Abstract
Macrophage migration inhibitory factor (MIF), a proinflammatory cytokine, is considered an attractive therapeutic target in multiple inflammatory and autoimmune disorders. In addition to its known biologic activities, MIF can also function as a tautomerase. Several small molecules have been reported to be effective inhibitors of MIF tautomerase activity in vitro. Herein we employed a robust activity-based assay to identify different classes of novel inhibitors of the catalytic and biological activities of MIF. Several novel chemical classes of inhibitors of the catalytic activity of MIF with IC50 values in the range of 0.2–15.5 μm were identified and validated. The interaction site and mechanism of action of these inhibitors were defined using structure-activity studies and a battery of biochemical and biophysical methods. MIF inhibitors emerging from these studies could be divided into three categories based on their mechanism of action: 1) molecules that covalently modify the catalytic site at the N-terminal proline residue, Pro1; 2) a novel class of catalytic site inhibitors; and finally 3) molecules that disrupt the trimeric structure of MIF. Importantly, all inhibitors demonstrated total inhibition of MIF-mediated glucocorticoid overriding and AKT phosphorylation, whereas ebselen, a trimer-disrupting inhibitor, additionally acted as a potent hyperagonist in MIF-mediated chemotactic migration. The identification of biologically active compounds with known toxicity, pharmacokinetic properties, and biological activities in vivo should accelerate the development of clinically relevant MIF inhibitors. Furthermore, the diversity of chemical structures and mechanisms of action of our inhibitors makes them ideal mechanistic probes for elucidating the structure-function relationships of MIF and to further determine the role of the oligomerization state and catalytic activity of MIF in regulating the function(s) of MIF in health and disease.
Keywords: Biophysics, Covalent Regulation, Enzyme Inhibitors, Inflammation, Protein Structure, Ebselen, Disulfide Mutant, Macrophage Migration Inhibitory Factor, Suicide Inhibitors
Introduction
Macrophage migration inhibitory factor (MIF)2 was discovered in the 1960's as a T-lymphocyte product that inhibits the random migration of macrophages during delayed-type hypersensitivity responses (1, 2). Two decades later, a human MIF gene was cloned (3). Yet, the biological activity of MIF remained ambiguous until the production of bioactive MIF and anti-MIF antibodies (4). Various biological activities have been attributed to MIF, which is recognized as a major regulator of inflammation and a central upstream mediator of innate immune responses (5, 6). MIF has broad regulatory properties and is considered as a critical mediator of multiple disorders including inflammatory and autoimmune diseases such as rheumatoid arthritis (7, 8), glomerulonephritis (9, 10), diabetes (11), atherosclerosis (12), sepsis (13–15), asthma (16, 17), and acute respiratory distress syndrome (18). Furthermore, recent studies have highlighted a role for MIF in tumorigenesis. Human cancer tissues, including skin, brain, breast, colon, prostate, and lung-derived tumors were observed to overexpress MIF, and MIF levels correlated with tumor aggressiveness and metastatic potential (19, 20). Therefore, MIF is considered a viable therapeutic target for treating inflammatory diseases and neoplasia.
In addition to its physiologic and pathophysiologic activities, MIF is known to act as a tautomerase. This activity was initially discovered during the investigation of melanin biosynthesis (21). Subsequent studies revealed that MIF catalyzes the tautomerization of d-dopachrome methyl ester (22–24) and bears high structure, but not sequence, homology with bacterial tautomerases 4-oxalocrotonate-tautomerase, 5-carboxymethyl-2-hydroxymuconate isomerase, and chorismate mutase (25–27). X-ray crystallography, NMR, and biophysical solution studies demonstrate that MIF exists predominantly as a homotrimer (28, 29). The tautomerase active site is formed at the monomer-monomer interfaces within the trimer and involves residues from two adjacent subunits. The N-terminal proline (Pro1) with an unusual pKa (5.6 ± 0.1) (24) is commonly conserved in all of these tautomerases and is essential for MIF tautomerase activity. Covalent modification of Pro1 or its replacement by serine, alanine, or glycine totally abolishes the tautomerase activity of MIF (23, 30). p-Hydroxyphenylpyruvate and phenylpyruvate were identified as potential tautomerase substrates (31). However, the kinetic parameters and the separate localization of these substrates from MIF suggest that these molecules are not physiological substrates of MIF.
Although the relationship between the catalytic activity and biological function of MIF is not yet fully understood, targeting MIF tautomerase activity using small-molecule inhibitors has emerged as an attractive strategy for inhibiting MIF proinflammatory activity and attenuating its biological activity in vitro and in vivo (32, 33). The first MIF inhibitors were reported in 1999 while trying to elucidate the mechanism of MIF tautomerase activity by testing the inhibitory effect of various structure analogues of its substrate, d-dopachrome methyl ester (34). Since then, different classes of tautomerase inhibitors have been developed and were later shown to modulate biological activities of MIF mediated by both its ability to act on intracellular and extracellular signaling pathways (33, 35). As of today, 11 distinct chemical classes of MIF inhibitors have been developed (36) using different approaches, including (i) active site-directed targeting; (ii) rational drug design, i.e. screening molecules that share structure similarity with known MIF tautomerase substrates and inhibitors; and (iii) virtual high throughput screening and computer-assisted drug design approaches. The majority of the inhibitors described to date exert their effects either by competing with the substrate for the catalytic site (e.g. ISO-1 and OXIM11) or via covalent modification of the catalytic Pro1 residue (NAPQI (37) and 4-iodo-6-phenylpyrimidine (4-IPP) (33)). For example, Senter and colleagues (37) identified a class of acetaminophen derivatives (NAPQI), which form a covalent complex with MIF by reacting with the catalytic proline residue. NAPQI was shown to block the ability of MIF to override the immunosuppressive effect of dexamethasone on LPS-induced TNF production by monocytes. A series of MIF inhibitors based on modifications of the scaffold of (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester (ISO-1) were developed (38–41). ISO-1 and some of its derivatives were shown to block MIF tautomerase activity, inhibit TNF secretion from macrophages upon stimulation with LPS, and increase survival in models of sepsis (35, 42).
Recent studies from our group demonstrate that subtle conformational changes induced by mutations distal to the active site result in significant inhibition of MIF tautomerase activity (43, 44). These findings indicate that “allosteric” inhibition of MIF enzymatic activity is possible, but may not be realized by current efforts focused on targeting the active enzymatic site of MIF using rational drug design strategies. Motivated by these findings, we sought to develop a high throughput screening (HTS) assay that would facilitate the identification of different classes of MIF inhibitors, including novel irreversible, competitive, and allosteric inhibitors, as well as molecules that may inhibit MIF by blocking and/or disrupting its oligomerization (i.e. trimer formation). To achieve this goal, we developed a robust tautomerase activity-based HTS assay and screened two chemical libraries containing a total of 15,440 compounds. Twelve novel classes of MIF inhibitors were identified with IC50 values in the range of 0.2–15.5 μm. Using structure-activity studies, and a battery of biochemical and biophysical methods, we were able to define the mechanism of action for each of the three classes of inhibitors. These results and their implications for developing therapeutic strategies targeting MIF and elucidating the biochemical and structural basis underlying its activities in health and disease are presented and discussed.
EXPERIMENTAL PROCEDURES
Chemical Libraries
The NINDS Custom Collection II library from Microsource Discovery Systems, Inc. and the Maybridge library were tested. These libraries were composed of 1,040 and 14,400 biologically active chemical molecules, respectively. The compounds were arrayed in 384-well plates at a final concentration of 10 μm and a final DMSO concentration of 1%.
Compounds Used for Follow-up Studies
All hits generated from the Maybridge library were purchased from Maybridge. Hexachlorophene (HCLP) and its analogues (dichlorophene, bithionol, bis(2-hydroxyphenyl)methane, 2,2′-diaminodiphenyl sulfide, 4,4′-dichlorobenzophenone, 2,2′-sulfinyl-bis(4,6-dichlorophenol), 3,4-dihydroxy benzophenone, igrasan, benzophenone, and emodin) were purchased from Sigma and Fluka and were of the highest purity available, whereas the analogue MDPI 894 was purchased from Molecular Diversity Preservation International (MDPI), Basel, Switzerland.
Expression and Purification of Human MIF and Its Mutants (C56S, C59S, C80S, and N110C)
MIF was expressed by heat shock transformation of the BL21/DE3 Escherichia coli strain (Stratagene) with the bacterial expression vector pET11b containing the human (MIF) gene under control of the T7 promoter. Four hours post-induction, the cells were harvested, re-suspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 50 mm KCl, 5 mm MgAc, 0.1% NaN3), sonicated, and centrifuged at 14,000 × g for 20 min. The clarified cell lysate was filtered, injected onto a MonoQ anion exchange column (HiPrep 16/10 Q FF, GE Healthcare), and eluted with a linear NaCl gradient in the elution buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl). The flow-through fractions containing MIF were pooled and loaded onto a Superdex 75 16/60 (HiLoad 16/60, Superdex 75, GE Healthcare) gel filtration column. Fractions corresponding to MIF were combined, dialyzed against 1× PBS, and filtered through a 0.2-μm filter. Recombinant MIF used for cellular studies was subjected to LPS removal as described previously (45). Briefly, bacterial cell lysate was injected onto an anion exchange column. The flow-through fractions containing MIF were applied to C8 Sep-Pak cartridges (Waters Associates) and MIF was eluted with 60% acetonitrile/water, lyophilized, and renaturated. The LPS content was quantified using the chromogenic Limulus amoebocyte assay.
MIF mutants (C56S, C80S, and N110C) were expressed and purified as described for the wild type human MIF. C59S and C56S/C59S mutants were expressed and purified as previously described by Kleemann et al. (46). Uniformly 15N-labeled protein samples were prepared for NMR experiments by culturing the bacteria in M9 minimal medium containing 15N-ammonium chloride (1 g/liter) as the only nitrogen source.
Assay Design to Screen for Potential Inhibitors of MIF Tautomerase Activity
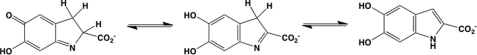
The assay principle is based on measuring the absorbance of d-dopachrome methyl ester (red), which is transformed by enzymatically active MIF to the enol form that is colorless (Fig. 1). In the absence of MIF, the keto form of the substrate shows an absorbance at 475 nm that diminishes upon addition of MIF (supplemental Fig. S1). Keto-enol tautomeric conversion of d-dopachrome methyl ester by MIF was optimized in cuvette format as described previously (23). A fresh stock solution of d-dopachrome methyl ester was prepared through oxidation of l-3,4-dihydroxy phenylalanine methyl ester with sodium meta-periodate. l-3,4-Dihydroxy-phenylalanine methyl ester (4 mm) was first diluted in 5 ml of distilled water, then the appropriate amount of sodium metaperiodate was added to a final concentration of 8 mm. The solution was mixed, incubated for 5 min at room temperature, protected from light and then kept on ice. d-Dopachrome methyl ester stability over time was evaluated at different temperatures. Complete stability was only observed when the substrate was stored at 4 °C. However, a decrease in absorbance was easily noticed at room temperature or 37 °C (supplemental Fig. 1A).
FIGURE 1.
MIF catalyzed tautomerization of d-dopachrome.
The tautomerase enzymatic activity was measured in 50 mm potassium phosphate buffer, 0.5 mm EDTA, pH 6.5. MIF (100 nm) dialyzed in 1× PBS was added to the cuvette and the decrease in absorbance at 475 nm was followed for 3 min using a CARY 100 Bio UV-visible spectrophotometer. To perform this assay in an HTS format, MIF (500 nm, 6 μl) was added to 384-well plates (Nunc, Black wall, clear bottom) containing 50 mm potassium phosphate buffer, 0.5 mm EDTA, pH 6, and the compounds of interest at a final concentration of 10 μm. MIF and compounds were mixed and incubated for a fixed time period of 15 min at room temperature before initiating the reaction by addition of a fresh stock solution of d-dopachrome methyl ester, which was prepared as described above (final reaction volume = 80 μl). The plates were centrifuged for 2 min at 2000 × g to remove air bubbles and the absorbance was measured at 475 nm using a Saphire II Tecan reader (supplemental Fig. S2A). As a positive control, we initially used ISO-1 and later switched to ebselen (10 μm), which gave complete inhibition of the enzymatic activity of MIF. The entire assay was performed using the Biomek 3000 Laboratory Automation Work station from Beckman Coulter, and the entire procedure was carried out in the dark. All assays were carried out in duplicate and hit compounds were retested individually to eliminate any false positives.
Data Analysis
The Z′ value was calculated according to the method developed by Zhang et al. (47),
 |
where S.D. positive control is the standard deviation (n = 16) of the positive control (with 10 μm ebselen), S.D. negative control is the standard deviation (n = 24) of the negative control (MIF in the absence of inhibitor), mean positive control is the mean (n = 16) of the positive control (with 10 μm ebselen), and the mean of the negative control is the mean (n = 24) of the negative control (MIF in the absence of inhibitor).
IC50 and Ki,app Assessment
For the determination of IC50 values, the d-dopachrome was used as a substrate; the hit compounds were distributed in 384-well plate at 12 dilutions starting from 0 to 50 μm. One percent DMSO was used as a final concentration in the reaction buffer; the assay was performed in triplicate as described above and each sample was evaluated in duplicate.
Ki,app values were obtained by plotting the relative initial velocities as a function of the inhibitor concentration. MIF was preincubated with 12 concentrations of hit compounds ranging from 0 to 50 μm for 15 min, followed by the addition of 2 mm HPP and the absorbance at 300 nm was measured for 3 min. Data were fitted to the simple inhibition expression using Sigmaplot software: Vi = (V0/(1 + (I/Ki,app)n))+ bkg; where V0, velocity at [I] = 0 and n is the Hill coefficient (48). The percent of inhibition was calculated relative to the DMSO vehicle control and blank control as: % inhibition = 100 − (100 × (test compound value − average of blank control)/(average of DMSO control − average of blank control)).
MALDI-TOF MS Measurements to Examine Possible Protein Modifications
Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) was performed to examine any inhibitor-induced protein modifications. The matrix solution was prepared by dissolving 14 mg/ml of sinapinic acid solution in 0.1% trifluoroacetic acid/acetonitrile (1:1). A thin matrix layer was generated on the mirror-polished target using a gel loading tip. One microliter of sample (15 μm MIF incubated with 10 μm hit compounds for 1 h) was mixed with 1 μl of sinapinic acid matrix solution, and 0.8 μl of this mixture was deposited on top of the thin layer and allowed to air dry. The samples were analyzed with a 4700 MALDI-TOF/TOF instrument (Applied Biosystems).
Protein Digestion Mass Spectrometry Analysis to Identify the Modified Residue(s)
Twenty microliters of MIF sample previously incubated for 1 h with inhibitors at 10 μm were digested overnight with trypsin, 1:50 (Promega). The digestion was stopped with 1 μl of 10% formic acid and stored at 4 °C until further use. The samples were analyzed by MALDI-MS on a 4700 MALDI-TOF/TOF instrument (Applied Biosystems) or an Axima CFR plus instrument (Shimadzu) without further purification. One microliter of sample was mixed with 1 μl of 2,5-dihydroxybenzoic acid (20 mg/ml in 1% phosphoric acid/acetonitrile (1:1)), and 0.8 μl of this mixture was deposited on the target and allowed to air dry.
Probing the Effects of the Hit Compounds on MIF Oligomerization by Analytical Ultracentrifugation
Analytical ultracentrifugation experiments were performed using purified and dialyzed MIF samples (10 μm) preincubated with ebselen for 1 h at an inhibitor concentration corresponding to 100% inhibition. Radial UV scans were recorded on a Beckman Optima XL-A at a wavelength of 277 nm. Sedimentation velocity experiments were carried out at 20 °C using 380–400 μl of protein solution. Data were recorded at rotor speeds of 50,000 rpm in continuous mode, with a step size of 0.003 cm. The experimentally determined partial specific volume of 0.765 ml/mg was used to calculate the molecular weight of wild-type MIF (49). The sedimentation velocity profiles were analyzed as a c(s) distribution of the Lamm equation using SEDFIT (50). To obtain the molecular weights, molar mass distributions c(M) were obtained by transforming the corresponding c(s) using SEDFIT.
MIF Aggregation Studies
Wild type human MIF at a concentration of 15 μm in 1× PBS was incubated with different ebselen concentrations ranging from 0.01 to 1 mm for 1 h at room temperature, and then filtered with a 0.2-μm filter (Millipore). The retentate was solubilized in 0.1% SDS. Both the supernatant and the resolubilized aggregates were analyzed in a 15% SDS gel. To further examine cysteine-induced aggregation by ebselen, either the N110C stable cross-linked trimer or the alkylated wild type human MIF or cysteine mutants were tested.
Cysteine Alkylation
To determine whether some of the hit compounds induce their effect via modification of cysteine residues (i.e. Cys56, Cys59, and Cys80), all free thiols were blocked by covalent modification with maleimide (3,3′-N-[e-maleimidocaproic acid)hydrazide, trifluoroacetic acid salt (EMCH)) (Pierce). Fifteen micromolar wild type or mutant forms of human MIF in 1× PBS, pH 7.4, was incubated with 10 mm EMCH for 1 h at room temperature. The unreacted EMCH was removed using PD-10 desalting columns (Pierce). The collected fractions were analyzed in an SDS-PAGE gel and the concentration was assessed using a UV spectrophotometer.
Nuclear Magnetic Resonance (NMR) Spectroscopy
NMR spectra were acquired at 27 °C on a Bruker Avance III 600 MHz NMR spectrometer. 500 μm wild type human MIF samples were prepared in 20 mm Na2HP04, 0.5 mm EDTA, 0.02% NaN3, pH 7.0, and 10% D2O. A specific amount of each compound was prepared in DMSO and mixed with the above sample to give a final 1% DMSO concentration in the total sample volume. Two-dimensional 1H-15N heteronuclear single quantum coherence (HSQC) spectra were recorded using 48 scans per increment with 256 × 1024 complex data points in F1 and F2 dimensions and relaxation delay of 1.0 s. Spectra were processed with Topspin (Bruker Biospin, Germany) and NMRPipe (51). Visualization and manipulation was performed using Sparky 3.110.3 Averaged chemical shift deviation was calculated with Equation 2.
 |
Biological Assays
Glucocorticoid Overriding Assay
Mouse RAW 264.7 macrophages (TIB-71, ATCC) were grown in RPMI 1640 medium containing 2 mm glutamine and 10% fetal calf serum. MIF was incubated with the inhibitors for 15 min at room temperature. RAW 264.7 macrophages (5 × 104 cells/well in 96-well plates) were preincubated for 1 h with 10−7 m dexamethasone, dexamethasone plus murine MIF (100 ng/ml; 8 nm), or dexamethasone plus MIF and Hit compounds at 10 μm before the addition of 100 ng/ml of Salmonella minnesota Ultra Pure LPS (List Biologicals Laboratories). TNF in cell culture supernatants collected after 4–8 h was measured by enzyme-linked immunosorbent assay (BD Biosciences).
Phosphorylation of AKT by MIF
Confluent HeLa cells (CCL-2) were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum. Cells were incubated for 2 h with human MIF (50 ng/ml; 4 nm) in the presence of hit compounds at final concentrations of 1 and 10 μm. Phosphorylation of AKT at Ser473 was measured using the Alpha screen SureFire phosphokinase kit, according to the protocol provided by the manufacturer (PerkinElmer Life Sciences) (53).
MIF-triggered Chemotaxis Assay
Chemotactic assays were performed with primary human endothelial progenitor cells (EPCs). EPCs were isolated from the mononuclear cell fraction obtained by density gradient centrifugation from human blood essentially as previously described (54). Briefly, “buffy coats” were obtained from volunteers in accordance with the local ethics committee. CD34+ cells were enriched from mononuclear cells by Biocoll density gradient centrifugation and CD34-specific magnetic separation. CD34+ cells were plated on fibronectin-coated 6-well plates and cultured in endothelial growth medium (MV2). Endothelial progenitor cells were harvested on day 14 and their identity verified by fluorescence-activated cell sorter analysis to stain for lectin-fluorescein isothiocyanate, DiI-conjugated acLDL, CD31, and VEGFR-2 (KDR) at a rate of >90%. EPC chemotaxis was evaluated using Transwell cell migration chambers in combination with FluoroBlok inserts (BD Biosciences; 8-μm pore size) in a 24-well plate format. Lower chambers contained 10 ng/ml of recombinant human MIF or (100 ng/ml; 0.8 nm) LPS in medium containing 0.5% bovine serum albumin as the chemoattractant. Calcein-stained (Calbiochem) EPCs were preincubated with 10 μm compound and placed into the upper chambers (50,000 cells per insert). Fluorescence signals representing the migration of calcein-stained cells into the bottom chambers were determined 3 h later using a fluorescence microplate reader. As day 14 EPCs express CD14, LPS (100 ng/ml) was used as a positive control. The percentage of cells migrated to the lower chamber was determined after 3 h.
Compound Toxicity Studies Using MTT Assay
RAW 264.7 macrophages (5 × 104 cells/well in 96-well plates) were incubated with 3, 10, and 20 μm of each compound. Twenty-four hours later, toxicity was assessed using the MTT assay (Sigma). Absorbance was recorded at 405 nm with 590 nm as a reference wavelength.
RESULTS
High Throughput Screening Identifies 18 Novel Inhibitors of MIF Tautomerase Activity
The tautomerase assay is a well established method to assess MIF-specific enzymatic activity, and has yielded reproducible results in different laboratories (22, 23, 55). We optimized the conditions for performing this assay in a high throughput (384-well plates) format. Two known non-physiological MIF substrates (d-dopachrome and phenylpyruvate) were evaluated. d-Dopachrome was chosen for screening because it absorbs at 475 nm, a wavelength at which small molecules do not absorb, which is not the case for HPP (300 nm). As a positive control, we initially used ISO-1, a known inhibitor of the enzymatic activity of MIF (35). A validation assay was performed to test the screening conditions in an automated manner and determine the robustness and reproducibility of the assay. Intra-plate and inter-plate variations were determined using single point measurements and Z′ values of 0.74 and 0.91 were obtained.
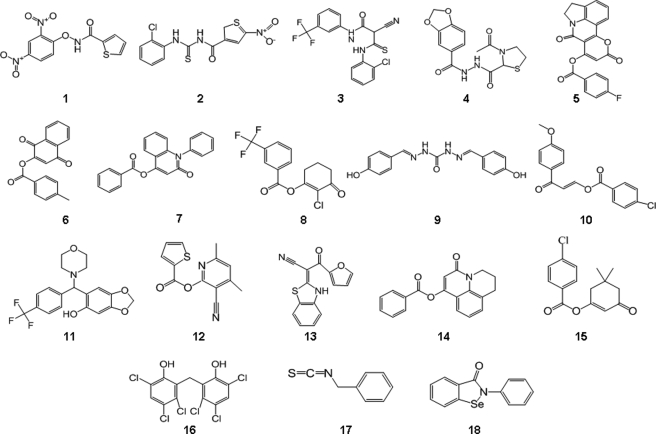
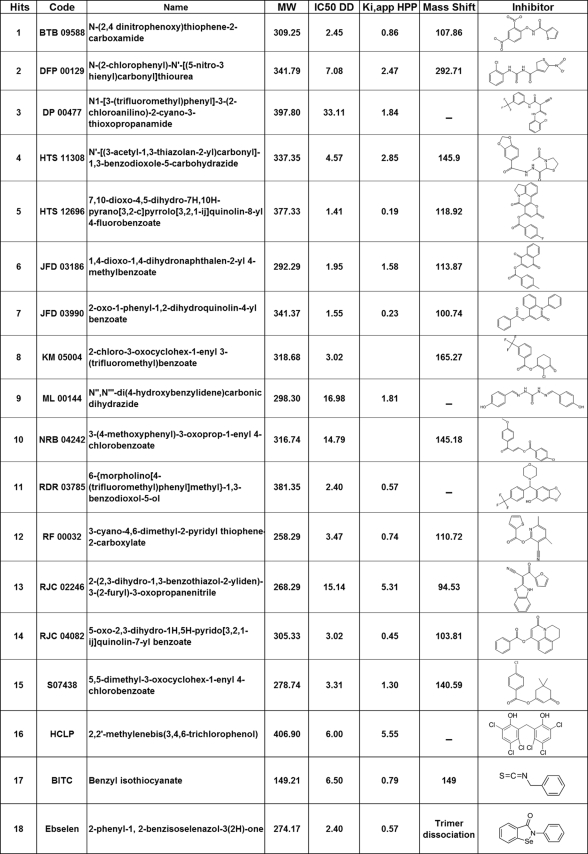
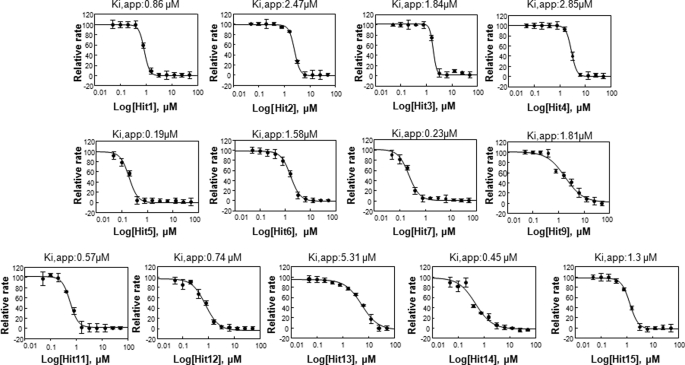
An FDA-approved library (NINDS Custom Collection II) composed of 1,040 bioactive compounds containing a diverse set of drugs, 85% of which are marketed drugs with a wide range of therapeutic usage including anti-inflammatory and analgesia, and the Maybridge library composed of 14,400 compounds were screened. The compounds were dissolved in DMSO to yield a final concentration of 10 μm. DMSO alone at a final concentration of up to 2% had no effect on MIF tautomerase activity (supplemental Fig. S1B). The inhibitor screening assay was performed as follows: MIF was added to 384-well plates containing the compounds in reaction buffer and incubated for 15 min at room temperature. The enzymatic reaction was initiated by adding the d-dopachrome substrate. In the positive control wells, with no enzyme, or with compounds only, the absorbance of the substrate at 475 nm was ∼0.9–1, whereas in the presence of enzyme the absorbance rapidly decreased (∼3 min) to an OD value of ∼0.1 (supplemental Fig. S2A). In our preliminary screens, ebselen was identified as a potent inhibitor, which fully inhibited MIF tautomerase activity at a concentration of 10 μm, making it more effective than ISO-1. Thus, ebselen was used as a positive control (10 μm) in all subsequent screens. The Z′ value for the FDA and Maybridge library screenings were 0.74 and 0.91, respectively (supplemental Fig. S2B). These data suggested that the assay is quite robust and reproducible. The hit compounds demonstrating greater than 50% inhibition of MIF activity were retested manually to exclude false positives. Of the 15,440 compounds tested, only 18 were found to be active (Fig. 2 and Table 1) and demonstrated more than 70% inhibition in the primary assay. To determine the IC50 values, we performed concentration-dependent studies by testing 12 different dilutions ranging from 0 to 50 μm and obtained IC50 values in the range of 0.2–15.5 μm (Fig. 3).
FIGURE 2.
High throughput screening of the NINDS and Maybridge libraries resulted in the identification of 18 novel inhibitors. Chemical structures of the primary and validated hits.
TABLE 1.
Summary table of Maybridge hit compounds code, name, structure, IC50, Ki,app and the mass shift observed by mass spectrometry upon 30 min incubation with MIF covalent modifiers
FIGURE 3.
Ki,app obtained by plotting relative initial velocities as a function of inhibitor concentration and data fitting to the simple inhibition expression: Vi = (V0/(1 + (I/Ki,app)n)) + bkg, where V0, velocity at [I] = 0, and n the Hill coefficient. Each data point represents the mean ± S.D., n = 4. The data were analyzed using Sigma plot software and each experiment was done in triplicate.
MIF Antagonists: Mode of Action
Inhibition of the tautomerase activity of MIF can occur via at least five different mechanisms: 1) binding to the active site; 2) allosteric inhibition; 3) covalent modification of active site residues; 4) disruption of the active site through compound-induced dissociation of the active trimer; and 5) stabilization of the MIF monomer and prevention of its re-association to form the active trimer. The inhibitors identified in our HTS were subjected to biochemical and biophysical studies to understand their mechanism of action.
The Compounds 1, 2, 4, 5, 6, 7, 8, 10, 12, 13, and 14 Inhibit MIF Tautomerase Activity by Selective Modification of the Catalytic N-terminal Proline Residue
To determine whether MIF inhibitors covalently modify MIF, the compounds were incubated for 1 h at room temperature with 15 μm human MIF (in 25 mm Tris buffer, pH 7.4) and subjected to analysis by MALDI-TOF mass spectrometry. Upon incubation with 10 μm of corresponding compounds 1, 2, 4, 5, 6, 7, 8, 10, 12, 13 and 14, we observed a shift in the monomeric molecular mass consistent with a single modification of MIF (supplemental Fig. S3). To identify the site of covalent modifications, the modified MIF was subjected to proteolytic digestion by trypsin and peptide mapping by mass spectrometry. In the case of compounds 2, 6, 8, and 12, we observed a peptide fragment with molecular mass corresponding to that comprising N-terminal residues PMFIVNTNVPR (molecular mass = 1287.6 Da) plus one molecule of inhibitor. MS/MS analyses and sequencing of the modified peptide fragments revealed that modification by these compounds takes place exclusively at the N-terminal proline residue.
Dissociation Constant (KD) Reveals a Strong Binding Behavior for HCLP and Compound 9
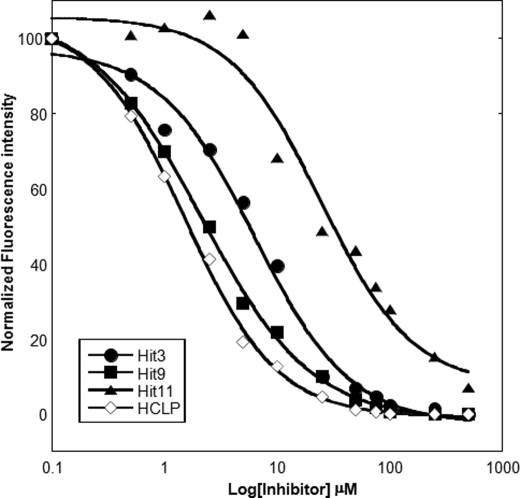
No covalent modifications were observed when compounds 3, 9, 11, and HCLP were incubated with MIF, suggesting that the inhibitory activity of these compounds is mediated by non-covalent interactions with the protein. To evaluate the strength of binding, the KD for each compound was measured using fluorescence spectroscopy. MIF at 5 μm was incubated with increasing concentrations of compounds and binding was evaluated by monitoring changes in the tryptophan emission maximum upon MIF binding to compounds. The maximum fluorescence emission was plotted as a function of compound concentration and normalized. HCLP and compound 9 showed KD values of 1.5 ± 0.1 and 2.11 ± 0.15 μm, respectively. Compounds 3 and 11 exhibited KD values of 6.28 ± 1.02 and 24.45 ± 6.74 μm, respectively (Fig. 4).
FIGURE 4.
Dissociation constant (KD) measurements for HCLP and compounds 3, 9, and 11 determined by monitoring MIF tryptophan fluorescence quenching at 295 nm as a function of increasing concentration of MIF inhibitors.
Elucidating the Structural Basis Underlying the Inhibitory Activity of Ebselen, HCLP, and Compounds 3, 9, and 11 Using NMR
To determine the structural basis for the inhibition of human MIF enzymatic activity, NMR titrations were performed with ebselen, HCLP, and compounds 3, 9, and 11.
Ebselen
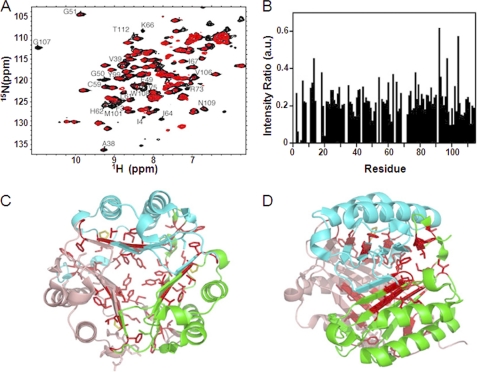
An equimolar mixture of MIF with ebselen resulted in severe resonance broadening throughout the whole sequence, indicative of conformational exchange (Fig. 5A). Most drastic effects were seen for residues 3–5, 38–39, 49–51, 61–62, 64, 99, 101, 106–108, and 112. Most of these residues are in the subunit-subunit interface. The β-sheet core of a monomeric subunit that consists of four β-strands, as well as the three β-strands coming from the other two subunits are highly affected by ebselen (Fig. 5, B and C), which supports the idea that a stable trimer dissociates into monomeric units upon destabilization by ebselen. In agreement with the sedimentation velocity experiments (see below), the drop in signal intensity could be due to a combination of both conformational changes and aggregation of the dissociated monomeric subunits into larger moieties that are beyond the detection limit of liquid state NMR. Among the three cysteines of MIF, only Cys59 displays a minor chemical shift, such an effect is observed for neither Cys56 nor Cys80 (supplemental Fig. S4A). Additionally, the only residue that has surface accessibility is Cys59 (supplemental Fig. S4, B-D), suggesting a role for this residue in ebselen interaction. No change is observed for the α-helices.
FIGURE 5.
Structural basis for the inhibition of MIF enzymatic activity by ebselen using NMR. A, the two-dimensional 1H-15N HSQC reference spectrum (black) was recorded with MIF in the presence of 1% DMSO. The addition of ebselen to MIF at equimolar concentrations (red spectra) resulted in resonance broadening. B, changes in NMR signal intensities in a 1H-15N HSQC upon addition of ebselen. C and D, residues with peak intensity ratios below 0.18 in B are highlighted in red on the three-dimensional structure of MIF (PDB code 1GD0). C and D are related to each other by 90°. Cyan, green, and pink show the three different subunits of MIF. The catalytically active P1 residue is indicated in yellow.
HCLP
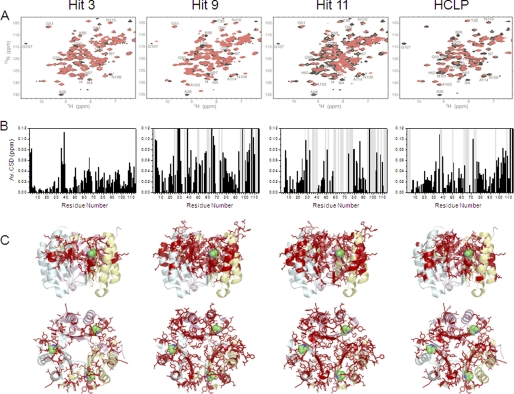
Titration of MIF with HCLP results in both resonance broadening and chemical shift changes (Fig. 6A), indicative of an intermediate exchange regime in the NMR time scale. The residues that are most affected by HCLP are 3–5, 35–38, 64, 67, 95, 107–110, 112, and 114 (Fig. 6B). Similar to the MIF inhibitor benzyl isothiocyanate (56) and HPP (31), these residues cluster around Pro1. Among these residues, Ile64 and Tyr95 are found in the active site.
FIGURE 6.
Structural basis for the inhibition of MIF enzymatic activity by compounds 3, 9, and 11 using NMR. A, the two-dimensional 1H-15N HSQC reference spectrum of MIF (black) was recorded in the presence of 1% DMSO. Addition of the three hit compounds to MIF at equimolar concentrations (red) resulted in NMR chemical shift changes and signal broadening. In case of compounds 9 and 11, a set of new NMR resonances appeared at equimolar ratio. Titration data of HCLP are given for comparison. B, averaged 1H-15N chemical shift deviation observed for backbone resonances of MIF in two-dimensional 1H-15N HSQC NMR spectra upon addition of compounds 3, 9, 11, and HCLP at equimolar ratio (shown in A). Residues affected by slow conformational exchange are indicated by gray bars. C, residues with averaged 1H-15N chemical shift changes above 0.02 ppm are highlighted on the three-dimensional structure of MIF (PDB code 1GD0) using a stick model representation. Cyan, yellow, and pink depict the different subunits of MIF.
Compounds 3, 9, and 11
Backbone amide signals of MIF were monitored for hit compounds 3, 9, and 11. An equimolar mixture of MIF and compound 3 resulted in chemical shift deviations averaged on 1H and 15N dimensions in residues 2–3, 35–39, 49–51, 63–66, 94–96, 107–110, and 112–113 (Fig. 6, A and B). Many of these residues are located in the subunit-subunit interface and enzymatic active site (Fig. 6C). Upon addition of compound 9, more than 0.75 molar ratio relative to MIF, several peaks in the HSQC analysis disappeared, indicating intermediate exchange of conformation. These peaks reappeared with an increasing molar ratio of compound 9, which implies a higher affinity of this inhibitor to MIF. Addition of compound 9 to MIF also induced a chemical shift deviation, but with two sequential changes: first a milder interaction followed by conformational changes in MIF. Up to a 0.5 molar ratio of compound 9, resonances from residues 2–3, 35–37, 50, 58, 60, 61, 65–66, 75, 81, 93, 95–96, 98, 107–108, and 110–111, which correspond to the subunit-subunit interface and enzymatic active site were observed (Fig. 6, B and C). Although compound 11 had the lowest IC50 value in the MIF tautomerase assay, its binding to MIF resulted in an even larger number of residues affected by slow conformational exchange, including residues 2–3, 35–37, 58–60, 60, 65–68, 72–78, 97–102, 105–106, and 112–114 (Fig. 6B). The residue stretches affected by slow conformational exchange are located in the subunit-subunit interface and close to the active site (Fig. 6C).
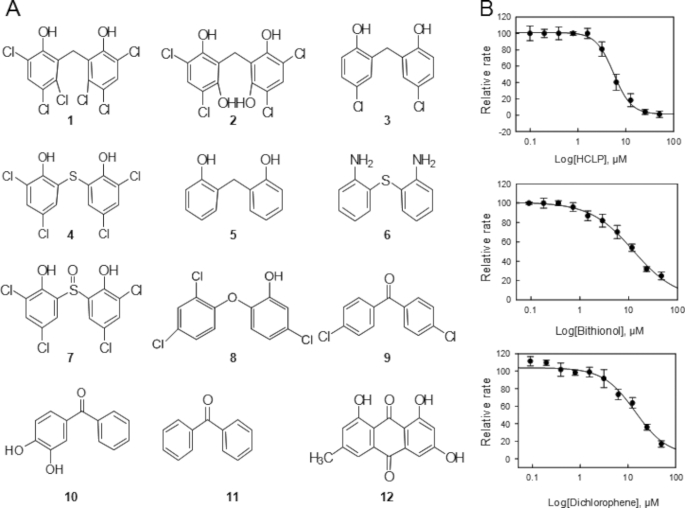
Structure-Activity Relationship Studies for Hexachlorophene
Hexachlorophene did not lead to covalent modification of MIF. Fortunately, several analogues of HCLP are commercially available. Therefore, we performed structure-activity relationship studies to better understand its mechanism of action and to identify the chemical groups responsible for evoking tautomerase inhibition. Eleven analogues of HCLP (Fig. 7A) were selected and tested. Only MDPI894 (IC50, 8 μm), bithionol (IC50, 6.5 μm and Ki,app, 4.74 ± 0.85 μm), and dichlorophene (IC50, 15 μm and Ki,app, 6.57 ± 0.54 μm) were observed to exhibit significant inhibition of MIF compared with HCLP with IC50, 3.5 μm, and Ki,app, 2.21 ± 0.39 μm (Fig. 7B). Compound 5, bis(2-hydroxyphenyl)methane, which lacks any of the chlorine substitutions in the phenol rings is inactive. Interestingly, the addition of only two chlorine atoms at the 3,3′ positions, dichlorophene, was sufficient to transform this molecule into an active inhibitor of MIF, albeit less active than HCLP. Removal of either the hydroxyl groups or the 3,3′ chlorine atoms results in loss of HCLP inhibitory activity. Together, the structure activity results shown in Fig. 7 highlight the importance of the 6,6′ hydroxyl groups and 3,3′ chloride atoms for MIF inhibition. Furthermore, our results suggest that the position of the remaining chlorine atoms on the aromatic rings as well as the nature of the bridging atom influence the potency of HCLP. The inhibitory activity of bithionol demonstrates that replacing the bridging carbon atom of HCLP by a sulfur and removal of the chlorine atoms at the 2,2′ positions reduce the inhibitory potency of HCLP. Substitution of the bridging carbon by a sulfoxide results in loss of inhibitory activity despite the presence of the chlorine atoms at the 3,3′, and 5,5′ positions, suggesting that some conformational flexibility between the two phenol rings is crucial to allow these inhibitors to adopt the right conformation in the active site.
FIGURE 7.
Structure-activity relationship studies for hexachlorophene. Chemical structures of selected HCLP analogues that were tested in the tautomerase assay. A, 1, HCLP; 2, MDPI894; 3, dichlorophene; 4, bithionol; 5, bis(2-hydroxyphenyl)methane; 6, 2,2′-diaminodiphenyl sulfide; 7, 2,2′-sulfinyl-bis(4,6-dichlorophenol); 8, irgasan; 9, 4,4′-dichlorobenzophenone; 10, 3,4-dihydroxybenzophenone; 11, benzophenone; 12, emodin. B, Ki,app of HCLP, bithionol, and dichlorophene.
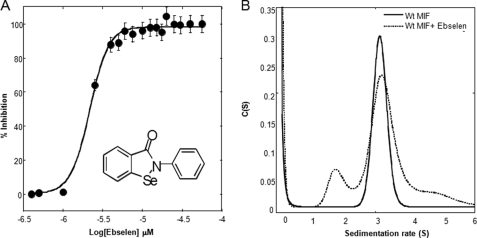
Sedimentation Velocity Demonstrates That Only Ebselen Induces Trimer to Monomer Dissociation
To determine whether any of the antagonists functioned by disrupting the MIF trimer, we compared the quaternary structure distribution of human MIF in the presence or absence of inhibitors by sedimentation velocity analytical ultracentrifugation. Human MIF preincubated with DMSO as a control sedimented predominantly as a single species with a sedimentation coefficient and molecular mass corresponding to that of the trimer (s = 3.15 ± 0.1 S and 36,373 kDa) (Fig. 8A). In the presence of ebselen, at a concentration corresponding to the IC50 (3 μm, Fig. 8A), we observed an additional sedimenting species with an average s value of 1.7 ± 0.2 S and a molecular mass of 12,428 kDa, indicative of the presence of monomeric MIF (Fig. 8B). Studies at higher ebselen concentrations were not possible due to ebselen-induced aggregation and precipitation of MIF (Fig. 9A). None of the other inhibitors was shown to disrupt the MIF trimer.
FIGURE 8.
A, IC50 determination of ebselen using d-dopachrome methyl ester as a substrate. Every data point represents an average of four replicates. B, sedimentation velocity profiles of wild-type (Wt) human MIF in the presence or absence of ebselen. 10 μm MIF in 1× PBS was preincubated with 3 μm ebselen for 1 h at room temperature. Sedimentation coefficient distributions were obtained by analysis of the sedimentation profiles using the C(s) distribution as a variant of Lamm equation solutions. Calculations were performed using SEDFIT software (50).
FIGURE 9.
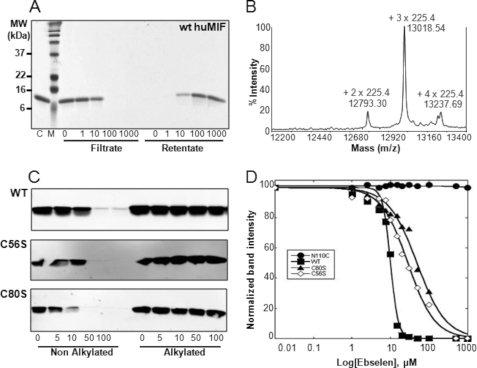
Ebselen induces MIF aggregation via interaction with cysteine. A, wild-type human MIF dialyzed against 1× PBS incubated with different ebselen concentrations (0, 1, 10, 100, and 1000 μm) for 120 min and filtered with a 0.2-μm filter by centrifugation at 13,000 × g for 5 min; both the filtrate and the retentate, which were resolubilized in 1% SDS, were analyzed in a 15% SDS gel. B, MALDI-TOF spectrum demonstrating that the three cysteine residues of MIF were alkylated upon co-incubation with 10 mm maleimide for 1 h. C, ebselen induces the aggregation of only non-alkylated forms of wild type (WT), C56S, and C80S MIF. Cystein alkylation of each protein abolishes ebselen-induced aggregation of MIF. Ten micromolar wild type, C56S, and C80S MIF were alkylated with 10 mm maleimide. The alkylated and the non-alkylated proteins were incubated with 0, 5, 10, 50, and 100 μm ebselen for 1 h at room temperature and centrifuged for 15 min at 13,000 × g, and the supernatant were analyzed in a 15% SDS gel by Western blot using the anti-MIF antibody (Zymed Laboratories Inc. at 1:20,000). D, comparison of aggregation propensity as determined by quantification of the band intensity of remaining soluble MIF (wild-type, C80S, C56S, and N110C) as a function of ebselen concentration.
Ebselen Inhibits MIF Tautomerase Activity by Inducing Cysteine-mediated Modification, Trimer Dissociation, and Aggregation of MIF
Ebselen is a well known anti-inflammatory and antioxidant drug (57, 58). The therapeutic effects of ebselen have been linked to its peroxidase activity; it has high chemical reactivity toward hydroperoxides and thiols. Ebselen has been used as a probe for characterizing cysteine-targeted oxidation of thioredoxin (59). In bovine systems, free ebselen quickly reacts with thiols to form two metabolites, an ebselen-glutathione adduct and an ebselen-cysteine adduct (60). MIF monomer has three cysteine residues at positions 56, 59, and 80. Previous studies showed that mutating or alkylating the cysteine residues at positions 56 or 59 led to a reduced conformational stability of MIF (61).
To determine whether ebselen inhibition of MIF is mediated by covalent modification of specific cysteine residues, mass spectrometry analyses were carried out on MIF samples preincubated with increasing concentrations of ebselen (0–10 μm) for 1 h at room temperature. MALDI-TOF analysis revealed a peak with a m/z = 12620.48, which corresponds to monomeric MIF with one bound molecule of ebselen (molecular mass of 274 Da). However, we also observed that ebselen induced rapid aggregation and precipitation of MIF. To quantify the degree of ebselen-induced MIF aggregation, the samples were centrifuged at high speed (13,000 × g) and the supernatants and pellets were collected and analyzed in SDS-PAGE gels. Fig. 9A demonstrates that the amount of MIF remaining in solution decreased as the concentration of ebselen increased, with complete precipitation of MIF at ebselen concentrations of ≥15 μm.
Blocking Cysteine Residues by Alkylation or Chemical Cross-linking Prevents Ebselen-induced MIF Aggregation
MIF contains three free cysteines at positions 56, 59, and 80. To assess whether ebselen-induced aggregation is mediated via covalent modification of specific cysteine residue(s), wild-type, C56S, and C59S MIF were expressed, purified, and alkylated for 1 h at room temperature using 10 mm maleimide. In the case of wild-type MIF, a major peak of 13,020 Da, corresponding to monomeric MIF with three alkylated cysteine residues (molecular mass of maleimide is 225.4 Da) could be detected by MS, confirming total alkylation of MIF under the native conditions (Fig. 9B). After removal of excess maleimide, alkylated MIF was incubated for 1 h at room temperature with 100 μm ebselen and the sample mixtures were centrifuged at 13,000 × g for 15 min. The supernatant and the re-suspended pellet (in the original working volume) were then analyzed in a 15% SDS gel to quantify the amount of soluble and precipitated protein. As expected, no aggregation could be observed for the alkylated wild type MIF, and likewise not for the C59S and C56S cysteine mutants (Fig. 9C), whereas the non-alkylated forms of all three proteins underwent complete aggregation. The C56S/C59S double mutant behaved similarly to wild type MIF, indicating that covalent modification through Cys80 is sufficient to induce the aggregation and precipitation of MIF. To verify that ebselen reacts with cysteine 80, both wild-type and the C80S MIF mutant were incubated separately with ebselen and the samples were analyzed by MALDI-TOF mass spectrometry (supplemental Fig. S5). Addition of ebselen resulted in a mass shift corresponding to the addition of a single molecule of ebselen to the wild type protein. A minor peak with an m/z of 12,894.19 Da corresponding to the addition of two molecules of ebselen was also observed. Upon tryrptic digestion, we observed a mass shift by 274 Da in a peptide fragment comprising residues 78–86 (78LLCGLLAER86) in the case of wild type MIF but not the C80S mutant (supplemental Fig. S5). This observation combined with the MS/MS fragmentation patterns (data not shown) suggest that ebselen modification of MIF occurs at cysteine 80. Although our mass spectrometery data demonstrated that ebselen modifies MIF at multiple cysteine residues (supplemental Fig. S5), our peptide coverage did not allow us to detect peptide fragments comprising the other cysteine residue, Cys56 and Cys59. Therefore, we were not able to determine more precisely the extent of modification at each of these residues.
Ebselen-induced Aggregation of MIF Is Mediated by Disruption of the Trimer
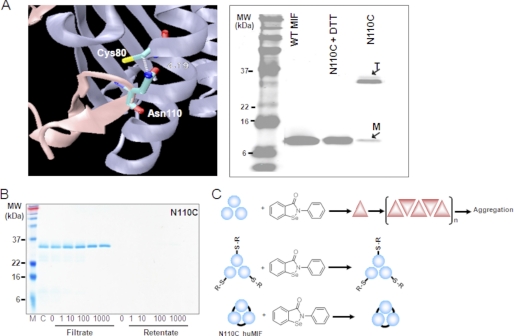
Based on our experience, the MIF trimer is very stable, whereas the monomer is extremely unstable and undergoes rapid aggregation upon dissociation from the trimer. Attempts to populate the monomer using inter-subunit disruption mutations and chemical agents were unsuccessful in destabilizing the trimer or led to the expression of (presumably monomeric) MIF in inclusion bodies.4 These findings are consistent with the absence of any reports in the literature demonstrating the accumulation of monomeric MIF or existence of stable monomeric MIF mutants. In our hands, the presence of monomer is always associated with accumulation of higher molecular weight aggregates of MIF (43). These findings suggest that ebselen-induced aggregation could be mediated by the induction of trimer-to-monomer dissociation and subsequent aggregation of the unstable monomeric species (Fig. 10). To test this hypothesis further, we designed and produced a disulfide-linked MIF mutant, which forms a stable trimer upon oxidation. This mutant was generated by replacing the C-terminal residue asparagine 110 with cysteine (N110C) to form an intermolecular disulfide bond between Cys110 and Cys80 at the monomer-monomer interfaces of the trimer (Fig. 10A). N110C undergoes spontaneous oxidation to form inter-subunit disulfides resulting in the formation of a stable cross-linked MIF trimers as the predominant species in solution under physiological conditions. Interestingly, co-incubation of the disulfide-linked N110C MIF trimer with increasing concentrations of ebselen had no effect on trimer stability. We did not observe any MIF aggregation even after longer incubations (Figs. 9D and 10B). The MIF band intensity of the control sample (N110C only) was equivalent to the bands corresponding to samples preincubated with various ebselen concentrations (Fig. 10B), suggesting that trimer stabilization via disulfide formation involving Cys80 prevents ebselen-induced MIF dissociation and aggregation.
FIGURE 10.
Ebselen-induced aggregation of MIF is mediated by disruption of the trimer. A, a ribbon diagram of the interface between two MIF monomers illustrating the positions of Cys80 and Asn110, which was mutated to cystine (N110C) to engineer the intersubunit disulfide bridge. The N110C cross-linked MIF migrates as a stable trimer in SDS gel, but disassociates into monomer in the presence of 10 mm dithiothreitol. B, N110C cross-linked MIF trimer does not undergo aggregation upon addition of ebselen up to a concentration of 1 mm. C, schematic depiction illustrating the mechanism of ebselen-induced aggregation of MIF. Reaction of ebselen with MIF induces conformational changes and trimer disassociation resulting in the population of monomeric MIF, which is highly prone to aggregation. Cysteine alkylation and/or trimer stabilization via intersubunit disulfide cross-linking prevents ebselen-induced dissociation and aggregation of MIF.
MIF Tautomerase Inhibitors Display Low Toxicity on RAW 264.7 Macrophages
The relative cytotoxicity of the investigated tautomerase inhibitors of MIF was analyzed by incubating RAW 264.7 macrophages for 24 h with 3, 10, and 20 μm of the various compounds. The toxicity was then evaluated using the MTT assay. A very low cytotoxicity was seen with all inhibitors at 3 and 10 μm, except for benzyl isothiocyanate and inhibitors 2 and 8 at 10 μm that resulted in ∼25% cytotoxicity. At 20 μm, MIF inhibitors 12, 13, and 15 and HCLP showed an increased toxicity to 30–40% (supplemental Fig. S7). Further studies are required to determine whether the increase in MTT toxicity by these compounds is mediated by blocking MIF, which is known to be anti-apoptotic.
MIF Tautomerase Inhibitors Significantly Inhibit MIF-mediated Overriding of Glucocorticoid Activity
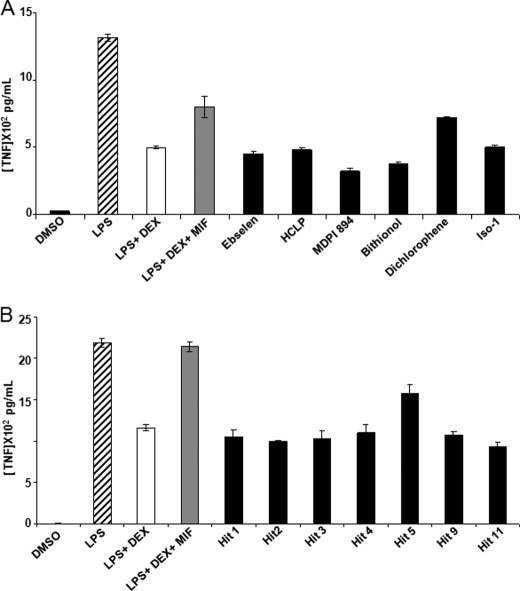
An important feature of the biology of MIF is its ability to override the immunosuppressive effects of glucocorticoids on proinflammatory cytokine production (62, 63). Therefore, we examined whether the MIF tautomerase inhibitors identified in this study interfered with the immunoregulatory function of MIF on RAW 264.7 macrophages. RAW 264.7 macrophages were preincubated for 1 h with dexamethasone (100 nm) with or without recombinant MIF (100 ng/ml; 8 nm) and ebselen, HCLP, and the HCLP analogues bithionol, MDPI894, and dichlorophene as well as the inhibitors 1, 2, 3, 4, 5, 9, and 11 (all used at 10 μm) prior to stimulation with LPS (100 ng/ml). TNF production was quantified in cell culture supernatants collected 4 h after stimulation. Treatment of RAW 264.7 cells with MIF overcame dexamethasone inhibition of TNF production. Ebselen, HCLP, bithionol, and MDPI894, but not dichlorophene, fully inhibited the capacity of MIF to override the immunosuppressive effect of dexamethasone on LPS-induced TNF production (Fig. 11A). Dichlorophene only slightly (25%) inhibited MIF overriding capacity, in line with the concept that the catalytic site of MIF is important for MIF counter-regulatory activity and that dichlorophene was a less potent inhibitor of MIF tautomerase activity compared with ebselen, HCLP, bithionol, and MDPI894. Bithionol and MDPI894 seemed to revert the MIF effect below the level of dexamethasone + LPS, which could be attributed to their ability to inhibit endogenous MIF as well. Similarly, inhibitors 1, 2, 3, 4, 5, 9, and 11 abrogated the MIF-mediated dexamethasone overriding activity (Fig. 11B). Taken together, these results demonstrate that compounds that inhibit MIF tautomerase activity were also efficient in inhibiting the overriding activity of MIF on glucocorticoids, suggesting that these two activities are linked.
FIGURE 11.
Overriding of glucocorticoid-mediated inhibition of cytokine production. RAW 264.7 macrophages were preincubated for 1 h with or without dexamethasone (Dex, 100 nm), MIF (100 ng/ml), and A, ISO-1 (50 μm), ebselen, HCLP, bithionol, MDPI894, and dichorophene; or B, compounds 1, 2, 3, 4, 5, 9, and 11 (all at 10 μm) before stimulation with 100 ng/ml of LPS. Cell-free supernatants were collected after 4 h to quantify the concentrations of TNF. Data shown are mean ± S.D. of triplicate samples from one experiment and are representative of two independent experiments.
Ebselen and HCLP Inhibit MIF-mediated AKT Phosphorylation
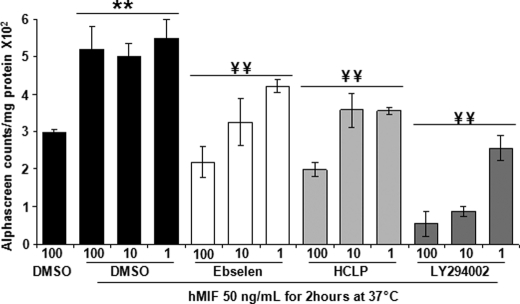
Previous studies have shown that MIF has anti-apoptotic activity and that it promotes tumor cell survival through activation of the AKT pathway (64). Exogenously added recombinant MIF led to an enhancement of phosphoinositide 3-kinase-dependent AKT phosphorylation in NIH3T3 and HeLa cells (64). To probe the effect of ebselen and HCLP on MIF-mediated AKT phosphorylation, we quantified the level of phosphorylated AKT in HeLa cells incubated for 2 h with or without MIF (50 ng/ml) in the presence or absence of 1, 10, and 100 μm MIF inhibitors, or DMSO as a control. The level of phosphorylated AKT was quantified using the Alpha screen technology. As shown in Fig. 12, MIF increased the level of phosphorylated AKT by 1.5-fold in HeLa cells, an effect that was fully inhibited by ebselen and HCLP and the control phosphoinositide 3-kinase inhibitor LY294002. Of note, ebselen, HCLP, and LY294002 dose-dependently decreased phosphorylated AKT levels above control values, which may suggest that endogenous MIF plays a role in sustaining AKT phosphorylation in resting HeLa cells.
FIGURE 12.
MIF-mediated activation of AKT phosphorylation and inhibition with selected MIF inhibitors. HeLa cells were incubated for 2 h with or without 50 ng/ml of recombinant human MIF in the presence or absence of ebselen, HCLP, or LY294002 (1, 10, and 100 μm). AKT phosphorylation was measured using the Alpha screen technology. Data are mean ± S.D. of triplicate samples from two representative experiments. **, p < 0.01 compared with DMSO; and ¥¥, p < 0.01 compared with MIF plus DMSO used at equal concentration using the Student-Neuman-Keuls test.
Ebselen Modulates MIF-triggered Chemotactic Migration Responses
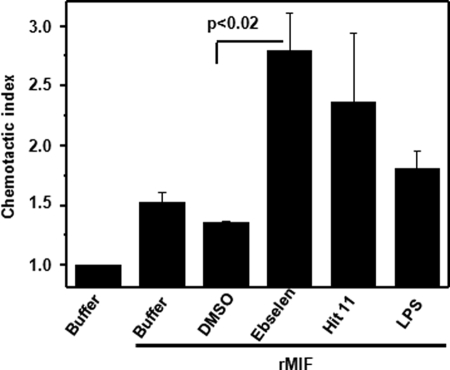
MIF has been found to promote monocyte chemotactic migration through activation of its receptor CXCR2 (54, 65). The oligomeric MIF species responsible for this effect is still unknown. More recently, MIF has also been shown to potently enhance the chemotactic migration of EPCs. Day 14-differentiated EPCs representing late outgrowth EPCs express CD14 and thus have both endothelial cell and monocyte-like properties. EPCs and monocytes play an important role in angio- and vasculogenic processes and atherogenic inflammation, respectively, and MIF has been implicated to play a role in the corresponding EPC and leukocyte recruitment processes. To begin to address potential effects of the tautomerase inhibitors on these inflammatory and angiogenic activities of MIF, MIF-mediated EPC chemotaxis was studied in the presence and absence of a selection of the compounds. Recombinant MIF or LPS as a control were added to the lower chamber of a Transwell device, whereas EPCs and compounds were added to the upper chamber. Directed chemotactic migration was followed over a time interval of 3 h. MIF led to a 1.53-fold enhancement of the chemotactic index. Inhibitors alone did not affect EPC chemotaxis. Surprisingly, ebselen pretreatment of EPCs markedly and significantly promoted migration of EPCs (chemotactic index = 2.8). Thus, ebselen exhibited a hyperagonistic effect. HCLP acted in a hyper-agonistic manner as well, but this effect did not reach statistical significance. The MIF inhibitors 1, 2, 3, 4, 5, and 9 neither inhibited nor enhanced MIF-directed EPC migration (Fig. 13).
FIGURE 13.
Ebselen acts as a hyper-agonist of MIF-mediated chemotaxis. Calcein-stained day-14 EPCs were preincubated with 10 μm ebselen or HCLP, added to the upper chamber of a transwell device, and subjected to a chemotactic gradient of 10 ng/ml of MIF added to the lower chamber. Migration was followed for 3 h. PBS and 0.1% DMSO were used as negative controls. The chemotactic index is plotted, with the number of cells migrated toward the buffer set to a value of 1.0. Data are mean ± S.D. of three independent experiments performed with day-14 EPCs and were confirmed further with n = 2 additional experiments with day-7 EPCs (data not shown). Statistical significance for the hyper-agonistic effect of ebselen is indicated (p < 0.02).
DISCUSSION
Given the role of the quaternary structure and the C-terminal region in modulating the catalytic activity of MIF, we can envision several mechanisms by which small molecules could inactivate MIF tautomerase activity: 1) binding to the active site; 2) allosteric inhibition; 3) covalent modification of active site residues; 4) disruption of the active site through compound-induced dissociation of the tautomerase-active trimer; and 5) stabilization of the MIF monomer and prevention of its re-association to form the active trimer. However, current efforts aimed at developing small molecule inhibitors of MIF are focused primarily on rational structure-based design approaches targeting the active site. Although these efforts have resulted in the discovery of several classes of active site inhibitors of MIF and have advanced our knowledge regarding the role of MIF in inflammatory disease, the chemical space explored by this approach is limited and molecules that modulate MIF activity via alternative mechanisms are likely to have been overlooked.
Based on the premise that HTS of chemical libraries should facilitate the exploration of biologically relevant chemical spaces, we developed an activity-based HTS assay that enables the identification of different classes of potent and selective inhibitors of MIF, thus facilitating the exploration of a much larger chemical space. This approach complements current strategies focused on targeting the tautomerase active site using structure-based design strategies. In addition to active site inhibitors, the use of the activity-based HTS assay should facilitate the identification of molecules that inactivate or modulate the activity of MIF by alternative mechanisms, including allosteric inhibition, destabilization/dissociation of the active trimer, or covalent modification of the catalytic N-terminal proline residue.
Despite decent reproducibility of the tautomerase assay, there have been no reports in the literature of MIF HTS tautomerase assays. Initial efforts to develop HTS were abandoned due to the reported instability of the d-dopachrome substrate (66), and most recent efforts have relied primarily on in silico HTS approaches focused on the catalytic site of MIF (66, 67). We investigated the stability of the substrate and found screening conditions under which the substrate is stable and suitable for HTS. As a proof of concept, we first screened a library of 1040 FDA-approved compounds containing a diverse set of drugs, 85% of which are marketed in a wide range of therapeutic areas and include anti-inflammatory and analgesic drugs. We reasoned that the identification of biologically active compounds with known toxicity, pharmacokinetic properties, and biological activities in vivo should accelerate the development of clinically relevant MIF inhibitors. Second, we screened the Maybridge library containing 14,400 compounds. Different classes of MIF inhibitors that exert their effects via distinct mechanisms were discovered and validated using a battery of biochemical and biophysical methods.
Active Site-directed Irreversible Inhibitors of MIF
Blocking MIF activity via covalent modification of the N-terminal catalytic Pro1 residue was previously seen with NAPQI (37). A mass shift was detected in rat MIF extracted from liver post-NAPQI administration. NAPQI-modified MIF failed to override the effect of the glucocorticoids dexamethasone (37). However, due to its toxicity no further clinical studies were performed with NAPQI (42). Another covalent modifier, which was identified by virtual screening, called 4-iodo-6-phenylpyrimidine, was also shown to inhibit MIF tautomerase activity via irreversible modification at the N-terminal proline (33). We recently reported a new class of isothiocyanate-based covalent modifiers of Pro1 in MIF and showed that they selectively modify catalytically active forms of MIF and block CD74 receptor binding and MIF-mediated phosphorylation of AKT. Herein, we report the identification and characterization of several novel irreversible inhibitors of MIF, the majority of which have potency in the range of 0.2 to 5.55 μm (Table 1). The 12 covalent modifiers can be grouped into four different classes of reactive electrophiles that react with the secondary amine of the N-terminal proline resulting in amide bond formation and loss of MIF tautomerase activity.
Non-covalent Inhibitors of MIF
In addition to covalent modifiers, our screening identified four novel chemical classes (compounds 3, 9, 11, and HCLP) of reversible MIF inhibitors with IC50 values ranging from 0.2 to 5.5 μm. The slow conformational exchange observed for compound 9, but not for compound 3, suggests a higher affinity of compound 9 for MIF, in agreement with the KD values. NMR titration demonstrated a chemical shift deviation with two sequential changes: a mild interaction followed by a conformational change of MIF. Increasing molar ratios of compound 9 induces intermediate exchange of conformation, which implies higher affinity of compound 9 for MIF. On the other hand, upon titration with compounds 3 or 11, many residues located in the subunit-subunit interface and enzymatic active site are shifted that may explain a competition with the substrate. Taken together, similar residues of human MIF are involved in the interaction with HCLP and compounds 3, 9, and 11, indicating that these four inhibitors have a similar binding mode.
Inhibition of MIF Enzymatic Activity May Not Be Sufficient to Block Its Cellular Functions
Our goal is to identify novel inhibitors that target specific or multiple activities of MIF, such as its tautomerase activity, CD74 receptor binding, and/or its interaction with other proteins. However, the notion that many of the cellular functions of MIF, such as receptor binding, do not involve tautomerase activity (68), combined with observations from x-ray structures showing small changes in MIF structure upon binding to inhibitors (69, 70), suggest that inhibition of the enzymatic activity of MIF alone is not sufficient to block all of its adverse effects. Similarly, neutralization with anti-MIF antibodies does not appear to affect the enzymatic activity of MIF.5 Therefore, discovering novel compounds that can target both activities by inhibiting the enzymatic activity and blocking CD74 receptor binding or binding to its CXC receptors of MIF, should constitute a more effective strategy for interfering and/or neutralizing MIF activities. Given that oligomerization, more specifically trimer formation, is essential for MIF tautomerase activity and its binding to CD74, we hypothesized that this targeting could be achieved using molecules that interfere with MIF oligomerization or induce trimer dissociation. In fact, dissociation of the MIF trimer by ebselen resulted in disruption of critical proinflammatory actions of MIF (Fig. 12). As CD74 also forms a trimeric structure and because MIF-stimulated AKT phosphorylation is mediated by interactions with CD74, this implies that disruption of MIF trimers could interfere with MIF/CD74-dependent inflammatory and survival pathways. In contrast, MIF trimer-disrupting compounds acted in a hyper-agonistic fashion in MIF/CXCR2-mediated recruitment processes, indicating that such inflammatory pathways might capitalize on the use of the MIF monomer rather than the trimer. One might thus hypothesize that in such settings the MIF monomer rather than the trimer is crucial. This hypothesis can be tested using the N110C mutant (Fig. 10), which forms stable cross-linked trimer in solution.
Although modulating protein-protein interactions using small molecules remains challenging due to the extensive nature of interactions at the interfaces of interacting proteins, our studies suggest an alternative strategy for disrupting protein-protein interactions by specific targeting and modification of key protein residues. Using single site mutagenesis and thiol-specific alkylation strategies, we demonstrated conclusively that covalent modification of MIF by ebselen occurs at multiple cysteine residues and induces MIF dissociation, misfolding, and aggregation. To test this hypothesis, we generated a stable disulfide-linked trimeric mutant of MIF (N110C) and showed that this mutant is modified by ebselen, but does not undergo aggregation. These findings are consistent with ebselen-induced trimer-to-monomer dissociation as the rate-limiting step. Induction of aggregation upon covalent modification of cysteines has been reported as a common feature of the P450 enzymes (71). Covalent modification of tubulin and the P450 apoprotein by benzyl isothiocyanate results in inhibition of tubulin polymerization, loss of P450 activity, and aggregation of both proteins in a dose- and time-dependent manner. We hypothesized that covalent modification of cysteine residues in MIF may induce conformational changes that result in the exposure of hydrophobic surfaces, which ultimately leads to protein aggregation. To determine whether MIF aggregation induced by ebselen is mediated by formation of disulfide bonds or misfolding of the proteins, we evaluated the solubility and reversibility of these aggregates upon treatment with reducing agents and/or SDS. We observed that addition of dithiothreitol or other reducing agents such as Tris(2-carboxyethyl)phosphine results only in partial recovery of monomeric MIF (supplemental Fig. 6A). However, nearly complete recovery is observed when the MIF aggregates are re-solubilized in SDS. These findings suggest that MIF aggregation is driven by noncovalent interactions and not simply disulfide-mediated polymerization of the protein. This is consistent with the fact that stable monomeric MIF have not been reported and MIF aggregation is observed under conditions that destabilize the trimer.
Ebselen, 2-phenyl-1,2-benzisoselenazol-3(2H)-one, is known to act as an antioxidant and possess anti-inflammatory properties. In addition, several studies have shown that ebselen is neuroprotective in different animal and cellular models of neurodegenerative diseases (e.g. Parkinson disease (72–74)) and glutamate-induced neurotoxicity (74). Our findings that ebselen modifications occurs at the Se-S bond is consistent with previous studies, which have documented that ebselen inhibition of several enzymes and receptors is mediated by directly reacting with thiol groups resulting in a modification of their oxidation state and/or conformational properties. Ebselen interacts with cysteine residues in proteins resulting in the formation of disulfide bond formation and an ebselen diselenide product (57). Among the proteins that have been shown to be inhibited by mechanisms involving direct ebselen modifications of cysteine residues are zinc metallothionein (75), glutathione S-transferase (76), NADPH oxidase (77), protein kinase C (78), inositol 1,4,5-trisphosphate receptor (79), N-methyl-d-asparatate receptor, and membrane calcium-ATPase (80). Consistent with this mechanism, studies in our laboratory demonstrate that the ebselen-induced aggregation of MIF is blocked in the presence of dithiothreitol and GSH (supplemental Fig. S6B). While this article was under review, a study by Terentis et al. (81) demonstrated that ebselen is a potent inhibitor of indoleamine 2,3-dioxygenase. The activity of ebselen was shown to be mediated by direct interaction with multiple cysteine residues of indoleamine 2,3-dioxygenase, which results in significant structural changes that alter the active site heme and substrate binding to indoleamine 2,3-dioxygenase. Similar to our observations with MIF, alkylation of cysteine residues or removal of indoleamine 2,3-dioxygenase-bound ebselen, via addition of reducing agents, abolished the inhibitory activity of ebselen (supplemental Fig. S6).
Although, some of the antioxidant and neuroprotective properties of ebselen have been attributed to its ability to mimic the activity of glutathione (GSH) peroxidase (82, 83), the mechanisms underlying its anti-inflammatory activity remain unknown. The data presented in this article are consistent with previous reports and provide additional evidence that ebselen interaction with inflammatory proteins goes beyond simply modification of these proteins. Further studies are required to determine whether ebselen modulation of the activities of MIF contributes to its anti-inflammatory activity. Studies in animal models as well as in humans have shown that ebselen exhibits low toxicity (52, 58) and is currently in clinical trials for the treatment of ischemic stroke (52) and subarachnoid hemorrhage (58).
In conclusion, we developed a simple, robust, and highly reproducible assay that allows for rapid and reliable screening of large chemical libraries to identify diverse and highly potent inhibitors of MIF enzymatic and biological activities. We described the discovery and characterization of 12 novel chemical classes of MIF inhibitors that exert their effects via distinct mechanisms. The mechanism of action of these inhibitors was determined and validated using a battery of biochemical and biophysical methods as well as by the generation of novel MIF mutants. The majority of the compounds identified displayed low IC50 values (0.2–6 μm) and toxicity, thus making them ideal lead candidates for follow-up medicinal chemistry efforts and in vivo studies. Importantly, all inhibitors demonstrated total inhibition of MIF-mediated glucocorticoid overriding. The two inhibitors ebselen and HCLP inhibit MIF-mediated AKT phosphorylation, whereas ebselen, a trimer-disrupting inhibitor, additionally acted as a potent hyper-agonist in MIF-mediated chemotactic migration. Some of the lead inhibitors are already approved drugs with known safety and oral bioavailability profiles. Our discovery of ebselen as the first example of small molecule inhibitor of MIF, which acts by selective modification and disruption of the trimer, reveal a novel mechanism for targeting protein-protein interactions. Furthermore, this discovery has significant implications for developing more effective drugs that could target multiple proinflammatory activities meditated by the MIF trimer.
Together, these results provide the foundation for future studies to determine the efficacy of these compounds in vivo and compare their efficacy to anti-MIF neutralizing antibodies using established experimental models of inflammatory and autoimmune diseases and tumorigenesis (35, 68). Furthermore, the diversity of chemical structures and mechanisms of action of our inhibitors makes them ideal mechanistic probes and diagnostic tools for elucidating the structure-function relationships of MIF and to further determine the role of the catalytic activity of MIF in regulating the function(s) of MIF in health and disease.
Supplementary Material
Acknowledgments
We thank Dr. Gerardo Turcatti, Pierre-Olivier Regamey, Dr. Damiano Banfi, and Dr. Marc Chambon from the EPFL biomolecular screening facility for advice and assistance in automated screening and data analysis. We also thank Dr. Marc Moniatte, Diego Chiappe, and Jérome Vialaret from the proteomic facility for assistance and Marlies Knaup-Reymond and Anne-Laure Schiesser from technical assistance. We thank Dr. Richard Bucala, Dr. Loay Awad, Dr. Sara Butterfield, and Dr. Hongqi Lue for valuable discussions.
This work was supported by grants from the Swiss Federal Institute of Technology Lausanne (to H. A. L., F. E., and H. O.-S.), Swiss National Science Foundation Grants 310000-110027 (to H. A. L.) and 3100-118266 (to T. C.), and German Research Council (DFG) Grants Be1977/4-1 and SFB542-A7 (to J. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
T. D. Goddard and D. G. Kneller (2010) SPARKY 3, University of California, San Francisco, CA.
F. El-Turk and H. A. Lashuel, unpublished results.
H. Ouertatani-Sakouhi and H. A. Lashuel, unpublished results.
- MIF
- migration inhibitory factor
- LPS
- lipopolysaccharide
- TNF
- tumor necrosis factor
- DMSO
- dimethyl sulfoxide
- ISO-1
- (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester
- HTS
- high throughput screening
- HCLP
- hexachlorophene
- MALDI-TOF
- matrix-assisted laser desorption ionization-time of flight
- MS
- mass spectrometry
- PBS
- phosphate-buffered saline
- EMCH
- 3,3′-N-[e-maleimidocaproic acid)hydrazide
- HSQC
- heteronuclear single quantum coherence
- EPC
- endothelial progenitor cell
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NAPQI
- N-acetyl-p-benzoquinone imine.
REFERENCES
- 1.Bloom B. R., Bennett B. (1966) Science 153, 80–82 [DOI] [PubMed] [Google Scholar]
- 2.David J. R. (1966) Proc. Natl. Acad. Sci. U.S.A. 56, 72–775229858 [Google Scholar]
- 3.Weiser W. Y., Temple P. A., Witek-Giannotti J. S., Remold H. G., Clark S. C., David J. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 7522–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhagen J., Calandra T., Mitchell R. A., Martin S. B., Tracey K. J., Voelter W., Manogue K. R., Cerami A., Bucala R. (1993) Nature 365, 756–759 [DOI] [PubMed] [Google Scholar]
- 5.Calandra T., Roger T. (2003) Nat. Rev. Immunol. 3, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lolis E., Bucala R. (2003) Expert Opin. Ther. Targets 7, 153–164 [DOI] [PubMed] [Google Scholar]
- 7.Leech M., Metz C., Hall P., Hutchinson P., Gianis K., Smith M., Weedon H., Holdsworth S. R., Bucala R., Morand E. F. (1999) Arthritis Rheum. 42, 1601–1608 [DOI] [PubMed] [Google Scholar]
- 8.Mikulowska A., Metz C. N., Bucala R., Holmdahl R. (1997) J. Immunol. 158, 5514–5517 [PubMed] [Google Scholar]
- 9.Lan H. Y., Bacher M., Yang N., Mu W., Nikolic-Paterson D. J., Metz C., Meinhardt A., Bucala R., Atkins R. C. (1997) J. Exp. Med. 185, 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown F. G., Nikolic-Paterson D. J., Hill P. A., Isbel N. M., Dowling J., Metz C. M., Atkins R. C. (2002) J. Am. Soc. Nephrol. 13, Suppl. 1, S7–13 [PubMed] [Google Scholar]
- 11.Yabunaka N., Nishihira J., Mizue Y., Tsuji M., Kumagai M., Ohtsuka Y., Imamura M., Asaka M. (2000) Diabetes Care 23, 256–258 [DOI] [PubMed] [Google Scholar]
- 12.Zernecke A., Weber C. (2010) Cardiovasc Res. 2, 192–201 [DOI] [PubMed] [Google Scholar]
- 13.Calandra T., Echtenacher B., Roy D. L., Pugin J., Metz C. N., Hültner L., Heumann D., Männel D., Bucala R., Glauser M. P. (2000) Nat. Med. 6, 164–170 [DOI] [PubMed] [Google Scholar]
- 14.Beishuizen A., Thijs L. G., Haanen C., Vermes I. (2001) J. Clin. Endocrinol. Metab. 86, 2811–2816 [DOI] [PubMed] [Google Scholar]
- 15.Emonts M., Sweep F. C., Grebenchtchikov N., Geurts-Moespot A., Knaup M., Chanson A. L., Erard V., Renner P., Hermans P. W., Hazelzet J. A., Calandra T. (2007) Clin. Infect Dis. 44, 1321–1328 [DOI] [PubMed] [Google Scholar]
- 16.Rossi A. G., Haslett C., Hirani N., Greening A. P., Rahman I., Metz C. N., Bucala R., Donnelly S. C. (1998) J. Clin. Invest. 101, 2869–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizue Y., Ghani S., Leng L., McDonald C., Kong P., Baugh J., Lane S. J., Craft J., Nishihira J., Donnelly S. C., Zhu Z., Bucala R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14410–14415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donnelly S. C., Haslett C., Reid P. T., Grant I. S., Wallace W. A., Metz C. N., Bruce L. J., Bucala R. (1997) Nat. Med. 3, 320–323 [DOI] [PubMed] [Google Scholar]
- 19.Bucala R., Donnelly S. C. (2007) Immunity 26, 281–285 [DOI] [PubMed] [Google Scholar]
- 20.Rendon B. E., Willer S. S., Zundel W., Mitchell R. A. (2009) Exp. Mol. Pathol. 86, 180–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M., Aman P., Grubb A., Panagopoulos I., Hindemith A., Rosengren E., Rorsman H. (1995) FEBS Lett. 373, 203–206 [DOI] [PubMed] [Google Scholar]
- 22.Rosengren E., Bucala R., Aman P., Jacobsson L., Odh G., Metz C. N., Rorsman H. (1996) Mol. Med. 2, 143–149 [PMC free article] [PubMed] [Google Scholar]
- 23.Bendrat K., Al-Abed Y., Callaway D. J., Peng T., Calandra T., Metz C. N., Bucala R. (1997) Biochemistry 36, 15356–15362 [DOI] [PubMed] [Google Scholar]
- 24.Swope M., Sun H. W., Blake P. R., Lolis E. (1998) EMBO J. 17, 3534–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chook Y. M., Gray J. V., Ke H., Lipscomb W. N. (1994) J. Mol. Biol. 240, 476–500 [DOI] [PubMed] [Google Scholar]
- 26.Subramanya H. S., Roper D. I., Dauter Z., Dodson E. J., Davies G. J., Wilson K. S., Wigley D. B. (1996) Biochemistry 35, 792–802 [DOI] [PubMed] [Google Scholar]
- 27.Stivers J. T., Abeygunawardana C., Mildvan A. S., Hajipour G., Whitman C. P. (1996) Biochemistry 35, 814–823 [DOI] [PubMed] [Google Scholar]
- 28.Suzuki M., Sugimoto H., Nakagawa A., Tanaka I., Nishihira J., Sakai M. (1996) Nat. Struct. Biol. 3, 259–266 [DOI] [PubMed] [Google Scholar]
- 29.Sun H. W., Bernhagen J., Bucala R., Lolis E. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5191–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swope M. D., Sun H. W., Klockow B., Blake P., Lolis E. (1998) J. Biol. Chem. 273, 14877–14884 [DOI] [PubMed] [Google Scholar]
- 31.Rosengren E., Aman P., Thelin S., Hansson C., Ahlfors S., Björk P., Jacobsson L., Rorsman H. (1997) FEBS Lett. 417, 85–88 [DOI] [PubMed] [Google Scholar]
- 32.Morand E. F., Leech M., Bernhagen J. (2006) Nat. Rev. Drug. Discov. 5, 399–410 [DOI] [PubMed] [Google Scholar]
- 33.Winner M., Meier J., Zierow S., Rendon B. E., Crichlow G. V., Riggs R., Bucala R., Leng L., Smith N., Lolis E., Trent J. O., Mitchell R. A. (2008) Cancer Res. 68, 7253–7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X., Bucala R. (1999) Bioorg. Med. Chem. Lett. 9, 3193–3198 [DOI] [PubMed] [Google Scholar]
- 35.Al-Abed Y., Dabideen D., Aljabari B., Valster A., Messmer D., Ochani M., Tanovic M., Ochani K., Bacher M., Nicoletti F., Metz C., Pavlov V. A., Miller E. J., Tracey K. J. (2005) J. Biol. Chem. 280, 36541–36544 [DOI] [PubMed] [Google Scholar]
- 36.Garai J., Lóránd T. (2009) Curr. Med. Chem. 16, 1091–1114 [DOI] [PubMed] [Google Scholar]
- 37.Senter P. D., Al-Abed Y., Metz C. N., Benigni F., Mitchell R. A., Chesney J., Han J., Gartner C. G., Nelson S. D., Todaro G. J., Bucala R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lubetsky J. B., Dios A., Han J., Aljabari B., Ruzsicska B., Mitchell R., Lolis E., Al-Abed Y. (2002) J. Biol. Chem. 277, 24976–24982 [DOI] [PubMed] [Google Scholar]
- 39.Dios A., Mitchell R. A., Aljabari B., Lubetsky J., O'Connor K., Liao H., Senter P. D., Manogue K. R., Lolis E., Metz C., Bucala R., Callaway D. J., Al-Abed Y. (2002) J. Med. Chem. 45, 2410–2416 [DOI] [PubMed] [Google Scholar]
- 40.Dagia N. M., Kamath D. V., Bhatt P., Gupte R. D., Dadarkar S. S., Fonseca L., Agarwal G., Chetrapal-Kunwar A., Balachandran S., Srinivasan S., Bose J., Pari K., B-Rao C., Parkale S. S., Gadekar P. K., Rodge A. H., Mandrekar N., Vishwakarma R. A., Sharma S. (2009) Eur. J. Pharmacol. 607, 201–212 [DOI] [PubMed] [Google Scholar]
- 41.Balachandran S., Rodge A., Gadekar P. K., Yadav V. N., Kamath D., Chetrapal-Kunwar A., Bhatt P., Srinivasan S., Sharma S., Vishwakarma R. A., Dagia N. M. (2009) Bioorg. Med. Chem. Lett. 16, 4773–4776 [DOI] [PubMed] [Google Scholar]
- 42.Dabideen D. R., Cheng K. F., Aljabari B., Miller E. J., Pavlov V. A., Al-Abed Y. (2007) J. Med. Chem. 50, 1993–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Turk F., Cascella M., Ouertatani-Sakouhi H., Narayanan R. L., Leng L., Bucala R., Zweckstetter M., Rothlisberger U., Lashuel H. A. (2008) Biochemistry 40, 10740–10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Turk F., Fauvet B., Min-kyu C., Ouertatani-Sakouhi H., Neri M., Cascella M., Rothlisberger U., Zweckstetter M., Lashuel H. A. (2010) Biochemistry, in press [Google Scholar]
- 45.Bernhagen J., Mitchell R. A., Calandra T., Voelter W., Cerami A., Bucala R. (1994) Biochemistry 33, 14144–14155 [DOI] [PubMed] [Google Scholar]
- 46.Kleemann R., Mischke R., Kapurniotu A., Brunner H., Bernhagen J. (1998) FEBS Lett. 430, 191–196 [DOI] [PubMed] [Google Scholar]
- 47.Zhang J. H., Chung T. D., Oldenburg K. R. (1999) J. Biomol. Screen 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 48.Liu M., Dobson B., Glicksman M. A., Yue Z., Stein R. L. (2010) Biochemistry 49, 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philo J. S., Yang T. H., LaBarre M. (2004) Biophys. Chem. 108, 77–87 [DOI] [PubMed] [Google Scholar]
- 50.Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi T., Sano K., Takakura K., Saito I., Shinohara Y., Asano T., Yasuhara H. (1998) Stroke 29, 12–17 [DOI] [PubMed] [Google Scholar]
- 53.Hurt S., Titus D. (2010) Optimization of an AlphaScreen® Sure Fire® Cell-based Assay to Measure Phosphorylation of Akt1/2/3 Isoforms at Ser-473, Application Note, PerkinElmer Life Sciences, Waltham, MA [Google Scholar]
- 54.Simons D., Grieb G., Hristov M., Pallua N., Weber C., Bernhagen J., Steffens G. (2010) J. Cell. Mol. Med., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishihira J., Fujinaga M., Kuriyama T., Suzuki M., Sugimoto H., Nakagawa A., Tanaka I., Sakai M. (1998) Biochem. Biophys. Res. Commun. 243, 538–544 [DOI] [PubMed] [Google Scholar]
- 56.Ouertatani-Sakouhi H., El-Turk F., Fauvet B., Roger T., Le Roy D., Karpinar D. P., Leng L., Bucala R., Zweckstetter M., Calandra T., Lashuel H. A. (2009) Biochemistry 41, 9856–9870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schewe T. (1995) Gen. Pharmacol. 26, 1153–1169 [DOI] [PubMed] [Google Scholar]
- 58.Saito I., Asano T., Sano K., Takakura K., Abe H., Yoshimoto T., Kikuchi H., Ohta T., Ishibashi S. (1998) Neurosurgery 42, 269–277; discussion 277–268 [DOI] [PubMed] [Google Scholar]
- 59.Sakurai T., Kanayama M., Shibata T., Itoh K., Kobayashi A., Yamamoto M., Uchida K. (2006) Chem. Res. Toxicol. 19, 1196–1204 [DOI] [PubMed] [Google Scholar]
- 60.Daiber A., Zou M. H., Bachschmid M., Ullrich V. (2000) Biochem. Pharmacol. 59, 153–160 [DOI] [PubMed] [Google Scholar]
- 61.Kleemann R., Kapurniotu A., Frank R. W., Gessner A., Mischke R., Flieger O., Jüttner S., Brunner H., Bernhagen J. (1998) J. Mol. Biol. 280, 85–102 [DOI] [PubMed] [Google Scholar]
- 62.Calandra T., Bernhagen J., Metz C. N., Spiegel L. A., Bacher M., Donnelly T., Cerami A., Bucala R. (1995) Nature 377, 68–71 [DOI] [PubMed] [Google Scholar]
- 63.Roger T., Chanson A. L., Knaup-Reymond M., Calandra T. (2005) Eur. J. Immunol. 35, 3405–3413 [DOI] [PubMed] [Google Scholar]
- 64.Lue H., Thiele M., Franz J., Dahl E., Speckgens S., Leng L., Fingerle-Rowson G., Bucala R., Lüscher B., Bernhagen J. (2007) Oncogene 26, 5046–5059 [DOI] [PubMed] [Google Scholar]
- 65.Bernhagen J., Krohn R., Lue H., Gregory J. L., Zernecke A., Koenen R. R., Dewor M., Georgiev I., Schober A., Leng L., Kooistra T., Fingerle-Rowson G., Ghezzi P., Kleemann R., McColl S. R., Bucala R., Hickey M. J., Weber C. (2007) Nat. Med. 13, 587–596 [DOI] [PubMed] [Google Scholar]
- 66.Orita M., Yamamoto S., Katayama N., Fujita S. (2002) Curr. Pharm. Des. 8, 1297–1317 [DOI] [PubMed] [Google Scholar]
- 67.Cournia Z., Leng L., Gandavadi S., Du X., Bucala R., Jorgensen W. L. (2009) J. Med. Chem. 52, 416–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fingerle-Rowson G., Kaleswarapu D. R., Schlander C., Kabgani N., Brocks T., Reinart N., Busch R., Schütz A., Lue H., Du X., Liu A., Xiong H., Chen Y., Nemajerova A., Hallek M., Bernhagen J., Leng L., Bucala R. (2009) Mol. Cell. Biol. 29, 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crichlow G. V., Lubetsky J. B., Leng L., Bucala R., Lolis E. J. (2009) Biochemistry 48, 132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orita M., Yamamoto S., Katayama N., Aoki M., Takayama K., Yamagiwa Y., Seki N., Suzuki H., Kurihara H., Sakashita H., Takeuchi M., Fujita S., Yamada T., Tanaka A. (2001) J. Med. Chem. 44, 540–547 [DOI] [PubMed] [Google Scholar]
- 71.Solano F., Hearing V. J., García-Borrón J. C. (2000) Neurotox. Res. 1, 153–169 [DOI] [PubMed] [Google Scholar]
- 72.Dhanasekaran M., Uthayathas S., Karuppagounder S. S., Parameshwaran K., Suppiramaniam V., Ebadi M., Brown-Borg H. M. (2006) Brain. Res. 1118, 251–254 [DOI] [PubMed] [Google Scholar]
- 73.Moussaoui S., Obinu M. C., Daniel N., Reibaud M., Blanchard V., Imperato A. (2000) Exp. Neurol. 166, 235–245 [DOI] [PubMed] [Google Scholar]
- 74.Porciúncula L. O., Rocha J. B., Boeck C. R., Vendite D., Souza D. O. (2001) Neurosci. Lett. 299, 217–220 [DOI] [PubMed] [Google Scholar]
- 75.Jacob C., Maret W., Vallee B. L. (1998) Biochem. Biophys. Res. Commun. 248, 569–573 [DOI] [PubMed] [Google Scholar]
- 76.Nikawa T., Schuch G., Wagner G., Sies H. (1994) Biochem. Pharmacol. 47, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 77.Wang J. F., Komarov P., Sies H., de Groot H. (1992) Hepatology 15, 1112–1116 [DOI] [PubMed] [Google Scholar]
- 78.Cotgreave I. A., Duddy S. K., Kass G. E., Thompson D., Moldéus P. (1989) Biochem. Pharmacol. 38, 649–656 [DOI] [PubMed] [Google Scholar]
- 79.Dimmeler S., Brüne B., Ullrich V. (1991) Biochem. Pharmacol. 42, 1151–1153 [DOI] [PubMed] [Google Scholar]
- 80.Ying J., Tong X., Pimentel D. R., Weisbrod R. M., Trucillo M. P., Adachi T., Cohen R. A. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 783–790 [DOI] [PubMed] [Google Scholar]
- 81.Terentis A. C., Freewan M., Sempértegui Plaza T. S., Raftery M. J., Stocker R., Thomas S. R. (2010) Biochemistry 49, 591–600 [DOI] [PubMed] [Google Scholar]
- 82.Wendel A., Fausel M., Safayhi H., Tiegs G., Otter R. (1984) Biochem. Pharmacol. 33, 3241–3245 [DOI] [PubMed] [Google Scholar]
- 83.Sies H. (1993) Free Radic. Biol. Med. 14, 313–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.