Abstract
Without guidance cues, cytoskeletal motors would traffic components to the wrong destination with disastrous consequences for the cell. Recently, we identified a motor protein, myosin X, that identifies bundled actin filaments for transport. These bundles direct myosin X to a unique destination, the tips of cellular filopodia. Because the structural and kinetic features that drive bundle selection are unknown, we employed a domain-swapping approach with the nonselective myosin V to identify the selectivity module of myosin X. We found a surprising role of the myosin X tail region (post-IQ) in supporting long runs on bundles. Moreover, the myosin X head is adapted for initiating processive runs on bundles. We found that the tail is structured and biases the orientation of the two myosin X heads because a targeted insertion that introduces flexibility in the tail abolishes selectivity. Together, these results suggest how myosin motors may manage to read cellular addresses.
Keywords: Actin, Fluorescence, Microscopic Imaging, Myosin, Single-molecule Biophysics, Fascin, Filopodia
Introduction
The essential role of cytoskeletal motor proteins in organizing cellular compartments and cargoes is widely accepted (1). However, many of the molecular details of the overall transport and organization process are poorly understood. These essential features include how motors engage cargoes at their source, how motors are activated once they engage cargo, how they choose the correct set of cytoskeletal tracks, how they disengage from cargo at their destination, and how they return to sources of cargo. Clearly, errors at any of these stages would lead to faults in cellular organization and misplaced cargoes.
To begin to address these questions we have focused on a particular motor protein, myosin X, due to its ability to navigate to a precise location within the cell (2). Myosin X is found at the mitotic and meiotic spindle, where it sets the orientation of the spindle relative to the substrate and influences spindle length (3, 4). However, myosin X is more commonly found in interphase cells at the tips of filopodia (5). These long, slender projections at the leading edge of migrating cells are involved in cell motility and environmental sensing. Myosin X transports components such as Ena/VASP and integrins that are found in the filopodial tip complex (2, 6). The early work by Berg et al. (5) demonstrated that myosin X reaches filopodial tips under its own power. Recently, we found that myosin X identifies filopodia by recognizing the fascin-bundled actin filaments found in the filopodial core (7). Myosin X takes long processive runs along these tightly bundled actin filaments while largely ignoring isolated actin filaments found throughout the cell. In contrast, myosin V does not distinguish between single actin filaments and bundled actin filaments when taking processive runs.
Here, we seek to identify the “selectivity module” that allows myosin X to recognize bundled actin filaments. Our first approach is to swap similar domains between the selective myosin X and the nonselective myosin V. In this approach, we are aided by the fact that both myosin V and myosin X have clearly recognizable lever arms, composed of repeating IQ domains that bind calmodulins. These lever arms are mechanical elements that are known to reorient and amplify smaller motions in the catalytic head domains of myosins during stepping. Because crystal structures of isolated lever arms are available, the ends of this domain are easy to recognize. In fact, many engineering studies of myosins alter the length of these lever arms or even replace them entirely with artificial, rigid structures (8–10). Thus, we were reasonably confident that we could generate functional myosins with altered selectivity properties through a domain swapping approach, where our cut points are located at each end of the lever arms.
We show through these domain swaps that the myosin X tail (post-IQ) is the most important contributor to selectivity. In fact, we find that every one of our chimeras with a myosin X tail has an enhanced run length on bundled actin filaments. This is a surprising result because the tail domains should be distant from the actin filaments where bundle recognition occurs. To address the role of this critical tail domain, we inserted a flexible polypeptide linker to disrupt its structure. We found that this targeted insertion abolishes bundle selectivity in myosin X. Our results point toward the presence of a novel tail domain structure, which positions the two motor domains of myosin X at the correct spacing and orientation to recognize an actin filament bundle. Moreover, we found a separate role for the myosin X head in the successful initiation of processive runs. Together, this work uncovers some of the structural features that allow molecular motors to select distinct tracks and ultimately arrange cellular contents with high fidelity.
EXPERIMENTAL PROCEDURES
Proteins
Fascin was prepared as previously described (11). Fascin-actin bundles were created by incubating 8 μm F-actin with 3 μm fascin for 2 days (4 °C, in F-buffer) to ensure fully formed and ordered bundles.
Myosin Constructs
The HMM2 forced dimer was previously prepared (7) containing the myosin X sequence from the N terminus to residue Leu-920, followed by residues 4–32 of GCN4-p1 to ensure dimerization, followed immediately by GFP2 and a FLAG tag. The myosin X HMM forced dimer construct was inserted into InsectDirect pBiEx3 vector (Novagen) modified to include the pBlueScript multiple cloning site (pBiEx3-BS), transfected into Sf9 insect cells, and purified by FLAG affinity chromatography. All constructs containing the myosin X IQ domains were coexpressed with the myosin X-specific light chain CALML3; constructs containing the myosin V IQ domains were coexpressed with MLC-1sa (both in pBiEx3-BS). Myosin X with a C tag (12) was similarly expressed and purified. This construct was used only for the single molecule three-bead trapping experiments because the affinity clamp serves as a superior handle compared with anti-GFP. Myosin V HMM-GCN4-YFP-FLAG construct with a native sequence from N terminus to residue Ile-1107 of chicken myosin VA, followed by an in-register GCN4 coiled-coil, was likewise expressed and purified in Sf9 cells.
Head, IQ, and tail chimeras between myosin X and myosin V were constructed with overlap extension PCR, using the forced dimer HMM DNAs as templates, to produce the desired combinations. Thus, all constructs here contain GCN4 at the C terminus of the coiled-coil domains (Leu-920 for myosin X tails and Ile-1107 for myosin V tails). Myosin X sequences used were: head domain to Glu-741, IQ from Val-742 to Ala-817, and tail from Ala-813. Myosin V sequences used were: head domain to Lys-766, IQ from Leu-767 to Lys-910, and tail from Ile-911. The constructs were inserted into pBiEx3-BS, transfected into Sf9 cells, and FLAG purified.
Forward and reverse primers encoding the (GSG)2 sequence (GGTAGTGGTGGTAGTGGT) were used for overlap extension PCR, introducing the six-amino acid flexible linker into the molecule at the desired location either before or after the single α-helix (SAH) domain. The SAH domain was designated to begin at Glu-818 where the VXV and XXV chimeras were spliced and end at Glu-861, 13 residues after previously identified SAH (13) to retain more charged residues RAQQEEAARKQRE that may contribute to the motif. The constructs were inserted into the pBiEx3-BS vector, transfected into Sf9 cells, and FLAG-purified as for myosin X.
All of the myosin constructs described here were verified by DNA sequencing of the pBiEx3-BS vector. Expressed myosins were initially characterized by SDS-PAGE (supplemental Fig. 1A), their ability to bind to actin in cosedimentation assays (supplemental Fig. 1B), and their actin-activated ATPase activity (supplemental Fig. 2 and supplemental Table 2).
Gliding Filament Assay
Motility assays were performed in flow chambers constructed of a glass slide, two strips of double-sided tape, and a nitrocellulose-coated coverslip. All reagents were prepared in assay buffer (AB) containing 25 mm imidazole, pH 7.5, 25 mm KCl, 1 mm EGTA, 4 mm MgCl2, and 10 mm dithiothreitol. Reagents were added to the flow chamber in 10-μl volumes in the following order: 50 ng/μl anti-GFP (Qbiogene), 1 mg/ml bovine serum albumin, myosin (variable concentration), AB, 100 nm tetramethylrhodamine-phalloidin actin, AB, and motility buffer. Motility buffer contains 2 mm ATP, 2 μm calmodulin, 0.86 mg/ml glucose oxidase, 0.14 mg/ml catalase, and 9 mg/ml glucose in AB.
Actin filaments were imaged in epifluorescence on a Zeiss Axiovert 200 by an Andor Luca CCD. Images were analyzed in ImageJ by manually tracking the leading or trailing ends of moving filaments.
Total Internal Reflection (TIRF) Microscopy Single-molecule Assay
We imaged myosin motility using a custom-built objective-type TIRF microscope. Images were collected with a 100×, 1.65 NA objective (Olympus) and an EMCCD camera (iXon; Andor Technologies). Frames were collected at 2 Hz. We prepared tetramethylrhodamine-phalloidin-stabilized actin filaments by mixing biotinylated G-actin (at Cys-374) with unbiotinylated G-actin at a 1:9 ratio before polymerization (14). Flow chambers were coated with neutravidin (0.5 mg/ml) and then blocked with bovine serum albumin (1 mg/ml). We applied to the flow cell either single filaments or fascin bundles and immobilized them through the biotin-neutravidin interaction. We applied motility buffer (as above) containing nanomolar myosin with 1 mg/ml bovine serum albumin and 1% Triton X-100 to further block nonspecific adsorption. Actin was imaged for four to six frames, then excitation was switched to the GFP channel to image motility. Overlaying the first few frames with the rest of the movie facilitated the identification of bundle ends. Construction and analysis of kymographs along the actin axis were used to track movement of single myosins. Data for spots that moved <200 nm or 1.5 s were discarded from analysis to eliminate misidentified diffusive events.
Optical Trap Assay for High Resolution Measurement of Processive Motors
We used a custom-built optical trapping/TIRF microscope, modeled after similar instruments previously described (15), with bead position detection performed in the condenser back focal plane. Detector responses were calibrated by raster scanning the bead through the detection area and calculating a fifth-order, two-dimensional polynomial response function (16). We decorated 0.5-μm-diameter beads with myosin X via anti-GFP antibodies. We introduced these beads into a flow chamber that had surface-attached fascin-actin bundles. The myosin density was chosen such that 10% of the beads bound or moved on surface attached fascin-actin bundles, establishing single-motor conditions. As the myosin engaged with actin undergoing forward motion, the trap position was adjusted to maintain a 1-pN force on the bead-myosin-actin interaction. The bead position trace represents the motion of the molecule along an actin bundle. A step-finder algorithm (17) was used to identify step-like transitions within the bead position trace.
We also used previously described optical trapping protocols and reagents (14, 18) in a three-bead geometry (19). Briefly, a flow cell made with bead-coated coverslips (1.5-μm-diameter silica; Bangs Labs) was incubated with the affinity clamp protein (12) followed by bovine serum albumin and nanomolar myosin X. The flow cell was washed with AB, and motility buffer was flowed in. Motility buffer contains 10 μm ATP, 2 μm calmodulin, 0.086 mg/ml glucose oxidase, 0.014 mg/ml catalase, 0.09 mg/ml glucose, biotinylated fascin-actin bundles stabilized with tetramethylrhodamine-phalloidin, 1-μm neutravidin beads (0.02% solids), and 1 μm phalloidin (Sigma). At the myosin densities used in these experiments ∼10% of beads or platforms showed motor activity. For this experiment the instrument generated multiple traps by rapidly chopping (20-kHz) trap position using a pair of orthogonal acoustooptical deflectors (Intraaction). Bead positions were detected with back focal plane detection (15), using two fiber-coupled laser diodes (785 and 850 nm) as probe beams and two spectrally separated duolateral position-sensitive diodes placed in planes conjugated to the back focal plane of the condenser. Bead positions were low pass antialias filtered at 1 kHz with a custom 4-pole Bessel filter and digitized at 2 kHz.
RESULTS
Rationale for Chimera Design
Because myosin X prefers bundled actin for processive motility and seems to function by straddling two actin filaments within the bundle (7), we expect that this myosin has specific structural or functional features which allow the leading head to recognize an adjacent filament while disfavoring binding along the same actin filament. One such feature is the lever arm. Each myosin family member has a variable number of IQ motifs that form the light chain-binding sites for calmodulin that comprise the lever arm (20). Myosin V produces long working strokes with its long lever, and truncations produce working strokes in proportion to the lever arm length (8, 9). Myosin X has a short, three-IQ lever arm, which is half of the six-IQ lever arm of the canonically processive myosin V. Part of this difference may be compensated by a SAH motif (13) that could extend the lever arm.
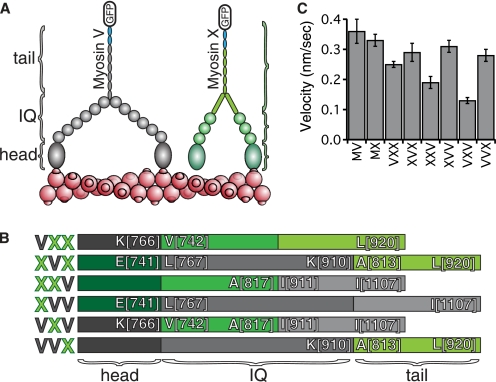
To dissect the domains responsible for selectivity, a combinatorial chimera design strategy was adopted. The experimental intent here was to convert myosin V HMM into a bundle-selective myosin and to abolish selectivity in myosin X HMM molecule by exchanging the head, the IQ, or tail (post-IQ through coiled-coil) domains. Using this strategy, six chimeric constructs were generated. The domain boundaries for these constructs were chosen to be immediately before the first and after the last IQ domains, which are easily identified within the sequences (Fig. 1, A and B). The bovine myosin X IQs span from Val-742 to Ala-817, and the chicken myosin V IQ spans from Leu-767 to Lys-910. To produce these recombinant myosins, the InsectDirect system was used to transiently transfect insect cells with the construct DNAs for accelerated production (21).
FIGURE 1.
Schematic of myosin V/myosin X chimeras. A, schematic of the domain organizations of HMM fragments of myosin V (gray) and myosin X (green), truncated after the predicted coiled-coil. Dimerization of the constructs is ensured by an in-register GCN4 coiled-coil at the C terminus of these constructs (blue). This GCN4 is followed by a GFP, used as a handle for gliding filament experiments as well as a means for visualizing individual motors. B, domain organization of the six possible combinations of the head, IQ, and tail regions between the two motors. Boundary residues of the splice sites are labeled using the chicken myosin V sequence and the bovine myosin X sequence. Each chimera is named according to a three-letter code (left). Each letter identifies the head, lever arm, and tail, respectively. C, gliding filament velocities of the constructs using single actin filaments indicating functional motors. Velocities are shown in nanometers/second, ±S.D., n = 50 filaments each.
Each of the six chimeras was successfully expressed and purified, and all of them moved actin filaments in the gliding filament assay (Fig. 1C), demonstrating that they have intact motor activity. The velocities measured in the gliding filament assay reflect the nature of the constructs. The two that are significantly slower than the others, VXV and XXV, contain a particularly poor combination of structural elements: myosin X IQ domains followed not by a SAH, but by a myosin V coiled-coil. This observation is in line with previous work where versions of myosin V with truncated lever arms are essentially nonprocessive (8–10). As we show below, these two slower chimeras have additional motility defects.
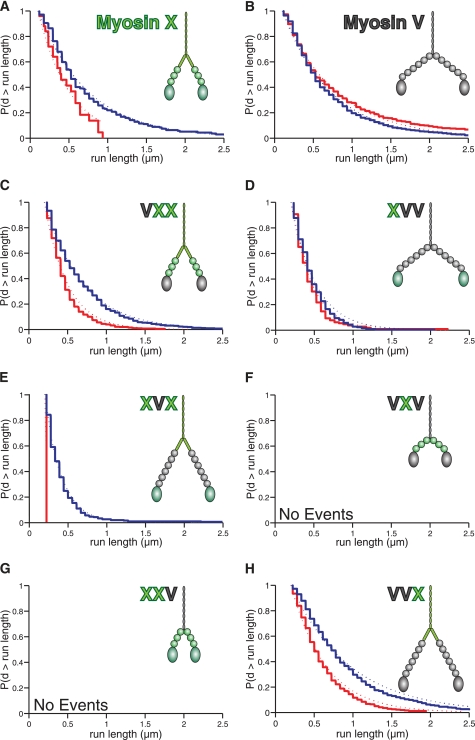
In the following three sections, we illustrate how each myosin domain affects the run length along single actin filaments versus bundles, a phenomenon that we call run length selectivity. In all cases, run lengths were determined using a TIRF motility assay (Fig. 2); selectivity was established by comparing run lengths along single filaments with bundled actin filaments.
FIGURE 2.
Run lengths of chimeras reveal a key role for the myosin X tail in selecting bundles. The domain composition of each construct is shown for visual clarity of their size. The Kaplan-Meier estimate of the run length survivor function is shown for each processive construct on fascin-actin bundles (blue) and single actin filaments (red) at 2 mm ATP. Events are left-truncated at 0.2 μm and are right-censored at track ends. Run lengths are estimated from single exponential fits to the empirical survivor function (dotted lines). Run length decay constants (±S.E.) are: A, myosin X, filaments: 0.17 ± 0.05 μm (n = 24); bundles: 0.63 ± 0.08 μm (n = 100). Data are from Ref. 7. B, myosin V, filaments: 0.66 ± 0.05 μm (n = 231); bundles: 0.57 ± 0.06 μm (n = 134). Data are from Ref. 7. C, VXX, filaments: 0.29 ± 0.01 μm (n = 779); bundles: 0.49 ± 0.02 μm (n = 571). D, XVV, filaments: 0.27 ± 0.02 μm (n = 129); bundles: 0.31 ± 0.02 μm (n = 334). E, XVX, filaments: not determined (n = 2); bundles: 0.21 ± 0.01 μm (n = 323). F, VXV: not determined. G, XXV: not determined. H, VVX, filaments: 0.45 ± 0.02 μm (n = 556); bundles: 0.76 ± 0.02 μm (n = 817). The VXV and XXV chimeras were nonprocessive on both structures. Note the longer run lengths on bundles for the constructs containing the myosin X tail. The differences in run lengths are significant for VXX and VVX (p = 6 × 10−21 and 2 × 10−20, respectively; using the Kolmogorov-Smirnov test) and apparent for XVX, despite the low number of observed events on single filaments. The mean velocities measured in this single-molecule TIRF assay are (nm/s ± S.D.): VXX, filaments: 320 ± 100; bundles: 280 ± 90. XVV, filaments: 350 ± 110; bundles: 320 ± 110. XVX, bundles: 280 ± 100. VVX, filaments: 290 ± 110; bundles: 300 ± 120.
Myosin X Catalytic Head Is Not Required for Run Length Selectivity
Here, we consider the possibility that the motor domain, defined as the N-terminal segment ending at the first IQ motif, may be specifically tuned via its kinetic cycle, actin binding properties, or power stroke geometry to favor processive runs on bundled filaments. The chimeras used to test this possibility are VXX and XVV, where the motor domains of the two myosins were exchanged.
We find that VXX is selective for bundled actin, although it is also processive on single filaments. The run length on fascin-actin bundles is almost double that on single filaments, 0.49 ± 0.02 μm versus 0.29 ± 0.01 μm (Fig. 2C). On the other hand, XVV had similar run lengths on both single actin filaments and fascin-actin bundles (0.27 ± 0.02 μm and 0.31 ± 0.02 μm, respectively; Fig. 2D). This indicates that the myosin X head alone does not impart selectivity on the molecule.
Length of Lever Arm Does Not Drive Run Length Selectivity
The next set of chimeras, VXV and XVX, tested the role of the number of IQ domains in run length selectivity. When we extend the lever arm of myosin X by three IQ motifs to generate XVX, we find that the run length selectivity for bundles remains intact. Although we obtained XVX run lengths of 0.21 ± 0.01 μm along fascin-actin bundles, only two runs were observed on single actin filaments, both of which fell below our minimum reliable detection threshold (200 nm; Fig. 2E). These results clearly indicate that the overall reach of myosin X is unimportant in its selectivity.
For VXV, we generated a motor that reduced the lever arm of myosin V by three IQ motifs. Although VXV is a functional myosin that glides filaments (Fig. 1C), it is rendered entirely nonprocessive on both types of actin structures (Fig. 2F). This result is consistent with previous truncation studies of myosin V, where a 2IQ domain construct was nonprocessive with a short power stroke (8–10). Thus, we can conclude that myosins with a shorter reach do not automatically select the bundled geometry.
Run Length Selectivity Requires Myosin X Tail
By the process of elimination, we arrive at the suggestion that the myosin X tail is responsible for run length selectivity. To establish the conclusion that the post-IQ region of myosin X is the “selectivity cassette,” we constructed two more chimeras, XXV and VVX. Replacing the myosin X tail region with the coiled-coil tail of myosin V (XXV) eliminates processivity of the molecule on both types of actin (Fig. 2G). This result suggests that some portion of the myosin X tail acts to extend the reach of the molecule, in line with the previously proposed role for the SAH domain following the IQ motifs of myosin X (13). The key role of the myosin X tail domain in run length selectivity is further confirmed by VVX, which is a myosin V molecule with the myosin X post-IQ region. This construct is almost twice as processive on bundled actin as on single filaments, 0.76 ± 0.02 μm versus 0.45 ± 0.02 μm (Fig. 2H).
Myosin X Head Is More Likely to Initiate Processive Runs on Bundles
Until now, we have considered how the relative run lengths on actin filaments versus bundles are altered in each of our chimeras. These run lengths are determined by the probability of terminating an already established processive run after each forward step. Here, we consider the probability that our chimeras successfully initiate a processive run. Surprisingly, we find that this initiation selectivity is a separate property that requires the myosin X head, rather than the tail.
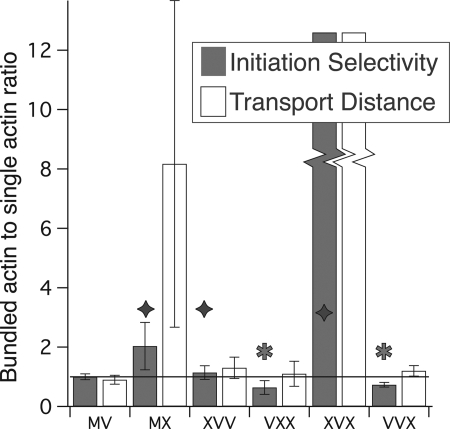
To facilitate these comparisons, we define “initiation selectivity” as a relative landing rate: the rate at which myosins land (with detectable processive movement) on bundles, divided by the landing rate on single filaments (Fig. 3). We use identical motor concentrations in these experiments, as we compare motility on single filaments and bundles as a matched pair using a single split stock of TIRF motility buffer and myosin. Thus, our measure of initiation selectivity is not compromised by errors in our motor stock concentration.
FIGURE 3.
Myosin X head domain is more successful at run initiation on bundled actin. Two dimensionless ratios are displayed. Gray bars represent the initiation selectivity for each processive construct. These initiation selectivities measure the relative rates of landing (with detectable movement) on bundles versus actin filaments and are given as [Rb·exp(0.2 μm/λb)]/[3.5·Rs·exp(0.2 μm/λs)] Here, λ is the run length, R is the measured initiation rate, and subscripts s and b denote single filaments and bundles, respectively. Initiation rates, R, are determined at identical motor concentrations and are calculated as the number of observed events per μm track length per second. The exponential factors correct for missed events that fall below our 0.2-μm detection threshold. We find that myosin V is 3.5 times more likely to initiate runs on the same length of bundles as on single actin. Therefore, we applied a factor of 3.5 in the denominator to set myosin V as a reference motor with an initiation selectivity of 1. The two motors with the largest initiation selectivities, or preference for bundled actin, are wild type MX and XVX. However, note that the initiation selectivity for XVX is large and indeterminate as there were only two events observed on actin filaments. Every construct with a myosin X head (diamonds) is more successful than wild type myosin V at run initiation on bundled actin, whereas myosin V head constructs (stars) are more successful on actin filaments. In the transport distance ratios (open bars), we show the relative distance traveled by each motor, given equal lengths of each type of track. To obtain these, we multiply the initiation selectivities by the run length ratio (λb/λs). Note that the wild type myosin X has the highest transport distance ratio, excluding the indeterminate XVX. Error bars indicate 95% confidence intervals.
Although there is a maximum of 20–25 actin filaments in fascin-actin bundles, we find that myosin V is only 3.5 times more likely to initiate a processive run per μm of bundles, relative to single actin filaments. This difference is expected, as there are many inaccessible myosin-binding sites in an actin bundle; many filaments are buried in the interior of the bundle, some sites are inaccessible because they face the coverslip surface, and other sites on the bundle surface are inaccessible due to the close spacing of the bundle filaments (see “Discussion”). Because myosin V shows no run length selectivity for bundles, it appears that it does not detect the higher local concentration of binding sites within the bundle. Therefore, we use myosin V as our reference for initiation selectivity. To this end, we divide our relative initiation rates by 3.5, to account for these structural differences between actin filaments and bundles. Initiation selectivities >1 indicate a myosin that prefers to start processive runs on bundles compared with myosin V, whereas initiation selectivities <1 likewise indicate a preference for single filaments.
We find that all constructs with a myosin X head prefer to initiate runs on bundles, with initiation selectivities >1 (Fig. 3). Likewise, all motors with a myosin V head prefer to initiate runs on single filaments, with initiation selectivities <1. The most bundle-selective motor is the wild type myosin X, or perhaps XVX. Because we were unable to measure a run length for XVX on single filaments, its initiation selectivity is an undetermined value that is >1.
When we combine this information with the run lengths determined in Fig. 2, to yield an effective transport distance along bundled actin relative to single filaments, we find that myosin X is the most successful at such selective transport. To obtain these relative distances, we multiplied the initiation selectivities by the ratio of bundle to single actin run lengths obtained in Fig. 2.
Structural Properties of the Tail Region Impart Fascin Bundle Selectivity
Evidence from the myosin V/myosin X chimeras implicates the post-IQ region of the molecule to be responsible for bundle selectivity in myosin X. The most prominent and unique feature of the myosin X tail domain is the SAH motif as mentioned earlier. We believe that this domain plays a major role in the myosin X selectivity of bundled actin. This domain may impose a structural constraint that hinders the second head from binding straight ahead along the same filament, instead biasing it to search for a binding site on a neighboring filament.
To determine the role of a structured region in imparting selectivity, free swivel mutants were designed. In these constructs a flexible linker either preceding or following the SAH domain was inserted (Fig. 4A). Such an approach was previously used to break the myosin V lever arm after the second IQ domain (9). The linker of choice was (GSG)2 as it has been previously used on anchoring domains on myosin VI (21). The introduction of flexibility into the system is motivated by myosin V, which contains a free swiveling joint between the two lever arms and before the coiled-coil domain, allowing for a spherical search for the most favorable binding site (1, 22). These swivel mutants will disrupt any structurally rigid elements in myosin X and, if our model is valid, result in a motor domain that is allowed to freely search for the next binding site. From the TIRF motility assays of the two constructs (Fig. 4D), it is clear that selectivity has essentially been abolished with similar run lengths on single filaments as on bundles (pre-SAH, 0.19 ± 0.01 μm and 0.27 ± 0.01 μm; and post-SAH, 0.23 ± 0.01 μm and 0.22 ± 0.03 μm, respectively). These constructs travel for much shorter distances than observed for wild type on bundles. This reduced run length may be the result of a disruption of a sensitive gating mechanism where the ATPase cycles of the two heads are regulated by internal stress as is common for processive myosins (23, 24). Consistent with this idea, we find that these swivels increase the actin-activated ATPase Vmax by ∼50%, suggesting that gating is partially disrupted due to the increased flexibility of the swivels (supplemental Fig. S2). The initiation selectivity also favors single actin filaments over fascin-actin bundles (Fig. 4C), indicating that binding configurations adopted on a bundle lead to more frequent run termination most likely due to disruption of head-to-head communication with binding along a single filament leading to a more strained arrangement.
FIGURE 4.
Free swivels disrupt selectivity on bundled actin. A, six-residue free swivel (GSGGSG) was inserted either before or after the SAH domain of myosin X (green letter). We define the C terminus of the SAH at the end of the charged region and preceding the start of the hydrophobic heptad repeat in the bovine sequence. Thus, we define the SAH domain to be somewhat shorter than previously defined: the entire tail ending before the pleckstrin homology domains (13). B, gliding filament velocities of the constructs indicate functional motor (±S.D., n = 50 filaments). Run lengths of the pre-SAH (C) and post-SAH (D) motors on bundles (blue) and single actin filaments (red) at 2 mm ATP are shown. Run lengths are plotted as in Fig. 2. Run length decay constants (±S.E.) are: pre-SAH, filaments: 0.19 ± 0.01 μm (n = 566); bundles: 0.27 ± 0.01 μm (n = 728). Post-SAH, filaments: 0.23 ± 0.01 μm (n = 204); bundles: 0.22 ± 0.03 μm (n = 96). See also supplemental Table S1. E, initiation selectivity and transport distance ratios for the swivel constructs indicate motors that favor processive runs on single filaments.
Optical Trapping Reveals Short Step Size on Bundles and Stepwise Detachment Behavior
If myosin X prefers to bind neighboring filaments within a bundle, the step size of myosin X may be <36 nm as is observed for myosins V and VI (25). Because there are many more available binding sites in a bundle, the myosin may be able to take advantage of natural repeats shorter than 36 nm. To measure the step size of this construct we employed a force clamp single-bead optical trapping assay (14, 26, 27) (Fig. 5). This experiment allows precise monitoring of the stepping behavior of wild type myosin X as it undergoes stepwise transitions along actin under a fixed force.
FIGURE 5.
Single-bead force feedback optical trapping indicates a short step size for myosin X. A fascin-actin bundle is adhered to the coverslip surface, and a polystyrene bead conjugated to motor is lowered to the track and allowed to interact, leading to processive motions detected on a position detector. Shown are histograms of step size distributions determined using a step-finder algorithm (17) along a bundle. Positive values are fit to a Gaussian distribution. Two stepping regimes are separated by load. A, under a <0.6-pN load the motor takes 17.9 ± 0.3-nm steps. B, under loads >0.6 and <1 pN, myosin X takes shorter 12.3 ± 0.2-nm steps (±S.E.). See also supplemental Fig. S3.
We were able to trap single myosins and observe their motions, which produced long and complex stepping patterns (supplemental Fig. S3). The back-and-forth motion of the molecule resembles myosin VI more than the regular stepping pattern produced by myosin V (27). Clear stepping behavior can be observed and analyzed using a previously developed step-finder algorithm (17). We find a variable step size under these conditions. For forces below 0.6 pN, myosin X is observed to step forward 17.9 ± 0.3 nm, whereas at forces from 0.6–1 pN the step size is smaller, 12.3 ± 0.2 nm (Fig. 5, A and B, respectively). This variability in stepping behavior may be the direct result of the dynamic nature of this molecule and its ability to adapt to the specific bundled actin environment on which it functions. This would allow myosin X to step successfully in regions such as the filopodia where the actin may be heterogeneous. Backward steps are also frequently observed as can be seen in the step size distributions (Fig. 5) as well as in the raw data traces (supplemental Fig. S3).
To observe further the behavior of this motor along a bundle, a more sensitive three-bead assay was used (19, 28). We constructed a dumbbell using fascin-actin bundles and lowered it onto a surface-attached myosin X. Here, the myosin was attached to the surface via an affinity clamp protein (12). The stepping behavior of the single myosin molecule as it engaged with actin resembles the traces obtained from single-bead experiments (Fig. 6), although we found that the detachment events were more clearly resolved. Interestingly, in addition to abrupt detachments we find numerous examples of stepwise detachment (red boxes in Fig. 6), which may represent the motor domains “slipping” under load while still associated with the bundle.
FIGURE 6.
Sample trapping traces of a three-bead fixed-trap assay illustrate complex single molecule stepping behavior of myosin X on bundled actin. Three behaviors are observed. Regular stepping is observed at the beginning of each event (following the dotted line) as well as in regions (blue) where the motor seems to recover forward stepping. The motor also spends periods (orange regions) with prominent forward and backward stepping behavior. In addition to sudden detachment events (arrows), this motor is observed to step/slide backward (red regions), allowing this motor to maintain interaction with actin while returning to zero load. The experimental geometry is shown in the inset.
DISCUSSION
Structured Tail Domain Is Responsible for Bundle Selection in Myosin X
The canonical processive myosin V walks in a tight pattern, using binding sites located every 36 nm along actin. While walking along a fascin-actin bundle, a myosin would have many more binding options, as it can also use sites along the adjacent actin filaments. Indeed, our initial work on myosin X showed that the spatial proximity of filaments was critically important for bundle selection. A collection of artificially bundled filaments was sufficient for the processive motility of myosin X, indicating that myosin X is capable of contacting multiple filaments while stepping (7). Thus, it seemed likely that myosin X used the additional binding sites to take shorter steps along the bundle (Fig. 7). Indeed, we have found that the processive step size is much shorter than the 36 nm favored on single actin filaments. Instead, we found a step size closer to 18 nm at zero to low loads (Fig. 5) (37). Such short, 18-nm steps are incompatible with walking along a single filament within the bundle because this would require a spiral path into the crowded bundle interior.
FIGURE 7.
Structural model of a fascin-actin bundle reveals isolated islands of accessible myosin-binding sites. A set of four actin filaments (light gray) bundled by fascin cross-linkers (dark gray) is shown. The barbed filament ends are directed away from the observer, as indicated by white arrows on the actin. These arrows also indicate the orientation of the myosin-binding sites. Green ellipsoids indicate a myosin X motor attached to the actin filament, with an ∼18-nm distance between the heads. The IQ orientations and the configuration of the tail are unknown and are shown as a black box. Although the components are shown to scale, we have taken some liberties with the shape and orientation of the actin and myosin to reduce the complexity of the model. Note that the length of the myosin head (∼8 nm) is longer than the gap between filaments (∼4 nm). The actin sites, or islands, shown in blue can accommodate a myosin head without steric clashes from the neighboring filament. We have determined this by building a complete atomic model in PyMOL, starting from the Holmes model (36), using a spacing of 12 nm between the filaments as we have observed in electron microscopy (7). The easiest processive path down this structure is to step from the front end of one island to the back of the next, skipping over the segments in gray. Note that the binding site in the middle of the island, pointing straight up, is a poor choice for processive stepping even though it is quite accessible because the step length must adjust up or down from ∼18 nm (center of mass) to reach the next favored position.
Myosin X is a 3IQ myosin, where the total length of the IQ domains is ∼10 nm. At first, we expected that bundle selection could be explained by the limited reach of a 3IQ myosin compared with the nonselective, 6IQ myosin V. This notion was quickly put to rest by our work on the lever arm chimeras (Fig. 2, E and F), which shows that the number of IQ domains has little impact on bundle selection. Our two lever-arm chimeras, XVX and VXV, certainly have an altered reach between the two heads, but this altered reach is insufficient to exchange the selectivity of myosin V and myosin X.
Based on our chimera run length studies shown in Fig. 2, we have also ruled out a prominent role for the myosin X head in the longer processive runs found on bundles. Instead, the myosin X tail is the critical region for selection. This result was most surprising, as the tail is presumably farthest of the three domains from the track. One potential explanation is that the tail domains position each myosin X head at an orientation and spacing that favor attachment to bundles and disfavor single filaments. To test this structured tail proposal, we engineered a free swivel at two different sites within the myosin X tail, both before and after the SAH domain. We reasoned that the additional flexibility in the tail should suffice to convert a myosin X tail into the functional equivalent of the myosin V tail. Myosin V is widely believed to contain similar flexibility at the junction of its two lever arms, just before the start of the coiled-coil tail (29). We found that the insertion of only six residues in the wild type myosin X sequence was sufficient to abolish bundle selectivity, as shown in Fig. 4.
It appears that the myosin X tail domain structure allows the motor domain to search effectively for binding sites along neighboring filaments in a bundle. One plausible role for the tail is to restrict the search space of the free head so that it cannot easily visit binding sites along the same filament and instead is directed to sites on adjacent filaments. This search space may be adapted to the specific filament spacing within a fascin-actin bundle. The post-IQ regions of other bundle-selective myosins may be tuned to the specific filament arrangements found in other bundled filament structures, in much the same manner that locomotives are matched to a specific gauge of railroad track. Additional work is needed to determine the exact nature of this structured region and how it directs the myosin X heads to the adjacent filament sites.
We routinely force dimerization of myosins with a stable coiled-coil domain placed in register with existing heptad repeats within the tail. The reason for this practice is that some myosins will fail to dimerize, even when they contain known coiled-coil domains (30). In certain cases, cargo binding regulates dimerization and function. This form of regulation may be general in cells, as it prevents myosins from engaging tracks and accumulating at their final destination without their cargoes (31). Moreover, myosin activity can be regulated by direct head-to-tail interactions in the absence of cargo (31, 32). Generally, these regulated forms are considered undesirable in mechanical studies, so tail truncations or substitutions are employed to activate the motors. However, note that our GCN4 dimerization domain is insufficient to force dimerization under the extremely low concentrations used here (28, 31). Therefore, we know that additional regions of the native myosin X sequence must contribute to the dimerization as well.
Role of SAH Domain
The SAH domains of myosin VI, VII, X, and Dictyostelium MyoM have attracted considerable recent attention. These unusual, highly charged sequences have been proposed to form isolated α-helices in solution. Isolated peptide fragments with the characteristic SAH domain pattern show a marked helical CD signal and thermally unfold without apparent cooperativity (13, 33). Some have proposed that this α-helix serves as a mechanical element that extends the lever arm of myosin VI and MyoM (33, 34). Indeed, myosins with engineered SAH domains show a longer working stroke at subpiconewton applied loads (33). The stiffness of these regions has been estimated to be one-tenth that of the equivalent length of a myosin IQ domain, which is an α-helix that is supported by its quaternary interactions with calmodulin (34).
Our results are consistent with the SAH domain extending the reach of the motor. In our 3IQ chimeras that lack the SAH domain (XXV and VXV), we find that processivity is abolished on all tracks. On the other hand, XXX and VXX have both the 3IQ domains and the SAH and are both functional processive myosins. Moreover, our swivel results are consistent with the SAH properly orienting the two heads, so that myosin X may select fascin-actin bundles. Swivels at either end of the lever arm disrupt selectivity, with the post-SAH swivel completely abolishing selectivity. Because swivels on either end of the SAH domain affect selectivity, the structured tail that properly orients the myosin X heads must extend through the SAH domain as well. The picture that emerges is one where the distal ends of the SAH domains are held in a relatively rigid tail structure at a fixed angle. This angle is propagated down to the proximal ends of the SAH domains and through the IQ domains, resulting at two heads positioned at an angle relative to one another. Indeed, the persistence length of the SAH domain has been estimated at 15 nm, longer than the 7-nm length of the SAH in myosin X, which means that the proximal and distal ends of the SAH point in roughly the same direction, even under thermal fluctuations (i.e. the tangent vectors of the wormlike chain SAH are highly correlated) (33). This is unlike the situation for myosin V, where a free head can explore all possible orientations relative to an actin-attached head.
Why is a SAH domain particularly useful in the tail of myosin X? Perhaps this unique domain also carries a unique set of mechanical features. The SAH structure may lie between that of an IQ/calmodulin lever arm (too rigid, adopting only a single orientation) and a random coil (too flexible, adopting all orientations). Some flexibility in this SAH domain may be required to allow both heads to bind simultaneously to a bundle and to continue processive runs.
Weakly Bound Search to Engage Fascin-Actin Bundle
To our surprise, we found that chimeras with a myosin X head were more likely to initiate processive runs on fascin-actin bundles. This initiation preference was independent of whether they contained the run length-selective tail domains. Normally, we would expect that the features that contribute to an enhanced run length would also contribute to the successful start of a processive run. However, as we will describe here, the first step of a myosin along a bundle is a unique situation, and the molecule must be able to adapt to the pattern of binding sites along the face of the bundle. We expect that the myosin X heads have kinetic features that allow them to adapt for successful runs on bundles.
A structural model of the surface of a fascin-actin bundle, along with an attached myosin X, is shown in Fig. 7. Note the presence of islands of accessible actin-binding sites, shown in blue. Given an ∼18-nm step at low load, the myosin must occasionally step from one island to the next. The most likely configuration of the two heads in such a step (walking hand over hand while using two filaments) is shown. These sites are particularly favorable, and if myosin X always stepped on the equivalent sites, it would take continuous, regular, ∼18-nm steps. However, note that the sites facing straight up, although accessible, do not fit with this stepping pattern. A motor that lands at these “up” sites may terminate a processive run prematurely.
We suspect that myosin X has kinetic and structural features that allow it to recognize the sites at the head and tail of the actin islands, as opposed to the unfavorable sites facing up. One property, a structured tail, has already been discussed. However, the second property may be just as important for starting a processive run. We expect that the motor, on its initial encounter with an actin bundle, samples several configurations before locking to the bundle in an orientation where both heads can engage. Consistent with this proposal, Kovács et al. (35) have shown that single heads of myosin X populate the weak binding states of actin. Moreover, in our optical trapping results we have seen many examples of stepwise detachments (Fig. 6, red), which may represent similar attempts of the motor to bind to the track while sampling the weakly bound states.
We expect that myosin V heads are particularly poor for such a weak search process. As a highly processive transporter on single filaments, myosin V is adapted to engage the actin track immediately, even with a single head. Single filaments do not present the same dead ends to a myosin that is adapted to take 36-nm steps, so immediate binding is not a problem for myosin V. Thus, chimeric molecules such as VXX are more likely to engage the bundle incorrectly and terminate the run, even though the run lengths of the few successful runs are longer on the bundles.
Optimal Bundle-selective Myosin?
At first glance, there are few structural features for a myosin to use to distinguish a single filament from an actin filament bundle. There are additional sites on neighboring filaments, but any molecule with a degree of flexibility should be able to switch tracks on a bundle while walking. However, nature seems to have selected for a myosin with an optimal set of features for identifying fascin-actin bundles within the cell. The move from a free swivel in the tail of the nonselective myosin V to a structured tail that restricts the diffusive search in the filopodial myosin X is one adaptation. The weak search behavior of the myosin X head is another important adaptation. Interestingly, when we examine the combined effect of these domains (shown in the transport distances in Fig. 3), we find that nature's solution is among the best: none of our chimeras (except, perhaps, XVX) improved the transport distance on bundles over the wild type myosin X.
Because the most important bundle-selection features reside in the head and tail sequences, the lever arm length is not a good predictor of selectivity. Because the tail structures are difficult to predict, we expect that other track-selective myosin classes are yet to be discovered.
Supplementary Material
Acknowledgment
We thank the Rock laboratory for technical assistance.
This work was supported by National Institutes of Health Grant GM078450 (to R. S. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Tables S1 and S2.
- HMM
- heavy meromyosin
- GFP
- green fluorescent protein
- AB
- assay buffer
- pN
- piconewton
- SAH
- single α-helix
- TIRF
- total internal reflection fluorescence.
REFERENCES
- 1.Ross J. L., Ali M. Y., Warshaw D. M. (2008) Curr. Opin. Cell Biol. 20, 41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sousa A. D., Cheney R. E. (2005) Trends Cell Biol. 15, 533–539 [DOI] [PubMed] [Google Scholar]
- 3.Woolner S., O'Brien L. L., Wiese C., Bement W. M. (2008) J. Cell Biol. 182, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wühr M., Mitchison T. J., Field C. M. (2008) Curr. Biol. 18, R912–R914 [DOI] [PubMed] [Google Scholar]
- 5.Berg J. S., Cheney R. E. (2002) Nat. Cell Biol. 4, 246–250 [DOI] [PubMed] [Google Scholar]
- 6.Tokuo H., Ikebe M. (2004) Biochem. Biophys. Res. Commun. 319, 214–220 [DOI] [PubMed] [Google Scholar]
- 7.Nagy S., Ricca B. L., Norstrom M. F., Courson D. S., Brawley C. M., Smithback P. A., Rock R. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9616–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell T. J., Morris C., Spudich J. A., Sweeney H. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14159–14164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakamoto T., Wang F., Schmitz S., Xu Y., Xu Q., Molloy J. E., Veigel C., Sellers J. R. (2003) J. Biol. Chem. 278, 29201–29207 [DOI] [PubMed] [Google Scholar]
- 10.Moore J. R., Krementsova E. B., Trybus K. M., Warshaw D. M. (2004) J. Muscle Res. Cell Motil. 25, 29–35 [DOI] [PubMed] [Google Scholar]
- 11.Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., Borisy G. G. (2006) J. Cell Biol. 174, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J., Nagy S. S., Koide A., Rock R. S., Koide S. (2009) Biochemistry 48, 11834–11836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knight P. J., Thirumurugan K., Xu Y., Wang F., Kalverda A. P., Stafford W. F., 3rd, Sellers J. R., Peckham M. (2005) J. Biol. Chem. 280, 34702–34708 [DOI] [PubMed] [Google Scholar]
- 14.Rock R. S., Rief M., Mehta A. D., Spudich J. A. (2000) Methods 22, 373–381 [DOI] [PubMed] [Google Scholar]
- 15.Visscher K., Gross S., Block S. (1996) IEEE J. Selected Topics Quantum Elect. 2, 1066–1076 [Google Scholar]
- 16.Lang M. J., Asbury C. L., Shaevitz J. W., Block S. M. (2002) Biophys. J. 83, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerssemakers J. W., Munteanu E. L., Laan L., Noetzel T. L., Janson M. E., Dogterom M. (2006) Nature 442, 709–712 [DOI] [PubMed] [Google Scholar]
- 18.Mehta A. D., Finer J. T., Spudich J. A. (1998) Methods Enzymol. 298, 436–459 [DOI] [PubMed] [Google Scholar]
- 19.Finer J. T., Simmons R. M., Spudich J. A. (1994) Nature 368, 113–119 [DOI] [PubMed] [Google Scholar]
- 20.Holmes K. C., Geeves M. A. (2000) Philos. Trans. R. Soc. Lond. B Biol. Sci. 355, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant Z., Altman D., Spudich J. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker M. L., Burgess S. A., Sellers J. R., Wang F., Hammer J. A., 3rd, Trinick J., Knight P. J. (2000) Nature 405, 804–807 [DOI] [PubMed] [Google Scholar]
- 23.Robblee J. P., Olivares A. O., de la Cruz E. M. (2004) J. Biol. Chem. 279, 38608–38617 [DOI] [PubMed] [Google Scholar]
- 24.Rosenfeld S. S., Sweeney H. L. (2004) J. Biol. Chem. 279, 40100–40111 [DOI] [PubMed] [Google Scholar]
- 25.Mehta A. D., Rock R. S., Rief M., Spudich J. A., Mooseker M. S., Cheney R. E. (1999) Nature 400, 590–593 [DOI] [PubMed] [Google Scholar]
- 26.Rock R. S., Rice S. E., Wells A. L., Purcell T. J., Spudich J. A., Sweeney H. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13655–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rief M., Rock R. S., Mehta A. D., Mooseker M. S., Cheney R. E., Spudich J. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9482–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rock R. S., Ramamurthy B., Dunn A. R., Beccafico S., Rami B. R., Morris C., Spink B. J., Franzini-Armstrong C., Spudich J. A., Sweeney H. L. (2005) Mol. Cell 17, 603–609 [DOI] [PubMed] [Google Scholar]
- 29.Vilfan A. (2005) Biophys. J. 88, 3792–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trybus K. M., Freyzon Y., Faust L. Z., Sweeney H. L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjea M., Llinas P., Kim H., Travaglia M., Safer D., Ménétrey J., Franzini-Armstrong C., Selvin P. R., Houdusse A., Sweeney H. L. (2009) Mol. Cell 35, 305–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y., Baboolal T. G., Siththanandan V., Chen M., Walker M. L., Knight P. J., Peckham M., Sellers J. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4189–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sivaramakrishnan S., Spudich J. A. (2009) J. Cell Biol. 187, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baboolal T. G., Sakamoto T., Forgacs E., White H. D., Jackson S. M., Takagi Y., Farrow R. E., Molloy J. E., Knight P. J., Sellers J. R., Peckham M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22193–22198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovács M., Wang F., Sellers J. R. (2005) J. Biol. Chem. 280, 15071–15083 [DOI] [PubMed] [Google Scholar]
- 36.Holmes K. C., Angert I., Kull F. J., Jahn W., Schröder R. R. (2003) Nature 425, 423–427 [DOI] [PubMed] [Google Scholar]
- 37.Ricca B. L., Rock R. S. (2010) Biophys. J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.