Abstract
Proper hedgehog (Hh) signaling is crucial for embryogenesis and tissue regeneration. Dysregulation of this pathway is associated with several types of cancer. The monoclonal antibody 5E1 is a Hh pathway inhibitor that has been extensively used to elucidate vertebrate Hh biology due to its ability to block binding of the three mammalian Hh homologs to the receptor, Patched1 (Ptc1). Here, we engineered a murine:human chimeric 5E1 (ch5E1) with similar Hh-binding properties to the original murine antibody. Using biochemical, biophysical, and x-ray crystallographic studies, we show that, like the regulatory receptors Cdon and Hedgehog-interacting protein (Hhip), ch5E1 binding to Sonic hedgehog (Shh) is enhanced by calcium ions. In the presence of calcium and zinc ions, the ch5E1 binding affinity increases 10–20-fold to tighter than 1 nm primarily because of a decrease in the dissociation rate. The co-crystal structure of Shh bound to the Fab fragment of ch5E1 reveals that 5E1 binds at the pseudo-active site groove of Shh with an epitope that largely overlaps with the binding site of its natural receptor antagonist Hhip. Unlike Hhip, the side chains of 5E1 do not directly coordinate the Zn2+ cation in the pseudo-active site, despite the modest zinc-dependent increase in 5E1 affinity for Shh. Furthermore, to our knowledge, the ch5E1 Fab-Shh complex represents the first structure of an inhibitor antibody bound to a metalloprotease fold.
Keywords: Antibodies, Biophysics, Calorimetry, Development, Protease, X-ray Crystallography, Sonic Hedgehog, Morphogen
Introduction
The secreted morphogens Sonic hedgehog (Shh),2 Indian hedgehog (Ihh), and Desert hedgehog (Dhh) are important for proper cellular differentiation during embryogenesis of vertebrates and some invertebrates, controlling left-right asymmetry, neural tube and limb patterning, branching morphogenesis, and bone formation (1). Although Hh signaling is mostly quiescent in adults, inappropriate pathway activity has been implicated in several cancer types (2, 3). Hh signals by binding to Patched1 (Ptc1), a 12-pass transmembrane protein. Hh binding of Ptc1 relieves inhibition of the 7-transmembrane protein Smoothened, allowing it to translocate to the cell surface or the primary cilium (4), which ultimately results in the activation of the Ci/Gli family of zinc finger transcription factors and expression of Hh-regulated genes (1).
Mature Hh ligand is produced by autoproteolytic removal of the C-terminal intein-like domain with concomitant addition of cholesterol at the C terminus followed by palmitoylation at the N terminus (5–9). Crystallographic studies have revealed that mammalian Hh ligands have a tetrahedrally coordinated Zn2+ and two Ca2+ cations with an overall topology similar to the MD clan of metalloproteases (10, 11). Despite possessing a metalloprotease-like protein fold with a pseudo-active site, Shh acts as a ligand for membrane-bound receptors rather than as an enzymatically active protease (12, 13).
Hh signaling is modulated by cell surface receptors such as the agonists Cdon (14, 15), Boc (16, 17), and GAS1 (18), as well as the antagonist Hedgehog-interacting protein (Hhip) (19). The structures of the N-terminal signaling domain of Shh (hereafter referred to as Shh, also known as Shh-N) bound to Hhip (20, 21) and Cdon (11) revealed that their respective binding sites are centered on the Shh pseudo-active site and the adjacent Ca2+-binding site. Biochemical and peptide mapping studies suggest that Ptc1 also interacts with the pseudo-active site groove (21). In contrast, the structure of Drosophila Hh with Ihog, the Drosophila ortholog of Cdon (22), shows that Ihog binds at a separate site away from either the Ca2+ site or the pseudo-active site groove.
The anti-Shh monoclonal antibody 5E1 (5E1) is a pathway antagonist that is widely used to study Hh signaling in both developmental biology (23–29) and cancer (3, 30–32). 5E1 was generated with mouse hybridoma technology using the rat Shh N-terminal domain as the antigen (33). 5E1 blocks binding of all three mammalian Hh ligands to Ptc1 with low nanomolar affinity, thereby inhibiting Hh signaling (21). Despite the wide use and extensive characterization of 5E1 in biological assays, a detailed understanding of the biochemical and structural aspects of the 5E1 interaction with Shh has been lacking. The 5E1-Shh interface has been probed using low resolution mapping strategies such as mutagenesis (12), labeling of residues (34), and tryptic protease protection mapping (21). These limited studies identified Ser177 and a peptide encompassing residues 158–178 of Shh as being involved in 5E1 recognition.
To better understand how 5E1 functions as a Hh pathway antagonist, we characterized the binding of a murine:human chimeric 5E1 Fab (ch5E1 Fab) to human Hh ligands and found that, like Hhip and Cdon, it has greater affinity in the presence of Ca2+ and Zn2+. Furthermore, we determined the x-ray crystal structure of ch5E1 Fab alone and in complex with human Shh and found that 5E1 blocks access to the pseudo-active site groove on Shh. Notably, the 5E1 epitope on Shh largely overlaps with the binding site of the natural Hh antagonist receptor Hhip, which we recently showed competes with Ptc1 for Shh binding (21). Thus, these data explain the molecular basis of 5E1 inhibition of the Hh-Ptc1 interaction.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of 5E1 Fab and Hh Ligands
The N terminus of the 5E1 mAb was sequenced by Edman degradation and used to design primers to isolate RNA encoding the antibody from the hybridoma cells by PCR. The variable heavy and light regions were separately subcloned into a human subtype III IgG backbone (that of trastuzumab (35)) in the pRK5 mammalian expression vector (Genentech, Inc.). ch5E1 IgG was expressed by transient co-transfection of heavy and light chains in Chinese hamster ovary cells using PS21 production media with 1% Ultra Low IgG fetal bovine serum (Invitrogen) and purified from the media using protein A-Sepharose (GE Healthcare) chromatography. After loading, the resin was washed with PBS and eluted with 0.1 m acetic acid at pH 2.7 followed by addition of 1.5 m Tris, pH 8.6, to a final pH of 5. Eluted ch5E1 IgG was concentrated and further purified by size exclusion chromatography on an S200 Sephadex in PBS. Chimeric 5E1 Fab fragments (ch5E1 Fab) were obtained by digestion with endoproteinase Lys C (Wako) in 0.1 m Tris, pH 8, with an enzyme:IgG ratio of 1:500 (w/w) for 1 h at 37 °C. The reaction was quenched with 1% acetic acid, diluted 10-fold, and loaded onto a HiTrap SP HP column (GE Healthcare) equilibrated with 50 mm sodium acetate, pH 5. The Fab and Fc fragments were eluted separately by running a 20 column volume gradient to 0.3 m NaCl.
Murine 5E1 Fab (m5E1 Fab) was isolated from full-length m5E1 IgG1 using the mouse IgG1 Fab preparation kit (Pierce) as described by the manufacturer. The murine Fab was then subjected to size exclusion chromatography using an S75 Sephadex column (GE Healthcare) at 30 cm/h flow rate in 20 mm HEPES, pH 7.2, and 150 mm NaCl.
Human Shh (residues 25–197), Ihh (residues 29–202), Dhh (residues 24–198), and rat Shh (residues 26–198 (43)), all lacking the N-terminal Hh cysteine to avoid aggregation, were expressed and purified essentially as described previously (21). Human and rat Shh with an additional C-terminal cysteine were expressed and labeled with EZ-Link Maleimide PEO2-Biotin (Thermo Scientific) as described previously (21). ch5E1 Fab was biotinylated with EZ-Link Sulfo-NHS-LC-Biotin according to the manufacturer's instructions (Thermo Scientific).
Flow Cytometry Analysis
Colorectal HT29 cells expressing endogenous Hh and COS cells stably transfected with human Shh (Shh-COS) were detached from plates using 1 mm EDTA in PBS at 37 °C. All following steps were carried out at 4 °C. After washing in growth media followed by PBS, cells were incubated in triplicate at the indicated concentrations of m5E1 or ch5E1 for 35 min in FACS buffer (3% fetal bovine serum in PBS; fetal bovine serum contains ∼3 mm Ca2+ (11)). After two washes in FACS buffer, the 5E1 signal was amplified with 1:100 biotinylated goat anti-mouse for m5E1 (catalog no. 115-065-071, Jackson ImmunoResearch) or anti-human for ch5E1 (catalog no. 109-065-003, Jackson ImmunoResearch) for 1 h prior to two washes and detection with 1:50 rhodamine streptavidin-phycoerythrin (catalog no. 016-110-084, Jackson ImmunoResearch). Live cells (∼60%) were sorted by propidium iodide exclusion and analyzed on a FACSCalibur machine (BD Biosciences).
Murine 5E1 and ch5E1 labeling of Shh-COS cells was directly detected with 15 μg/ml fluorescein isothiocyanate-conjugated goat anti-mouse Fc (catalog no. 109-096-098, Jackson ImmunoResearch) or anti-human Fc (catalog no. 115-096-071, Jackson ImmunoResearch), respectively, and cells were fixed in 2% paraformaldehyde prior to analysis. For competition experiments, Shh-COS cells were incubated with 0.1 μg/ml (∼0.69 nm) m5E1 or ch5E1 in the presence of competing antibody of the other species (or isotype controls), and the noncompeting antibody was detected with fluorescein isothiocyanate anti-mouse or anti-human Fc, respectively.
Embryo Immunofluorescence
SvEv129 mouse embryos at stage E10.5 were dissected in cold PBS, fixed in 4% paraformaldehyde for 40 min, and then washed three times for 5 min in PBS. Embryos were then subjected to an overnight buffer exchange into 30% sucrose in PBS at 4 °C. Equilibrated embryos were embedded in OCT compound, flash-frozen with dry ice/ethanol, and sectioned at 10 μm thickness onto microscope slides. Frozen sections were washed twice with PBS for 5 min and then blocked with blocking buffer (0.1% Triton X-100, 3% bovine serum albumin, 1% sheep serum in PBS) at room temperature for 20 min. The sections were first incubated overnight at 4 °C with 5 μg/ml m5E1 or ch5E1 in blocking buffer and then washed three times for 5 min in PBS and incubated with 1:400 Cy3-conjugated donkey anti-mouse or anti-human secondary antibodies, respectively (catalog no. 715-166-150 and 709-166-149, Jackson ImmunoResearch) in blocking buffer at room temperature for 1 h. After three 5-min PBS washes, slides were coverslip-mounted using 4′,6-diamidino-2-phenylindole-containing Vectashield (Vector Laboratories, Burlingame, CA).
Kinetic Binding Measurements by Biolayer Interferometry
Binding kinetics and affinities of ch5E1 Fab for various Hh ligands were measured by biolayer interferometry (ForteBio). ShhCysbiotin, human Shh, rat Shh, and ch5E1 Fab were dialyzed 1:4000 at 4 °C for 24 h against buffer A (20 mm HEPES, pH 7.2, 100 mm NaCl) containing 5 mm EDTA and 5 mm EGTA followed by dialysis (2× 1:4000 at 4 °C for 24 h) against buffer A to remove all bound divalent ions. Atomic absorption spectroscopy confirmed that recombinant Shh contains equimolar amounts of zinc, ∼99% of which is removed by dialysis.
Kinetic buffer (20 mm HEPES, pH 7.2, 100 mm NaCl, 0.1 mg/ml bovine serum albumin, 0.002% Tween 20) was used for binding experiments and was supplemented with 5 mm CaCl2 and/or 50 μm ZnSO4 or 5 mm EDTA to measure the dependence of binding affinity on calcium and/or zinc ions. Prior to immobilization on streptavidin biosensor tips (ForteBio), ShhCysbiotin was preincubated with kinetic buffer in the presence or absence of 50 μm ZnSO4. Binding measurements of immobilized Shh (charged with or without Zn2+) to ch5E1 Fab were then carried out in the presence or absence of 5 mm CaCl2. Alternatively, biotinylated ch5E1 Fab was immobilized on the biosensor tips, followed by binding of Hh ligands under various divalent metal ion conditions as above.
Biotinylated ligands (ch5E1 Fab or Hh ligand at 25 μg/ml in kinetic buffer) were captured by biosensors during a 5-min incubation, followed by a 5-min wash in kinetic buffer. The association and dissociation binding kinetics for unlabeled ch5E1 Fab or Hh ligand were measured for 15 min each. Binding affinities were independent of which protein was immobilized. The binding kinetics of seven different concentrations (2-fold serial dilution starting at 125 nm) of ligand in solution were measured and evaluated with ForteBio OctetRED Evaluation software 6.1 using a 1:1 binding model to derive kon, koff, and KD values.
Cell-based Hh Signaling Assay
Hh signaling was measured using stably transfected Gli-luciferase reporter S12 cells (36) as described previously (21). Briefly, 10,000 S12 cells/well were plated for 24 h, and 20,000 HT29 cells were then co-cultured on top for 24 h, followed by 24 h of serum starvation in the presence of various concentrations of m5E1, ch5E1, or isotype controls prior to luciferase measurement (HTS SteadyLite kit, PerkinElmer Life Sciences).
Isothermal Titration Calorimetry
ch5E1 and human and rat Shh were dialyzed against Calorimetry buffer (20 mm HEPES, pH 7.2, 150 mm sodium chloride) containing 5 mm EDTA and 5 mm EGTA, followed by dialysis against Calorimetry buffer to remove all divalent ions bound to the proteins. Interactions were measured using a VTC calorimeter (Microcal, GE Healthcare) at 30 °C. The sample cell was loaded with 2 μm 5E1, and a 20 μm solution of either human or rat Shh was added over 25 injections of 10 μl each. Calorimetry buffer was supplemented with 5 mm CaCl2 and/or 500 μm ZnSO4 to measure the effect of divalent metal ions on the protein interactions. The data were fit using the Microcal evaluation software using a single site model.
Competition with Hhip L2 Peptide
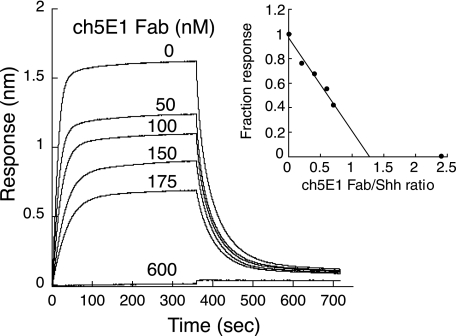
Competition binding experiments were measured by biolayer interferometry on an OctetRED (ForteBio) with cyclic Hhip L2 peptide (21) and ch5E1 Fab competing for Shh binding. Streptavidin biosensors (ForteBio) were loaded with biotinylated cyclic Hhip L2 peptide (25 μg/ml, 3 min) and then incubated with a solution containing 250 nm Shh and ch5E1 Fab at concentrations of 600, 175, 150, 100, 50, or 0 nm for 6 min followed by incubation in buffer for 6 min. Kinetic buffer with additional 5 mm CaCl2 was used throughout this experiment as described earlier. The fraction responses were determined by dividing the responses in the presence of ch5E1 Fab by that in the absence of ch5E1 Fab, each at 350 s after binding reached equilibrium.
Crystallization of ch5E1 Fab Alone and in Complex with Shh
A 1:1 complex of ch5E1 Fab with Shh was isolated from a mixture of ch5E1 Fab with excess Shh by gel filtration chromatography using an S200 Sephadex column in 20 mm HEPES, pH 7.2, 0.1 m NaCl, and 5 mm CaCl2 and subsequently concentrated to 15 mg/ml. Crystallization trials with this complex using the hanging drop vapor diffusion method at 19 °C resulted in crystals at both low and neutral pH. At low pH (∼pH 4.0), crystals were obtained by mixing the protein solution with an equal volume of reservoir solution (0.2 m (NH4)2SO4, 20% (w/v) PEG 4000) to form 0.3–2-μl drops, which were incubated over 500 μl of reservoir solution with a measured pH of 4.0. Crystals appeared within 1 week and were found to contain only ch5E1 Fab. A second crystallization condition was found by mixing the same ch5E1 Fab-Shh complex solution with an equal volume of reservoir solution at near neutral pH (0.1 m HEPES, pH 7.5, 22% (w/v) PEG 4000, 0.2 m Li2SO4) to form 0.3–2-μl drops, which were equilibrated over 500 μl of reservoir solution. Crystals appeared within 1 week and were found to contain the ch5E1 Fab-Shh complex.
Human Shh numbering is used throughout this report; rat and murine Shh numbers are one higher due to an extra amino acid in the signal sequence. ch5E1 chains are numbered according to standard Kabat format starting at 1 for the first amino acid of the mature heavy and light chains, and human Shh numbering starts at amino acid 25.
X-ray Crystallography
Crystallographic data were collected at ALS beamline 5.0.1 (Table 2) and processed with HKL2000 (HKL, Charlottesville, VA). Both structures were solved by molecular replacement using the program PHASER (37) followed by refinement with REFMAC5 (38) and model building in COOT (39). For the ch5E1 Fab, the structure of a variant of the 4D5 Fab (closely related to Protein Data Bank code 1FVE) was used as a search model. Initial maps revealed that only ch5E1 Fab was present in the crystal lattice likely due to the low pH of the crystallization conditions, which may have disfavored the complex. The ch5E1 Fab-Shh complex was solved using the refined ch5E1 Fab structure and the mouse Shh structure (Protein Data Bank code 1VHH) as search models. In contrast to the ch5E1 Fab structure, clear molecular replacement solutions and electron density were present for both ch5E1 and Shh. The final models exhibited excellent geometry, with 90.1, 9.1, 0.5, and 0.3% (ch5E1 Fab) and 90.0, 9.2, 0.6, and 0.2% (ch5E1 Fab-Shh complex) of residues in the most favored, additional allowed, generously allowed, and disallowed regions of a Ramachandran plot, respectively. All structure figures were made with PyMOL.
TABLE 2.
Solution binding measurements of human and rat Shh to ch5E1 Fab by isothermal titration calorimetry
| Hh ligand | Divalent metal cation | KD | ΔH | TΔS | ΔG |
|---|---|---|---|---|---|
| nm | kcal/mol | kcal/mol | kcal/mol | ||
| Shh | None | 7 | −13.6 | 2.3 | −11.3 |
| Shh | Zn2+ | 1.1 | −18.8 | 6.4 | −12.4 |
| Shh | Ca2+ | 1.8 | −14.7 | 2.6 | −12.1 |
| Shh | Zn2+, Ca2+ | 0.7 | −19.6 | 6.9 | −12.7 |
| Rat Shha | Zn2+, Ca2+ | 1.1 | −12.0 | 0.4 | −12.4 |
a Rat Shh refers to the sequence as described in Roelink et al. (43).
RESULTS
Functional Equivalence of m5E1 and ch5E1
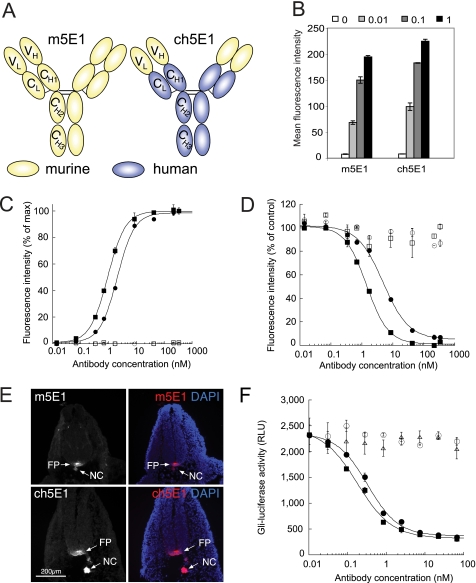
To facilitate recombinant expression of 5E1 for biochemical and structural studies, we cloned the variable heavy and light chains from the hybridoma-derived parental murine antibody (m5E1) and grafted them onto the human constant domains of trastuzumab (35) to create chimeric 5E1 (ch5E1; Fig. 1A). Recombinant ch5E1 was expressed in Chinese hamster ovary cells and purified by protein A and gel filtration chromatography. ch5E1 was comparable with the parental m5E1 antibody in recognizing cell surface Shh stably expressed in COS cells and endogenous Hh produced by HT29 cells as analyzed by flow cytometry (Fig. 1, B and C). As expected, both antibodies bind to the same site on Shh, because they compete with each other for Shh binding on the cell surface (Fig. 1D). ch5E1 also retained its specificity for Hh, staining the notochord, floor plate (Fig. 1E), and gut (data not shown) of developing mouse embryos by immunofluorescence, in agreement with previous data (40, 41). Importantly, ch5E1 blocked Hh signaling in S12 cells stimulated by endogenous Hh (IC50 of 0.18 nm) with similar potency to m5E1 (IC50 of 0.33 nm), as measured by inhibition of a Gli-luciferase reporter (Fig. 1F), and was equivalently active at inhibiting the growth of HT29 xenografts in vivo (data not shown).
FIGURE 1.
Chimeric 5E1 is functionally equivalent to murine 5E1. A, schematic of murine 5E1 (m5E1, yellow) IgG (33) and its chimeric counterpart (ch5E1), where the constant domains (CH1–3 and CL) have been replaced with the corresponding domains from the humanized antibody trastuzumab (blue) (35), leaving the variable light and heavy (VL and VH) domains of the murine 5E1 antibody intact. B, ch5E1 and m5E1 bind similarly to an endogenous Hh-expressing cell line. Flow cytometry analysis is shown of endogenous Hh in HT29 cells with the indicated concentrations of m5E1 or ch5E1 (in μg/ml). The means ± S.D. of triplicate reactions are plotted. C, ch5E1 (■) and m5E1 (●) bind similarly to stably transfected Shh-COS cells by flow cytometry analysis. The means ± S.D. of a representative duplicate experiment are shown. Isotype controls (chimeric IgG (□) or murine IgG1 (○)) show no appreciable binding. D, ch5E1 and m5E1 compete for cell surface Shh. The ability of increasing amounts of ch5E1 (■) or m5E1 (●) to compete with ∼0.69 nm (0.1 μg/ml) m5E1 for binding to Shh-expressing cells and vice versa as monitored by flow cytometry analysis is shown, normalized to 100% for no competitor after background subtraction. Isotype controls (murine IgG1 (○) or chimeric IgG (□)) are unable to compete for Shh binding. E, ch5E1 specifically detects Hh in the developing mouse embryo. E10.5 embryos were sectioned and stained with m5E1 (top) or ch5E1 (bottom), followed by Cy3-conjugated secondary antibodies (left panel and red in merged right panel) and 4′,6-diamidino-2-phenylindole (DAPI) (blue nuclear staining in right merged panels). ch5E1 is as specific as m5E1 in detecting Hh in the notochord (NC) and floor plate (FP). Scale bar is 200 μm (images taken at ×10 magnification). F, ch5E1 and m5E1 inhibit Hh signaling similarly. HT29 cells secreting Hh were co-cultured with S12 cells (C3H10T1/2 cells stably expressing a Gli-luciferase reporter (36)). Hh signaling was stimulated by serum starvation for 24 h in the presence of the indicated concentrations of ch5E1 (■), m5E1 (●), hIgG1 (▴), or mIgG1 (○) antibodies. The means ± S.D. of the luciferase signals (RLU; relative luminescence units) of triplicate measurements are plotted. This experiment is representative of multiple independent experiments.
Enhancement of 5E1 Binding to Hh with Divalent Cations
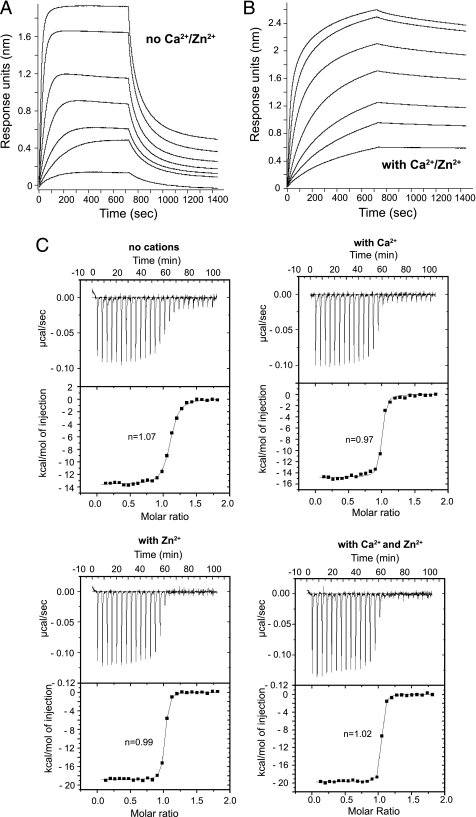
The requirements for high affinity interaction between ch5E1 and Hh were first characterized by biophysical methods. As Zn2+ and Ca2+ ions have been shown to improve Hh interactions with other binding partners (11, 20, 21), we tested whether the ch5E1 interaction with Shh was affected by either of these cations using biolayer interferometry. We isolated Fab fragments of ch5E1 and m5E1 using limited proteolysis and used these in all further experiments. Shh was immobilized to biosensors in buffer alone or buffer with Zn2+ and/or Ca2+. In the absence of any divalent ions, ch5E1 Fab bound to Shh with a KD of 7 nm (Table 1). The addition of Ca2+ favored the interaction, reducing the KD by ∼6-fold to 1.1 nm. The presence of Zn2+ in the buffer did not substantially change the affinity (KD = 4.8 nm), indicating that Zn2+ by itself is not a major modulator of the interaction under these conditions. However, addition of Zn2+ in the presence of Ca2+ further reduced the KD by ∼3.6-fold to 0.31 nm, resulting in an overall increase in affinity of ∼22-fold in the presence of both metal cations. This increase in affinity is primarily due to an ∼54-fold reduction in the dissociation rate constant (koff), which decreased from 35 × 10−4 to 0.65 × 10−4 s−1 upon addition of both Ca2+ and Zn2+ cations (Table 1 and Fig. 2, A and B). The association kinetics were less cation-dependent, exhibiting only a 2.4-fold decrease in kon in the presence of both cations (Table 1). We also tested the affinity of the Fab fragment of m5E1 for Shh both in the absence or presence of Ca2+ and Zn2+. The resulting KD values are comparable with those measured for ch5E1 (Table 1), indicating that the chimera recapitulates both the Shh binding affinity and cation dependence of the parental murine antibody.
TABLE 1.
Binding kinetics of Hh ligands to ch5E1 Fab in the presence and absence of divalent ions by biolayer interferometry
Binding measurements for human and rat Shh were carried out using immobilized Hh ligand and ch5E1 or m5E1 Fabs in solution. Binding measurements for human Ihh and human Dhh were carried out using immobilized ch5E1 and Hh ligand in solution as described under “Experimental Procedures.”
| Hh ligand | Divalent metal cation | KD | kon | koff |
|---|---|---|---|---|
| nm | m−1s−1 | s−1 | ||
| Shh | None | 6.94 (6.16)a | 5.07 × 105 | 35.2 × 10−4 |
| Shh | Zn2+ | 4.82 (5.23) | 5.71 × 105 | 27.5 × 10−4 |
| Shh | Ca2+ | 1.12 (1.46) | 2.58 × 105 | 2.89 × 10−4 |
| Rat Shhb | Zn2+, Ca2+ | 1.51 | 3.67 × 105 | 5.55 × 10−4 |
| Shh | Zn2+, Ca2+ | 0.31 (0.67) | 2.09 × 105 | 0.65 × 10−4 |
| Ihh | Zn2+, Ca2+ | 0.29 | 3.55 × 105 | 1.05 × 10−4 |
| Dhh | Zn2+, Ca2+ | 1.71 | 4.95 × 105 | 8.47 × 10−4 |
a Values in parentheses are for the parental murine 5E1 Fab.
b Rat Shh refers to the sequence as described in Roelink et al. (43).
FIGURE 2.
5E1 binding to Shh is enhanced by divalent cations. Biolayer interferometry sensorgrams of ch5E1 Fab binding to C-terminally biotinylated Shh on streptavidin-coated biosensors are shown in the absence (A) or presence (B) of Ca2+ and Zn2+. Sensorgrams of seven 2-fold serial dilutions of ch5E1 Fab starting at 125 nm are shown, where 125 nm results in the largest response. C, isothermal titration calorimetry of human Shh with ch5E1 Fab. Experiments were carried out in the absence or presence of Ca2+ (5 μm CaCl2) and/or Zn2+ (500 μm ZnSO4) as indicated. As expected, the stoichiometry of 5E1-Shh complex formation in solution derived from the ITC data indicates 1:1 binding in all cases.
The effect of Ca2+ and Zn2+ cations on binding affinity for Shh was confirmed by solution binding studies using isothermal titration calorimetry (ITC) (Table 2 and Fig. 2C). The affinity of ch5E1 for Shh in solution was 10-fold higher in the presence of both divalent ions (KD = 0.7 nm) than in their absence (KD = 7.0 nm), whereas Ca2+ or Zn2+ alone resulted in 4–6-fold higher affinities. The ITC data show that the interaction between Shh and ch5E1 is enthalpy-driven, consistent with an interface rich in hydrogen bonds and ionic interactions. The solution and biolayer binding results agree well with each other. Both methods indicate that 5E1 has enhanced affinity for Shh in the presence of Ca2+ and has greater binding affinity when both Zn2+ and Ca2+ are present.
Crystal Structure of Chimeric 5E1 Fab Bound to Shh
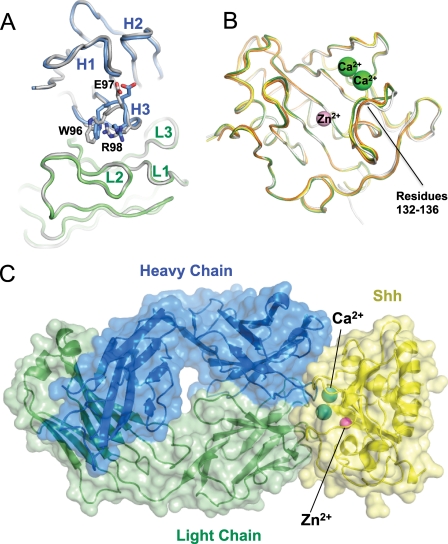
To better understand how Ca2+ and Zn2+ cations affect the ch5E1-Shh interaction and to compare the ch5E1 epitope to the binding sites of other known Hh pathway regulators, we determined the structure of ch5E1 Fab both alone and in complex with Shh at 1.90 and 1.83 Å, respectively (Table 3). The structure of the ch5E1 Fab fragment alone shows that it has canonical complementarity determining region (CDR) conformations with a compact CDR H3 loop (Fig. 3A). Binding Shh does not substantially alter the conformation of ch5E1, as superposing the structures of the bound and free ch5E1 results in a root mean square deviation of 0.34 Å on residues in the variable region. The maximal difference is a modest change of less than 1 Å in the backbone and side chains of the heavy chain CDR3 (H3) (Fig. 3A). Because of differences in the crystal packing environments, the elbow angle formed by the variable and the constant domains differs between the two structures, which results in a higher overall root mean square deviation of 1.1 Å when calculated over the entire ch5E1 Fab.
TABLE 3.
X-ray data collection and refinement statistics
| ch5E1 Fab | ch5E1 Fab-Shh complex | |
|---|---|---|
| Data collection | ||
| Space group | R3 | P212121 |
| Cell dimensions | ||
| a, b, c | 110.3, 110.3, 112.5 Å | 60.3, 90.5, 111.7 Å |
| Resolution | 50 to 1.90 Å (1.97 to 1.90 Å) | 50 to 1.83 Å (1.90 to 1.83 Å) |
| Rsym | 7.0 (48.2)a | 6.8 (46.4) |
| 〈I〉/〈σI〉 | 15.8 (2.3) | 16.4 (2.2) |
| Completeness | 100% (100%) | 98.8% (90.7%) |
| Redundancy | 3.2 (3.2) | 4.3 (2.6) |
| Refinement | ||
| Resolution | 50 to 1.90 Å | 50 to 1.83 Å |
| No. of reflections | 43,799 | 54,060 |
| Rwork/Rfree | 17.9, 22.4% | 17.6, 21.8% |
| No. of atoms | ||
| Protein | 3339 | 4563 |
| Solvent | 372 | 439 |
| Root mean square deviation | ||
| Bond lengths | 0.009 Å | 0.011 Å |
| Bond angles | 1.2° | 1.3° |
a Values in parentheses are for highest resolution shell.
FIGURE 3.
Structure of 5E1 bound to Shh. A, comparison of the structures of ch5E1 Fab alone and in complex with Shh. Shh-bound ch5E1 light and heavy chains are shown as C-α ribbons and colored green and blue, respectively; both heavy and light chains of the free ch5E1 Fab are colored gray. Residues in CDR H3 differing most upon binding to Shh (not pictured) are labeled. B, comparison of the structures of Shh free (white), bound to Cdon (orange), Hhip (green), or 5E1 (yellow). The bound divalent metal cations are shown as spheres (Zn2+ in pink and Ca2+ in green). C, complex between 5E1 and Shh. The Fab is colored as in A, and Shh is in yellow, with Zn2+ and Ca2+ colored as in B.
The structure of Shh is likewise not significantly affected by ch5E1 Fab binding. Superposing the structure of human Shh from the ch5E1-Shh complex on the structures of human Shh from the Hhip-Shh complex, murine Shh crystallized alone, or murine Shh bound to Cdon (Protein Data Bank codes 3HO5, 1VHH, and 3D1M, respectively) results in an r.s.m.d. of less than 0.25 Å for each pairwise comparison (Fig. 3B). The region involving residues 132–136 (human Shh numbering) differs most with changes of up to 1.6 Å in the backbone and more significant differences in the side chain conformation when compared with murine Shh bound to Cdon. These differences are likely due to distinct crystal packing environments in the Cdon complex, as the other structures agree well in this region aside from minor changes in side chain conformations due to the absence of Ca2+ in the murine Shh structure.
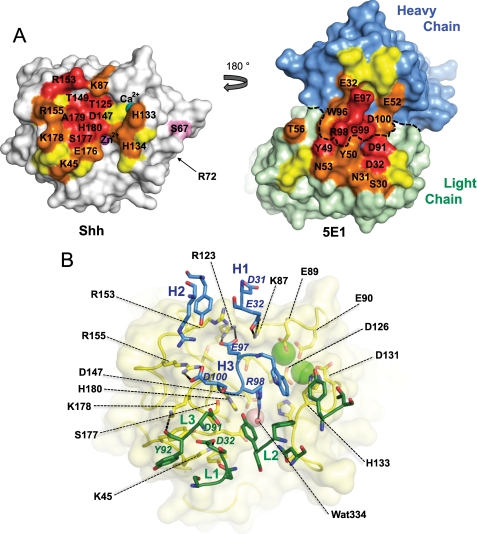
The interaction between ch5E1 and Shh is clearly defined in the structure of the ch5E1-Shh complex (Table 3 and Fig. 3C). The interface is evenly split between the paratope on the ch5E1 Fab and the epitope on Shh. The total buried surface area (∼1700 Å2) is typical for an antigen-Fab complex. Consistent with m5E1 being hybridoma-derived, the paratope on ch5E1 for Shh is formed by all six CDRs with heavy chain CDR3 (H3) and light chain CDR2 (L2) contributing most of the solvent-accessible surface area buried at the ch5E1-Shh interface (Fig. 4A). Notably, ch5E1 binds at the Shh pseudo-active site groove, which has been recently found to be the site of Hhip binding (20, 21). This binding site on Shh is a continuous surface in three-dimensional space formed by residues that are noncontiguous in the Shh linear sequence. The nonlinearity of the epitope likely explains the poor ability of 5E1 to recognize Hh by Western blotting (12) and formalin-fixed paraffin-embedded immunohistochemistry (FFPE IHC, data not shown) compared with its excellent reactivity by fluorescence-activated cell sorter, enzyme-linked immunosorbent assay, immunofluorescence, and in vivo activity (Fig. 1) (21, 31, 33, 40–42).
FIGURE 4.
Detailed views of the 5E1-Shh interaction. A, “open book” view of the 5E1-Shh interface. The surface of Shh (left, oriented as in Fig. 3B) and ch5E1 Fab (right) are colored according to buried surface area upon complex formation. Residues that bury 75–100, 50–75, or 25–50% of their solvent-accessible surface area on complex formation are colored red, orange, and yellow, respectively. Residues colored red or orange are labeled. The boundary between the heavy and light chains is indicated with a dotted line. Shh residue Ser67, which is a threonine in mouse and rat Shh, is colored pink. The location of Shh residue Arg72, corresponding to Hh residue Lys132 implicated in multimerization of Hh (44), is also indicated. B, 5E1-Shh interface. Shh is shown in yellow with a transparent molecular surface. Shh residues mentioned in the text are labeled with the exception of Asp95, Asp129, and Ala179, which are obscured in this orientation. 5E1 is colored with the heavy chain in blue and the light chain in green. CDR loops are labeled, and individual 5E1 side chains mentioned in the text are labeled in italic font. Hydrogen bonds and salt bridges mentioned in the text are shown as dashed lines. The Zn2+ and Ca2+ ions are shown as pink and green spheres, respectively.
Consistent with the enthalpy-driven affinity as determined by calorimetry, there is a high degree of charge complementarity in the ch5E1-Shh interface. Shh presents a basic patch formed by residues Lys45, Lys87, Arg123, Arg153, Arg155, and Lys178 (Fig. 4, A and B). This region interacts with an acidic patch on 5E1 formed by heavy chain residues Glu32, Glu97, and Asp100 and light chain residues Asp32 and Asp91 forming several salt bridges such as Shh Lys45 to Asp32 in light chain CDR1 (L1), Arg123 to Glu97 in H3, and Arg155 to Asp100 in H3 (Fig. 4B). Some of the interactions do not involve salt bridges but rather form more general electrostatic complementarity, as found with Arg153 from Shh, which binds in the vicinity of Asp31 and Glu32 from H1 and forms a hydrogen bond with the carbonyl of Asp31. Numerous additional hydrogen bonds are formed, including Lys178 from Shh to the backbone carbonyl of Tyr92 in L3.
Despite binding the Shh pseudo-active site groove, ch5E1 does not directly contact the Zn2+ cation. Instead, Arg98 from H3 interacts with Shh residue Asp147 and a water molecule (water 334), both of which participate in the Zn2+-coordination sphere (Fig. 4B). Similarly, ch5E1 does not contact the two bound Ca2+ cations in Shh but does contact the loops that surround and form the Ca2+-binding site, including van der Waals contacts between His133 from Shh, which packs against the Fab surface at the juncture of the light and heavy chains. In addition, the charge on the Ca2+ ion at least partially neutralizes the negatively charged Ca2+-coordinating residues from Shh (Glu89, Glu90, Asp95, Asp126, Asp129, and Asp131), thereby increasing the overall charge complementary between the basic regions in the Shh interface and the acidic ch5E1 surface.
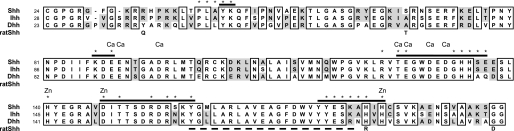
The ch5E1 structural epitope on Shh is consistent with previous efforts to map the m5E1 interface using function-driven strategies (12, 21, 34). These studies identified Ser177 and a peptide encompassing residues 158–178 as being involved in 5E1 recognition. In particular, the structure reveals that the loop region formed by residues 174–180 is in intimate contact with CDRs H3, L1, L2, and L3 with Shh residues Ser177, Ala179, and His180 almost completely sequestered from solvent upon complex formation (Fig. 4, A and B). The positively charged guanidinium group of Arg98 in CDR H3 is in van der Waals contact with the electron-rich aromatic imidazole ring of His180. This contact is particularly striking, as His180 was reported to be an arginine in the rat Shh used as the immunogen to generate 5E1 (see Fig. 5) (33). We measured the affinity of ch5E1 Fab for the rat Shh sequence as reported by Roelink et al. (43) by both biolayer interferometry and ITC and found it similar (1.1 nm versus 0.7 nm by ITC) to that of human Shh (Tables 1 and 2). Because current data base sequences suggest that rat and mouse Shh sequences are identical, differing from human Shh only at residue 67 (Ser in human and Thr in rat and mouse), we can only speculate as to whether the presence of arginine at position 180 in the antigenic material was fortuitous at targeting generation of 5E1 to the functionally important Shh pseudo-active site groove (11, 21).
FIGURE 5.
Sequence alignment of human Shh, Ihh, and Dhh. The amino acid sequences of the Shh, Ihh, and Dhh N-terminal signaling domains are aligned showing the conservation of residues in the 5E1 epitope (overlined). The dashed line highlights the 5E1-protected tryptic peptide previously shown to form part of the epitope (21). Residues that contact Hhip are indicated with an asterisk. Rat and mouse sequence numbers are one higher than human Shh due to an extra amino acid in the signal sequence. Thus, Ser67 of human Shh is Thr68 in rodent Shh; His180 is Arg181 in the rat Shh immunogen. Thr68 is listed below the alignment as are the residues in the rat Shh immunogen described by Roelink and co-workers (33, 43) that differ from current data base sequences for rat Shh. Residues whose side chains or backbones coordinate the Ca2+ ions are indicated by “Ca” and those that coordinate the Zn2+ ion by “Zn”.
As expected from the cross-reactivity of 5E1, the residues forming the 5E1 epitope are identical in human and murine Shh sequences with the species difference at residue 67 remote from this site (Ser67 is colored pink in Fig. 4A). Furthermore, Shh and Ihh are also identical within the ch5E1 epitope (Fig. 5), with the exception of S156N, consistent with their near identical binding affinities for the ch5E1 Fab (Table 1). In contrast, Dhh differs from Shh at four residues within the epitope (Fig. 5), consistent with the modestly weaker interaction between ch5E1 Fab and Dhh (Table 1). The key difference between Shh and Dhh is likely A179N, as the ch5E1-Shh interface is tightly packed around this residue and the increased bulk of the Asn side chain in Dhh may not be as well accommodated. The remaining differences between Shh and Dhh (S135A, E136Q, and K178R) should be easily tolerated by 5E1, as the side chains of these residues are either minimally involved or away from the epitope.
DISCUSSION
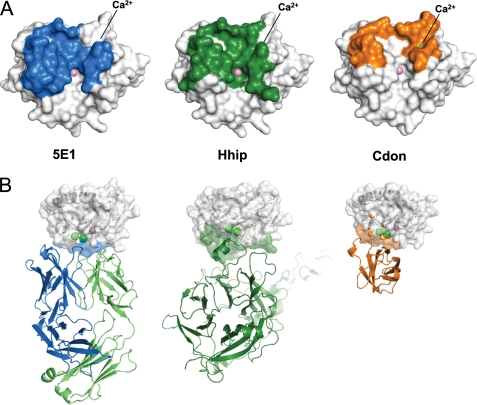
We show here the crystal structure of a chimeric Fab version of the neutralizing anti-Hh antibody 5E1 in complex with human Shh. Strikingly, the ch5E1-Shh interface is very similar in size and location to the Hhip-Shh interface (Fig. 6A) (20, 21). Although this is not completely unexpected as both 5E1 and Hhip are negative regulators of the Hh pathway, both competing for Hh binding and preventing its interaction with Ptc1 (21), the extent of the overlap was unanticipated. Despite the similarity of their interaction surfaces on Shh, Hhip and 5E1 use very different structural motifs to recognize Shh. The Hhip-Shh interface is dominated by a single Hhip loop (the Asp383-containing L2 loop), which binds in the Shh pseudo-active site groove and directly contacts the bound Zn2+ (Fig. 6B). This interaction is mediated by Asp383 of Hhip, which coordinates the Shh Zn2+ (20, 21), explaining the significant loss of binding affinity and decreased cellular activity upon chelation of Zn2+ or mutation of Asp383. 5E1 presents a very different composite interface formed by the six CDRs from the antibody heavy and light chains. In the 5E1-Shh complex, there are no direct contacts between the Shh Zn2+ and 5E1, hence the lesser dependence of 5E1 binding on this cation as compared with the Hhip-Shh interaction. Because 5E1 and Hhip both compete for the pseudo-active site groove despite the structural differences between their Shh-binding surfaces, we also measured the ability of ch5E1 Fab to compete with the much smaller Hhip L2 loop-derived cyclic peptide (Hhip L2 peptide), previously shown to bind to the Shh pseudo-active site (21). Competition binding experiments reveal ch5E1 Fab efficiently displaces Hhip L2 peptide from Shh (Fig. 7), consistent with overlapping binding sites.
FIGURE 6.
Comparison of the 5E1 epitope with the Shh receptor-binding sites. A, footprint of 5E1 (left panel, blue), Hhip (middle panel, green) (21), and Cdon (right panel, orange) (11) on the molecular surface of Shh; Zn2+ and Ca2+ are shown as pink and green spheres, respectively. The location of the Ca2+ is indicated as the ions are largely hidden by the Shh molecule surfaces. The footprint is defined as Shh residues within 4.2 Å of the binding partner. Shh is oriented as in Fig. 3B. B, comparison of the binding modes of Shh with 5E1 (left panel), Hhip (center panel, green), and Cdon (right panel, orange). 5E1, Hhip and Cdon are shown as C-α ribbons. The complexes are oriented such that the Shh pseudo-active site groove runs from left to right in the plane of the figure. The 5E1 and Cdon complexes are in the same orientation. The Hhip complex has been rotated slightly to improve the visibility of Hhip.
FIGURE 7.
Competition binding of ch5E1 Fab with Hhip L2 peptide binding to Shh. Biolayer interferometry sensorgrams of 250 nm Shh binding to biotinylated Hhip L2 peptide on streptavidin-coated biosensors are shown in the presence and absence of various concentrations of ch5E1 Fab. The inset shows the fraction response after binding at equilibrium versus the molar ratio of ch5E1 Fab/Shh. Because Shh and ch5E1 concentrations used are well above the KD value, ch5E1 effectively titrates Shh, having a molar ratio of 1.25 (x axis intercept), close to the predicted value of 1.0. At molar ratio 2.4, there is no observed binding of Shh to the Hhip L2 peptide.
Commensurate with the activity of ch5E1 in vivo (31), the ch5E1-binding site does not include areas of Shh known to be involved in multimerization. Specifically, Lys132 in Hh has been shown to be important for the multimerization and nanoscale organization of Hh necessary for long range signaling (44). The corresponding residue in human Shh is Arg72, which is ∼25 Å away from the 5E1-binding site (Fig. 4A).
The 10–20-fold increased affinity of 5E1 for Shh in the presence of Ca2+ and Zn2+, with calcium having a stronger enhancing effect on binding than zinc, was unexpected. Because the extracellular physiological concentrations of 1.3 mm Ca2+ and 12–17 μm Zn2+ (45, 46) exceed their respective KD values for Shh of ∼100 μm (11) and <100 pm (47), the two Shh cation-binding sites are likely fully occupied under physiological conditions and are thus unlikely to regulate Shh activity in vivo or affect 5E1 activity in a therapeutic setting. In addition, in studies using 5E1 in a Hh-expressing xenograft model of colorectal cancer, efficacious doses of 5E1 result in serum levels of 5E1 that are over 1000-fold higher than the KD for Shh for up to a week (Ref. 31 and data not shown). Nonetheless, the effect of calcium on 5E1 binding further highlights the importance of the Zn2+-containing pseudo-active site groove and adjacent Ca2+ site of Shh as being critical for Shh interactions with its binding partners (11, 20, 21). Specifically, Hh binding to the positive regulator Cdon is undetectable in the absence of Ca2+, although Hhip and 5E1 merely show an ∼10–25-fold increase in affinity for Hh in the presence of Ca2+.
The enhancement of 5E1 binding, rather than absolute dependence on Ca2+ observed with Cdon, is consistent with the structures of the Cdon-Shh, Hhip-Shh, and 5E1-Shh complexes. In the Cdon-Shh complex, the Ca2+ site and coordinating residues are buried in the Cdon-Shh interface (Fig. 6) (11). In contrast to the Cdon-Shh structure, the Hhip-Shh and 5E1-Shh interactions are centered on the Zn2+-containing pseudo-active site groove, whereas the Ca2+ site is at the periphery of these interfaces.
Although the importance of Ca2+ for Shh binding to 5E1 (and co-receptors) has only recently been elucidated, antibodies against other antigens have been identified whose binding occurs in a Ca2+-dependent manner (48–51). Perhaps the best known is the anti-FLAG M1 antibody, which is widely used for affinity purification of FLAG-tagged antigens on columns due to its elutability with the calcium chelator EGTA (52). Additionally, the noninhibitory monoclonal antibody BG6 binds to the serine protease C1r only when the protease contains bound Ca2+ (53). The antibody Q425 also requires calcium for high affinity interactions with CD4 (54), although in this particular case, the Ca2+-binding site is in the antibody CDRs rather than in the antigen, in contrast to the ch5E1 Fab-Shh complex.
To our knowledge, the ch5E1 Fab-Shh complex represents the first report of the experimental structure of an inhibitory antibody bound to an antigen with a metalloprotease fold. Although the monoclonal antibody DX-2400 (55) blocks the enzymatic activity of MMP-14 and shows strong anti-tumor activity in vivo, no structural characterization has been reported. A few structures are available for inhibitor Fabs bound to serine proteases. These structures show that the antibodies partially or completely block access to the protease substrate-binding cleft either by directly occluding the cleft or by altering the cleft in an allosteric manner (56–58). Thus 5E1, and even Hhip (20, 21), despite the presence of a pseudo-active rather than active site in Shh, share the active site blocking mechanism employed by some of the protease-neutralizing antibodies.
Taken together, these data further highlight the role of the Shh pseudo-active site groove as an important protein-protein interaction interface for regulating Hh signaling (13). Activation of the pathway occurs upon binding of Hh to Ptc1, but signaling is inhibited if Hh binds instead to the decoy receptor Hhip (itself up-regulated by Hh signaling, thus providing a negative feedback loop) or the exogenous blocking antibody 5E1, a potential therapeutic. The 5E1 epitope also overlaps with the binding site of the agonist co-receptor Cdon, and thus we predict that 5E1 should also compete with Cdon for binding to Shh (Fig. 6). It will therefore be intriguing to see if other Hh inhibitors such as the heparan chains of the glycoprotein glypican-3 (59) or the small molecule robotnikinin (60, 61) also exert their effects by targeting the Hh pseudo-active site groove.
Acknowledgments
We thank Kurt Schroeder for growing the mouse 5E1 hybridoma; Wendy Sandoval for N-terminally sequencing 5E1; Yongmei Chen and Yan Wu for making the ch5E1 mammalian expression construct; George Dutina, Mark Nagel, and Erin Harrod for expressing and purifying ch5E1; Richard Vandlen for isolation of ch5E1 Fab; Hua Tian for the mouse embryo; Steven Russell for atomic absorption spectroscopy, and Christine Yu for mounting and characterizing the crystals. We also gratefully acknowledge our collaborators at Curis, Inc. (Cambridge, MA), for providing the mouse 5E1 hybridoma. ALS beamline 5.0.1 and the Berkeley Center for Structural Biology are supported by the National Institutes of Health, NIGMS, and United States Department of Energy.
The atomic coordinates and structure factors (codes 3MXV and 3MXW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Shh
- Sonic hedgehog
- Hh
- hedgehog
- Dhh
- Desert hedgehog
- Ihh
- Indian hedgehog
- Ptc1
- Patched1
- Hhip
- Hedgehog-interacting protein
- CDR
- complement determining region
- ITC
- isothermal titration calorimetry
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Ingham P. W., McMahon A. P. (2001) Genes Dev. 15, 3059–3087 [DOI] [PubMed] [Google Scholar]
- 2.Jiang J., Hui C. C. (2008) Dev. Cell 15, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scales S. J., de Sauvage F. J. (2009) Trends. Pharmacol. Sci. 30, 303–312 [DOI] [PubMed] [Google Scholar]
- 4.Huangfu D., Anderson K. V. (2006) Development 133, 3–14 [DOI] [PubMed] [Google Scholar]
- 5.Lee J. J., Ekker S. C., von Kessler D. P., Porter J. A., Sun B. I., Beachy P. A. (1994) Science 266, 1528–1537 [DOI] [PubMed] [Google Scholar]
- 6.Porter J. A., Ekker S. C., Park W. J., von Kessler D. P., Young K. E., Chen C. H., Ma Y., Woods A. S., Cotter R. J., Koonin E. V., Beachy P. A. (1996) Cell 86, 21–34 [DOI] [PubMed] [Google Scholar]
- 7.Porter J. A., von Kessler D. P., Ekker S. C., Young K. E., Lee J. J., Moses K., Beachy P. A. (1995) Nature 374, 363–366 [DOI] [PubMed] [Google Scholar]
- 8.Porter J. A., Young K. E., Beachy P. A. (1996) Science 274, 255–259 [DOI] [PubMed] [Google Scholar]
- 9.Pepinsky R. B., Zeng C., Wen D., Rayhorn P., Baker D. P., Williams K. P., Bixler S. A., Ambrose C. M., Garber E. A., Miatkowski K., Taylor F. R., Wang E. A., Galdes A. (1998) J. Biol. Chem. 273, 14037–14045 [DOI] [PubMed] [Google Scholar]
- 10.Hall T. M., Porter J. A., Beachy P. A., Leahy D. J. (1995) Nature 378, 212–216 [DOI] [PubMed] [Google Scholar]
- 11.McLellan J. S., Zheng X., Hauk G., Ghirlando R., Beachy P. A., Leahy D. J. (2008) Nature 455, 979–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuse N., Maiti T., Wang B., Porter J. A., Hall T. M., Leahy D. J., Beachy P. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10992–10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maun H. R., Kirchhofer D., Lazarus R. A. (2010) Biol. Chem., in press [DOI] [PubMed] [Google Scholar]
- 14.Zhang W., Kang J. S., Cole F., Yi M. J., Krauss R. S. (2006) Dev. Cell 10, 657–665 [DOI] [PubMed] [Google Scholar]
- 15.Kang J. S., Gao M., Feinleib J. L., Cotter P. D., Guadagno S. N., Krauss R. S. (1997) J. Cell Biol. 138, 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenzen T., Allen B. L., Cole F., Kang J. S., Krauss R. S., McMahon A. P. (2006) Dev. Cell 10, 647–656 [DOI] [PubMed] [Google Scholar]
- 17.Kang J. S., Mulieri P. J., Hu Y., Taliana L., Krauss R. S. (2002) EMBO J. 21, 114–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen B. L., Tenzen T., McMahon A. P. (2007) Genes Dev. 21, 1244–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang P. T., McMahon A. P. (1999) Nature 397, 617–621 [DOI] [PubMed] [Google Scholar]
- 20.Bishop B., Aricescu A. R., Harlos K., O'Callaghan C. A., Jones E. Y., Siebold C. (2009) Nat. Struct. Mol. Biol. 16, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosanac I., Maun H. R., Scales S. J., Wen X., Lingel A., Bazan J. F., de Sauvage F. J., Hymowitz S. G., Lazarus R. A. (2009) Nat. Struct. Mol. Biol. 16, 691–697 [DOI] [PubMed] [Google Scholar]
- 22.McLellan J. S., Yao S., Zheng X., Geisbrecht B. V., Ghirlando R., Beachy P. A., Leahy D. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17208–17213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podlasek C. A., Barnett D. H., Clemens J. Q., Bak P. M., Bushman W. (1999) Dev. Biol. 209, 28–39 [DOI] [PubMed] [Google Scholar]
- 24.Wallace V. A., Raff M. C. (1999) Development 126, 2901–2909 [DOI] [PubMed] [Google Scholar]
- 25.Wallace V. A. (1999) Curr. Biol. 9, 445–448 [DOI] [PubMed] [Google Scholar]
- 26.Outram S. V., Varas A., Pepicelli C. V., Crompton T. (2000) Immunity 13, 187–197 [DOI] [PubMed] [Google Scholar]
- 27.Koyama E., Wu C., Shimo T., Pacifici M. (2003) Dev. Dyn. 226, 149–154 [DOI] [PubMed] [Google Scholar]
- 28.Berman D. M., Karhadkar S. S., Maitra A., Montes De Oca R., Gerstenblith M. R., Briggs K., Parker A. R., Shimada Y., Eshleman J. R., Watkins D. N., Beachy P. A. (2003) Nature 425, 846–851 [DOI] [PubMed] [Google Scholar]
- 29.Angot E., Loulier K., Nguyen-Ba-Charvet K. T., Gadeau A. P., Ruat M., Traiffort E. (2008) Stem Cells 26, 2311–2320 [DOI] [PubMed] [Google Scholar]
- 30.Huang S., He J., Zhang X., Bian Y., Yang L., Xie G., Zhang K., Tang W., Stelter A. A., Wang Q., Zhang H., Xie J. (2006) Carcinogenesis 27, 1334–1340 [DOI] [PubMed] [Google Scholar]
- 31.Yauch R. L., Gould S. E., Scales S. J., Tang T., Tian H., Ahn C. P., Marshall D., Fu L., Januario T., Kallop D., Nannini-Pepe M., Kotkow K., Marsters J. C., Rubin L. L., de Sauvage F. J. (2008) Nature 455, 406–410 [DOI] [PubMed] [Google Scholar]
- 32.Bailey J. M., Mohr A. M., Hollingsworth M. A. (2009) Oncogene 28, 3513–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ericson J., Morton S., Kawakami A., Roelink H., Jessell T. M. (1996) Cell 87, 661–673 [DOI] [PubMed] [Google Scholar]
- 34.Pepinsky R. B., Rayhorn P., Day E. S., Dergay A., Williams K. P., Galdes A., Taylor F. R., Boriack-Sjodin P. A., Garber E. A. (2000) J. Biol. Chem. 275, 10995–11001 [DOI] [PubMed] [Google Scholar]
- 35.Carter P., Presta L., Gorman C. M., Ridgway J. B., Henner D., Wong W. L., Rowland A. M., Kotts C., Carver M. E., Shepard H. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4285–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank-Kamenetsky M., Zhang X. M., Bottega S., Guicherit O., Wichterle H., Dudek H., Bumcrot D., Wang F. Y., Jones S., Shulok J., Rubin L. L., Porter J. A. (2002) J. Biol. 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D. Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 38.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D. Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 39.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40.Wijgerde M., McMahon J. A., Rule M., McMahon A. P. (2002) Genes Dev. 16, 2849–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu M., Lui V. C., Sham M. H., Pachnis V., Tam P. K. (2004) J. Cell Biol. 166, 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Incardona J. P., Lee J. H., Robertson C. P., Enga K., Kapur R. P., Roelink H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 12044–12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roelink H., Augsburger A., Heemskerk J., Korzh V., Norlin S., Ruiz i Altaba A., Tanabe Y., Placzek M., Edlund T., Jessell T. M., Dodd J. (1994) Cell 76, 761–775 [DOI] [PubMed] [Google Scholar]
- 44.Vyas N., Goswami D., Manonmani A., Sharma P., Ranganath H. A., VijayRaghavan K., Shashidhara L. S., Sowdhamini R., Mayor S. (2008) Cell 133, 1214–1227 [DOI] [PubMed] [Google Scholar]
- 45.Hofer A. M., Brown E. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 530–538 [DOI] [PubMed] [Google Scholar]
- 46.Arnaud J., Weber J. P., Weykamp C. W., Parsons P. J., Angerer J., Mairiaux E., Mazarrasa O., Valkonen S., Menditto A., Patriarca M., Taylor A. (2008) Clin. Chem. 54, 1892–1899 [DOI] [PubMed] [Google Scholar]
- 47.Day E. S., Wen D., Garber E. A., Hong J., Avedissian L. S., Rayhorn P., Shen W., Zeng C., Bailey V. R., Reilly J. O., Roden J. A., Moore C. B., Williams K. P., Galdes A., Whitty A., Baker D. P. (1999) Biochemistry 38, 14868–14880 [DOI] [PubMed] [Google Scholar]
- 48.Borowski M., Furie B. C., Bauminger S., Furie B. (1986) J. Biol. Chem. 261, 14969–14975 [PubMed] [Google Scholar]
- 49.Horn I. R., Moestrup S. K., van den Berg B. M., Pannekoek H., Nielsen M. S., van Zonneveld A. J. (1995) J. Biol. Chem. 270, 11770–11775 [DOI] [PubMed] [Google Scholar]
- 50.Huang M., Furie B. C., Furie B. (2004) J. Biol. Chem. 279, 14338–14346 [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi K., Yoshida M., Shinoda Y., Yazawa M., Yagi K. (1991) J. Biochem. 109, 551–558 [DOI] [PubMed] [Google Scholar]
- 52.Prickett K. S., Amberg D. C., Hopp T. P. (1989) BioTechniques 7, 580–589 [PubMed] [Google Scholar]
- 53.Ward S. L., Ingham K. C. (1992) Mol. Immunol. 29, 83–93 [DOI] [PubMed] [Google Scholar]
- 54.Zhou T., Hamer D. H., Hendrickson W. A., Sattentau Q. J., Kwong P. D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14575–14580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devy L., Huang L., Naa L., Yanamandra N., Pieters H., Frans N., Chang E., Tao Q., Vanhove M., Lejeune A., van Gool R., Sexton D. J., Kuang G., Rank D., Hogan S., Pazmany C., Ma Y. L., Schoonbroodt S., Nixon A. E., Ladner R. C., Hoet R., Henderikx P., Tenhoor C., Rabbani S. A., Valentino M. L., Wood C. R., Dransfield D. T. (2009) Cancer Res. 69, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 56.Farady C. J., Egea P. F., Schneider E. L., Darragh M. R., Craik C. S. (2008) J. Mol. Biol. 380, 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ganesan R., Eigenbrot C., Wu Y., Liang W. C., Shia S., Lipari M. T., Kirchhofer D. (2009) Structure 17, 1614–1624 [DOI] [PubMed] [Google Scholar]
- 58.Wu Y., Eigenbrot C., Liang W. C., Stawicki S., Shia S., Fan B., Ganesan R., Lipari M. T., Kirchhofer D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19784–19789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Capurro M. I., Xu P., Shi W., Li F., Jia A., Filmus J. (2008) Dev. Cell 14, 700–711 [DOI] [PubMed] [Google Scholar]
- 60.Stanton B. Z., Peng L. F., Maloof N., Nakai K., Wang X., Duffner J. L., Taveras K. M., Hyman J. M., Lee S. W., Koehler A. N., Chen J. K., Fox J. L., Mandinova A., Schreiber S. L. (2009) Nat. Chem. Biol. 5, 154–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng L. F., Stanton B. Z., Maloof N., Wang X., Schreiber S. L. (2009) Bioorg. Med. Chem. Lett. 19, 6319–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]