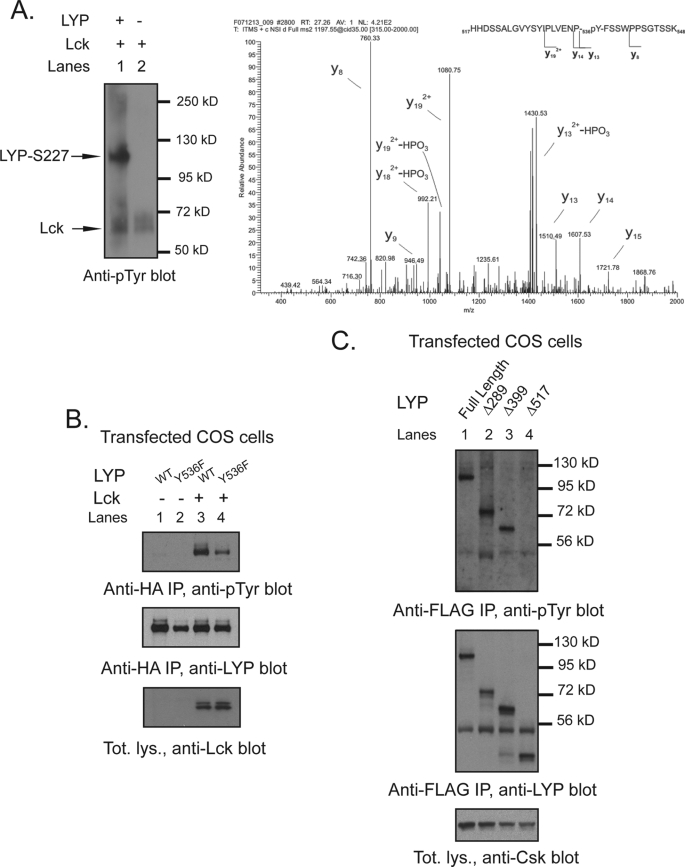
Hypothesizing that Lck is a major LYP kinase, we set out to map the Lck phosphorylation site(s) that are less phosphorylated in LYP-W620 than LYP-R620. We noticed that (i) N-terminal truncation of LYP up to aa 517 did not significantly affect the phosphorylation of LYP by Lck in COS cells (Fig. 6A) and (ii) the difference in TCR-induced phosphorylation between the two LYP variants was conserved after N-terminal truncation of the protein to aa 517 (Fig. 6B). We also noticed that transfected LYP2 was efficiently phosphorylated in T cells (data not shown). Because LYP2 lacks the last three C-terminal polyproline domains between aa 685 and 807 (11), we reasoned that a major Lck phosphorylation site affected by the R620W variation is likely located between aa 518 and 684. Thus, we mutagenized all five Tyr residues between aa 518 and 684 into Phe. Fig. 6 (C and D) shows that Y536F was the only mutation that abolished phosphorylation of Δ517-LYP-R620 by Lck in COS cells and the TCR-induced phosphorylation of Δ517-LYP-R620 in T cells. Interestingly, in the region between aa 518 and 684, Tyr536 is one of the only two Tyr residues that are highly conserved and the only one surrounded by a highly conserved aa motif (Fig. 6E). Analysis by Netphos (38) also indicated Tyr536 as the only putative Lck phosphorylation site in the aa 518–684 region (data not shown). Phospho-mass spectrometry analysis of recombinant LYP-S227 in vitro phosphorylated with Lck detected phosphate on Tyr536 (Fig. 7A), further supporting the idea that the 536 residue is a direct phosphorylation site for Lck. Phosphorylation of the Y536F mutant of full-length LYP by Lck in COS cells was dramatically decreased (Fig. 7B). We concluded that Tyr536 is a major Lck phosphorylation site in LYP. As mentioned, reduced phosphorylation of LYP-W620 by Csk could also play a role in the mechanism of action of the R620W mutation. However, N-terminal truncation of LYP at aa 517 abolished the phosphorylation of LYP by Csk in COS cells, suggesting that Tyr536 is not a Csk phosphorylation site (Fig. 7C).
FIGURE 6.
Tyr536 is a major Lck phosphorylation site of LYP. A and B, mapping by use of truncation mutants. A, anti-FLAG IPs were performed from lysates of COS cells transfected with FLAG-tagged full-length LYP-S227 (lanes 1 and 5) or the truncation mutants Δ288LYP (lanes 2 and 6), Δ399LYP (lanes 3 and 7), or Δ517LYP (lanes 4 and 8) alone (lanes 5–8) or together with Lck (lanes 1–4). B, JTAg cells were transfected with FLAG-tagged truncation mutants Δ517LYP-R620 (lanes 1, 3, and 5) or Δ517LYP-W620 (lanes 2, 4, and 6). Lanes 1–4, denaturing IPs were performed from lysates of unstimulated cells (lanes 3 and 4) or cells stimulated with C305 for 2 min (lanes 1 and 2), using anti-Tyr(P) Ab. Lanes 5 and 6 show total lysates. C and D, mapping of Tyr536 by site-specific mutagenesis. C, anti-FLAG IPs were performed from lysates of COS cells transfected with FLAG-tagged truncation mutants Δ517LYP-F526 (lanes 1 and 2), Δ517LYP-F528 (lanes 3 and 4), Δ517LYP-F536 (lanes 5 and 6), Δ517LYP-F577 (lanes 7 and 8), Δ517LYP-F578 (lanes 9 and 10), or Δ517LYP-WT (lanes 11 and 12) alone (lanes 2, 4, 5, 7, 9, and 11) or together with Lck (lanes 1, 3, 6, 8, 10, and 12). D, JTAg cells were transfected with FLAG-tagged truncation mutants Δ517LYP-F526 (lanes 1 and 2), Δ517LYP-F528 (lanes 3 and 4), Δ517LYP-F536 (lanes 5 and 6), Δ517LYP-F577 (lanes 7 and 8), Δ517LYP-F578 (lanes 9 and 10), or Δ517LYP-WT (lanes 11 and 12). IPs were performed from lysates of unstimulated cells (lanes 1, 3, 5, 7, 9, and 11) or cells stimulated with C305 for 2 min (lanes 2, 4, 6, 8, 10, and 12), using anti-FLAG M2 beads. E, Tyr536 is located within a highly conserved motif in the interdomain of LYP. Alignment of the aa 518–684 region of human LYP (Homo sapiens) with the homolog regions of LYP from mouse (Mus musculus), rat (Rattus norvegicus), and cow (Bos taurus) is shown. The alignment was performed using CLUSTALW2 (41). The figure shows the alignment in ALN/ClustalW2 format. Asterisks indicate identities, colons indicate conservative substitutions, and periods indicate semi-conservative substitutions. The underlined Ys indicate fully conserved Tyr residues in the region. The fully conserved motif around Tyr536 is highlighted in gray.
FIGURE 7.
Tyr536 is a direct phosphorylation site for Lck but not for Csk. A, detection of Tyr(P)536 by phospho-mass spectrometry. Recombinant LYP-S227 was in vitro phosphorylated with Lck, and the resulting protein mixture was separated by SDS-PAGE and stained by Coomassie. The LYP protein band was excised and digested in gel by trypsin. The resulting peptide mixture was analyzed by nanoLC-MS/MS. The panel shows the MS/MS spectrum of the peptide spanning residues 517–548 of LYP, showing fragment ions that unambiguously pinpoint phosphorylation of residue Tyr536. B, Tyr536 is a major Lck phosphorylation site. Anti-HA IPs were performed from lysates of COS cells transfected with LYP-WT and LYP-F536 alone (lanes 1 and 2) or with Lck (lanes 3 and 4). C, Tyr536 is an unlikely Csk phosphorylation site. Anti-FLAG IPs were performed from lysates of COS cells transfected with Csk together with FLAG-tagged full-length LYP-S227 (lane 1) or the truncation mutants Δ288LYP (lane 2), Δ399LYP (lane 3), and Δ517LYP (lane 4).
Y536F Mutation of LYP-R620 Induces Gain-of-Function Activity
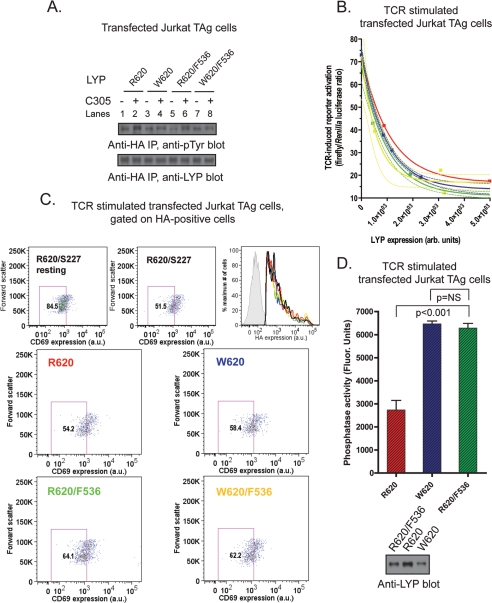
Fig. 8A shows that in T cells mutation of Tyr536 to Phe reduced the difference in phosphorylation between LYP-R620 and -W620. However, the Y536F mutation did not completely eliminate either the overall phosphorylation of LYP or the difference in phosphorylation between the two variants of LYP, suggesting that there is at least one additional site that is less phosphorylated in LYP-W620. Next, we assessed whether the mutagenesis of Tyr536 into a Phe in LYP-R620 could mimic at least in part the gain-of-function phenotype of LYP-W620. LYP-R620/F536 inhibited TCR signaling more efficiently than the wild type LYP-R620, as assessed by TCR-induced activation of an NFAT/AP1 luciferase reporter and induction of CD69 expression (Fig. 8, B and C). These data also show that the Phe536 mutation attenuated differences in TCR signaling inhibition between LYP-R620 and LYP-W620 in JTAg cells. Importantly, Fig. 8D shows that the Y536F mutation of LYP-R620 also led to increased LYP phosphatase activity. We concluded that Lck-mediated phosphorylation of LYP on Tyr536 plays an inhibitory role on the phosphatase activity and that reduced phosphorylation on Tyr536 leads to a gain-of-function that contributes to the phenotype shown by the autoimmune-predisposing LYP-W620. Reduced phosphorylation of LYP-W620 at additional sites that are targets of Lck or other kinase activities might well contribute to the same phenotype and should be investigated.
FIGURE 8.
Reduced phosphorylation of Tyr536 on LYP-W620 leads to gain-of-function inhibition of TCR signaling. A, Tyr536 is more phosphorylated in LYP-R620 than LYP-W620. Anti-HA IPs were performed from lysates of JTAg cells transfected with LYP-R620 (lanes 1 and 2), LYP-W620 (lanes 3 and 4), LYP-R620/F536 (lanes 5 and 6), or LYP-W620/F536 (lanes 7 and 8) constructs. The cells were left unstimulated (lanes 1, 3, 5, and 7) or stimulated with C305 for 2 min (lanes 2, 4, 6, and 8). B–D, phosphorylation on Tyr536 inhibits LYP in TCR signaling. B, activation of an NFAT/AP1 reporter. JTAg cells were co-transfected with a 3xNFAT/AP1 firefly luciferase reporter, a control Renilla luciferase reporter, and LYP-R620, LYP-W620, LYP-R620/F536, or LYP-W620/F536. The cells were stimulated for 7 h with OKT3 and then lysed, and luciferase activity was measured on lysates. The average ± S.D. stimulation-induced increase in the ratio between firefly and Renilla luciferase activities of lysates of cells transfected with LYP-R620 (red squares and line), LYP-W620 (blue triangles and line), LYP-R620/F536 (green circles and line), or LYP-W620/F536 (yellow squares and line) was plotted versus LYP expression in same lysates as assessed by anti-HA blot. The lines are nonlinear fitting of data to an exponential decay equation, and 90% confidence intervals are shown (dashed lines). The data are representative of two experiments with similar results. C, induction of CD69. JTAg cells were transfected with HA-LYP-R620, HA-LYP-W620, HA-LYP-R620/F536, HA-LYP-W620/F536, HA-LYP-R620/S227, or vector alone. The cells were left unstimulated or stimulated with OKT3 for 4 h and were co-stained with an AlexaFluor488-conjugated anti-HA antibody and an allophycocyanin-conjugated anti-CD69 antibody. Live cells were gated by forward and side scatter and further gated for CD69 expression by comparison with the lower half of activated cells transfected with catalytically inactive LYP (HA-LYP-R620/S227). The corresponding percentage of gated T cells is shown in each box. Levels of overexpression of LYP mutants are shown as histograms of AlexaFluor488 fluorescence of HA-positive cells transfected with HA-LYP-R620 (red), HA-LYP-W620 (blue), HA-LYP-R620/F536 (green), HA-LYP-W620/F536 (yellow), or HA-LYP-R620/S227 (black). The gray-shaded graph shows HA-negative cells. D, phosphorylation on Tyr536 inhibits the phosphatase activity of LYP. JTAg cells were transfected with HA-LYP-R620, HA-LYP-W620, or HA-LYP-R620/F536. Anti-HA IPs were performed from lysates of cells stimulated with C305 for 2 min to maximize phosphorylation of LYP, and phosphatase activity was assessed continuously using the 14LckpCAP394 peptide as substrate. The histogram shows the average activity ± S.D. of IPed LYP-R620 (red column), LYP-W620 (blue column), or LYP-R620/F536 (green column) normalized for LYP expression by densitometric scanning of anti-HA blots of fractions of the IPs (see bottom panel). The time of reaction was optimized to ensure initial rate conditions and avoid any significant auto-dephosphorylation of the phosphatase (data not shown). The data are representative of two experiments with identical results.
DISCUSSION
PTPN22 is currently classified as a “shared autoimmunity gene,” and its association with human autoimmunity is robust and population-independent (3). In Caucasian populations the contribution of PTPN22 to the genetic risk of autoimmunity is substantial: PTPN22 currently ranks in third place (after the human leukocyte antigen and the insulin genes) and in second place (after the human leukocyte antigen) in terms of single-gene contribution to the etiology of type 1 diabetes and rheumatoid arthritis, respectively (5).
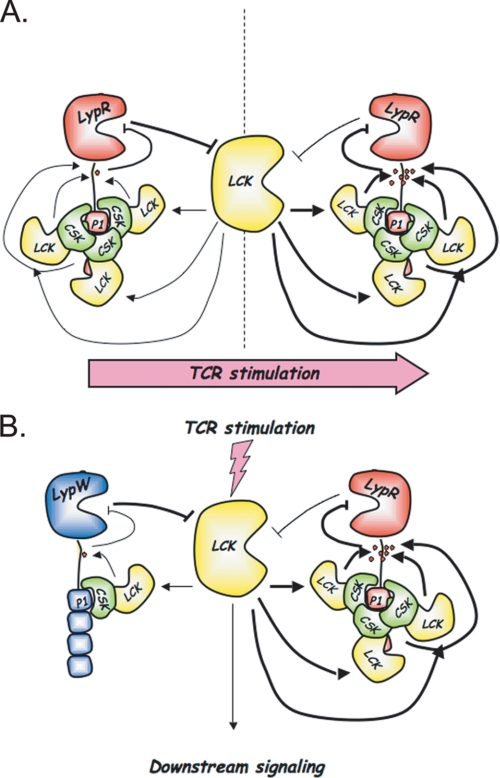
The R620W polymorphism does not result in alterations of PTPN22 mRNA levels in primary T cells (39), and the increased phosphatase activity is so far the only known functional consequence of the R620W genetic variation. Here we report the first study of the functional effects of the autoimmune-associated LYP-R620W variation at the molecular level. Our current working model is summarized in Fig. 9 and is based on several observations, which are discussed in detail below.
FIGURE 9.
Model of regulation of LYP and Lck activity by phosphorylation of LYP on Tyr536.
First, we found that LYP is phosphorylated on tyrosine at levels that are detectable in resting T cells and are strongly induced in the early phase of TCR signaling. We believe that Lck is a major LYP kinase in T cells, based on the following evidence: (i) knockdown of Lck reduced the phosphorylation of LYP; (ii) Lck was an efficient LYP kinase in vitro; and (iii) ZAP70 and Itk did not phosphorylate LYP, whereas phosphorylation of LYP was conserved in Fyn KO cells. Other kinases might well phosphorylate LYP on tyrosine residues in T cells, the most obvious candidate being Csk. Indeed, we also observed a reduction of LYP phosphorylation after Csk knockdown and in Csk KO cells.
Second, we showed that Lck forms a complex with LYP. This complex seems to be constitutive, although more experiments are needed to exclude an effect of TCR stimulation on the interaction between Lck and LYP. We believe that the interaction between LYP and Lck is dependent upon/facilitated by recruitment of Csk to the P1 motif of LYP and in turn facilitates the phosphorylation of LYP by Lck. This model is supported by the following evidence: (i) a truncation mutant of LYP missing the P1–P4 motifs did not show any interaction with Lck; (ii) knockdown of Csk reduced the interaction of LYP with Lck and the phosphorylation of LYP; and (iii) importantly, in T cells LYP-W620 showed reduced interaction with Lck and reduced basal and TCR-induced phosphorylation. Further investigation at the molecular level is needed to assess how Csk recruits Lck to LYP and whether Lck interacts with LYP or Csk. The data shown in Fig. 5F suggest that Csk might recruit Lck to LYP by inducing phosphorylation of LYP on a secondary site. Indirect recruitment of Lck through a phospho-site could also explain the reduced effect of the R620W mutation in our Lck co-precipitation assays compared with the Csk ones. Alternatively, it is possible that Csk recruits Lck to LYP through additional protein interactors. Considering the known inhibitory effect of Csk on Lck activity, Csk-mediated recruitment of Lck to LYP might contribute to keep LYP activity high in resting cells by increasing the dependence of LYP inhibition on TCR-induced Lck activation levels.
Third, we mapped Tyr536 in the interdomain of LYP as one of the major Lck phosphorylation sites and showed that phosphorylation of Tyr536 is reduced by the R620W mutation. Our hypothesis is that reduced phosphorylation on Tyr536 contributes to the gain-of-function phenotype of LYP-W620 and is supported by the following evidence: (i) a Y536F mutant of LYP-R620 showed gain-of-function inhibitory activity on TCR signaling and (ii) the Y536F mutation increased the phosphatase activity of LYP-R620 to levels close to LYP-W620. Little is currently known about the structure and function of the LYP interdomain, where the Tyr536 residue is located. Further experimental evidence is needed to assess whether Tyr(P)536 has a direct effect on the activity or requires/recruits further components of the LYP protein complex.
LYP is well known to negatively regulate Lck activity in effector T cells through dephosphorylation of Tyr394 in the catalytic domain of the kinase (20). Our data now suggest that Lck in turn regulates LYP activity through phosphorylation of the inhibitory Tyr536 residue in the interdomain of the phosphatase. Mutation of Tyr536 to Phe leads to gain-of-function inhibition of T cell activation, supporting the idea that reciprocal inhibition between LYP and Lck is an important modulator of TCR signaling. The data suggest that after TCR engagement, increased phosphorylation of LYP by Lck acts as a positive feedback loop, by further boosting the activation of Lck (Fig. 9A). By reducing tonic inhibition of signaling by LYP at the time of the initial wave of kinase activation, this feedback system might be critically important to ensure correct TCR signal propagation. Moreover, in resting T cells the phosphorylation levels of LYP could modulate the T cell activation threshold.
By interfering with the formation of the complex between LYP and Csk, the autoimmune-associated LYP-R620W polymorphism causes reduced interaction between LYP and Lck, with subsequent reduced phosphorylation of LYP on Tyr536 and gain-of-function inhibition of TCR signaling (Fig. 9B). Such reduced feedback between Lck and LYP likely plays a role in mediating the gain-of-function signaling inhibition of LYP-W620. However, the contribution of other mechanisms cannot be excluded at present. First of all, there are additional tyrosine residues that are less phosphorylated in LYP-W620 compared with LYP-R620. In addition, the polymorphism could affect the interaction of LYP with unknown proteins and/or its recruitment to specific subcellular fractions/compartments. Reduced co-precipitation with Lck might not be sufficient to explain all of the reduction in phosphorylation of LYP-W620 compared with LYP-R620. Also, Lck might not be the only kinase acting on the sites that are less phosphorylated in LYP-W620. Possible effects of the R620W variation on the activity of Csk are also worthy of further investigation, because they could contribute to the gain-of-function inhibition of signaling observed in carriers of LYP-W620.
In conclusion, we reported here the first molecular model for the gain-of-function phenotype of the LYP-W620 variant. Our model is supported by several lines of evidence but also leaves some open questions. For example, our data suggest that Csk induces indirect inhibition of LYP activity in T cells, a scenario in apparent disagreement with the results of previous studies (8, 9), which concluded that PEP and Csk act synergistically as TCR signaling inhibitors. Functional differences between human LYP and mouse PEP could underlie this discrepancy. For example, because Tyr536 is located in one of the regions with the lowest overall homology between human and mouse LYP (Ref. 11; see also Fig. 6E), it is possible that phosphorylation of Tyr536 has different effects on the activity of LYP and PEP. Alternatively, it is possible that the stoichiometry of the interaction between PTPN22 and Csk is different between human and mouse cells. Because multiple TCR signaling regulators are known to interact with the Csk-SH3 domain in T cells (for example see Ref. 40), competition/compensation phenomena and the relative affinity of the various protein-protein interactions involved should be taken into account when interpreting results obtained in overexpression systems.
Acknowledgments
We are grateful to Tomas Mustelin and Torkel Vang for sharing plasmids and to Amy Barrios for providing the pCAP peptide.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI070544 (to N. B.). This work was also supported by Juvenile Diabetes Research Foundation Grant 1-2005-342. This is manuscript 1298 from the La Jolla Institute for Allergy and Immunology.
This work is dedicated to the memory of Dr. Alceo Floris.
E. Fiorillo, V. Orrú, S. M. Stanford, Y. Liu, M. Salek, N. Rapini, A. D. Schenone, P. Saccucci, L. G. Delogu, F. Angelini, M. L. M. Bitti, C. Schmedt, A. C. Chan, O. Acuto, and N. Bottini, unpublished data.
- LYP
- lymphoid tyrosine phosphatase
- Ab
- antibody
- CAP
- coumaryl amino propionic acid
- Csk
- c-Src tyrosine kinase
- DTT
- dithiothreitol
- EF
- elongation factor
- hu
- human
- HA
- hemagglutinin
- IP
- immunoprecipitation
- IPed
- immunoprecipitated
- Itk
- interleukin-2-inducible T cell kinase
- KO
- knock-out
- mo
- mouse
- pCAP
- phosphocoumaryl amino propionic acid
- PEP
- PEST-enriched phosphatase
- PTK
- protein-tyrosine kinase
- PTP
- protein-tyrosine phosphatase
- PV
- pervanadate
- TCR
- T cell receptor
- aa
- amino acid(s)
- LC
- liquid chromatography
- MS
- mass spectrometry
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Bottini N., Musumeci L., Alonso A., Rahmouni S., Nika K., Rostamkhani M., MacMurray J., Meloni G. F., Lucarelli P., Pellecchia M., Eisenbarth G. S., Comings D., Mustelin T. (2004) Nat. Genet. 36, 337–338 [DOI] [PubMed] [Google Scholar]
- 2.Begovich A. B., Carlton V. E., Honigberg L. A., Schrodi S. J., Chokkalingam A. P., Alexander H. C., Ardlie K. G., Huang Q., Smith A. M., Spoerke J. M., Conn M. T., Chang M., Chang S. Y., Saiki R. K., Catanese J. J., Leong D. U., Garcia V. E., McAllister L. B., Jeffery D. A., Lee A. T., Batliwalla F., Remmers E., Criswell L. A., Seldin M. F., Kastner D. L., Amos C. I., Sninsky J. J., Gregersen P. K. (2004) Am. J. Hum. Genet. 75, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottini N., Vang T., Cucca F., Mustelin T. (2006) Semin. Immunol. 18, 207–213 [DOI] [PubMed] [Google Scholar]
- 4.Gregersen P. K., Lee H. S., Batliwalla F., Begovich A. B. (2006) Semin. Immunol. 18, 214–223 [DOI] [PubMed] [Google Scholar]
- 5.Todd J. A., Walker N. M., Cooper J. D., Smyth D. J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S. F., Payne F., Lowe C. E., Szeszko J. S., Hafler J. P., Zeitels L., Yang J. H., Vella A., Nutland S., Stevens H. E., Schuilenburg H., Coleman G., Maisuria M., Meadows W., Smink L. J., Healy B., Burren O. S., Lam A. A., Ovington N. R., Allen J., Adlem E., Leung H. T., Wallace C., Howson J. M., Guja C., Ionescu-Tîrgovişte C., Simmonds M. J., Heward J. M., Gough S. C., Dunger D. B., Wicker L. S., Clayton D. G. (2007) Nat. Genet. 39, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vang T., Congia M., Macis M. D., Musumeci L., Orrú V., Zavattari P., Nika K., Tautz L., Taskén K., Cucca F., Mustelin T., Bottini N. (2005) Nat. Genet. 37, 1317–1319 [DOI] [PubMed] [Google Scholar]
- 7.Hill R. J., Zozulya S., Lu Y. L., Ward K., Gishizky M., Jallal B. (2002) Exp. Hematol. 30, 237–244 [DOI] [PubMed] [Google Scholar]
- 8.Cloutier J. F., Veillette A. (1999) J. Exp. Med. 189, 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gjörloff-Wingren A., Saxena M., Williams S., Hammi D., Mustelin T. (1999) Eur. J. Immunol. 29, 3845–3854 [DOI] [PubMed] [Google Scholar]
- 10.Wu J., Katrekar A., Honigberg L. A., Smith A. M., Conn M. T., Tang J., Jeffery D., Mortara K., Sampang J., Williams S. R., Buggy J., Clark J. M. (2006) J. Biol. Chem. 281, 11002–11010 [DOI] [PubMed] [Google Scholar]
- 11.Cohen S., Dadi H., Shaoul E., Sharfe N., Roifman C. M. (1999) Blood 93, 2013–2024 [PubMed] [Google Scholar]
- 12.Cloutier J. F., Veillette A. (1996) EMBO J. 15, 4909–4918 [PMC free article] [PubMed] [Google Scholar]
- 13.Rieck M., Arechiga A., Onengut-Gumuscu S., Greenbaum C., Concannon P., Buckner J. H. (2007) J. Immunol. 179, 4704–4710 [DOI] [PubMed] [Google Scholar]
- 14.Aarnisalo J., Treszl A., Svec P., Marttila J., Oling V., Simell O., Knip M., Körner A., Madacsy L., Vasarhelyi B., Ilonen J., Hermann R. (2008) J. Autoimmun. 31, 13–21 [DOI] [PubMed] [Google Scholar]
- 15.Sakaguchi N., Takahashi T., Hata H., Nomura T., Tagami T., Yamazaki S., Sakihama T., Matsutani T., Negishi I., Nakatsuru S., Sakaguchi S. (2003) Nature 426, 454–460 [DOI] [PubMed] [Google Scholar]
- 16.Siggs O. M., Miosge L. A., Yates A. L., Kucharska E. M., Sheahan D., Brdicka T., Weiss A., Liston A., Goodnow C. C. (2007) Immunity 27, 912–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahmouni S., Vang T., Alonso A., Williams S., van Stipdonk M., Soncini C., Moutschen M., Schoenberger S. P., Mustelin T. (2005) Mol. Cell. Biol. 25, 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito K., Williams S., Bulankina A., Höning S., Mustelin T. (2007) J. Biol. Chem. 282, 15170–15178 [DOI] [PubMed] [Google Scholar]
- 19.Kim J. S., Raines R. T. (1993) Protein Sci. 2, 348–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasegawa K., Martin F., Huang G., Tumas D., Diehl L., Chan A. C. (2004) Science 303, 685–689 [DOI] [PubMed] [Google Scholar]
- 21.Kung P., Goldstein G., Reinherz E. L., Schlossman S. F. (1979) Science 206, 347–349 [DOI] [PubMed] [Google Scholar]
- 22.Vang T., Abrahamsen H., Myklebust S., Enserink J., Prydz H., Mustelin T., Amarzguioui M., Tasken K. (2004) Eur. J. Immunol. 34, 2191–2199 [DOI] [PubMed] [Google Scholar]
- 23.Methi T., Ngai J., Mahic M., Amarzguioui M., Vang T., Tasken K. (2005) J. Immunol. 175, 7398–7406 [DOI] [PubMed] [Google Scholar]
- 24.Shaw J. P., Utz P. J., Durand D. B., Toole J. J., Emmel E. A., Crabtree G. R. (1988) Science 241, 202–205 [DOI] [PubMed] [Google Scholar]
- 25.Weiss A., Stobo J. D. (1984) J. Exp. Med. 160, 1284–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein P. L., Lee H. M., Rich S., Soriano P. (1992) Cell 70, 741–750 [DOI] [PubMed] [Google Scholar]
- 27.Schmedt C., Saijo K., Niidome T., Kühn R., Aizawa S., Tarakhovsky A. (1998) Nature 394, 901–904 [DOI] [PubMed] [Google Scholar]
- 28.Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 29.Mitra S., Barrios A. M. (2005) Bioorg. Med. Chem. Lett. 15, 5142–5145 [DOI] [PubMed] [Google Scholar]
- 30.Mitra S., Barrios A. M. (2007) Anal. Biochem. 370, 249–251 [DOI] [PubMed] [Google Scholar]
- 31.Taylor J. R. (1997) An Introduction to Error Analysis, 2nd Ed., pp. 45–92, University Science Books, Sausalito, CA [Google Scholar]
- 32.Read R. D., Bach E. A., Cagan R. L. (2004) Mol. Cell. Biol. 24, 6676–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., Stanford S. M., Jog S. P., Fiorillo E., Orrú V., Comai L., Bottini N. (2009) Biochemistry 48, 7525–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tailor P., Gilman J., Williams S., Couture C., Mustelin T. (1997) J. Biol. Chem. 272, 5371–5374 [DOI] [PubMed] [Google Scholar]
- 35.Lu W., Gong D., Bar-Sagi D., Cole P. A. (2001) Mol. Cell 8, 759–769 [DOI] [PubMed] [Google Scholar]
- 36.Aoki N., Ueno S., Mano H., Yamasaki S., Shiota M., Miyazaki H., Yamaguchi-Aoki Y., Matsuda T., Ullrich A. (2004) J. Biol. Chem. 279, 10765–10775 [DOI] [PubMed] [Google Scholar]
- 37.Straus D. B., Weiss A. (1992) Cell 70, 585–593 [DOI] [PubMed] [Google Scholar]
- 38.Blom N., Gammeltoft S., Brunak S. (1999) J. Mol. Biol. 294, 1351–1362 [DOI] [PubMed] [Google Scholar]
- 39.Nielsen C., Barington T., Husby S., Lillevang S. T. (2007) Genes Immun. 8, 131–137 [DOI] [PubMed] [Google Scholar]
- 40.Davidson D., Cloutier J. F., Gregorieff A., Veillette A. (1997) J. Biol. Chem. 272, 23455–23462 [DOI] [PubMed] [Google Scholar]
- 41.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]