Abstract
Activation of the pro-drug isoniazid (INH) as an anti-tubercular drug in Mycobacterium tuberculosis involves its conversion to isonicotinyl-NAD, a reaction that requires the catalase-peroxidase KatG. This report shows that the reaction proceeds in the absence of KatG at a slow rate in a mixture of INH, NAD+, Mn2+, and O2, and that the inclusion of KatG increases the rate by >7 times. Superoxide, generated by either Mn2+- or KatG-catalyzed reduction of O2, is an essential intermediate in the reaction. Elimination of the peroxidatic process by mutation slows the rate of reaction by 60% revealing that the peroxidatic process enhances, but is not essential for isonicotinyl-NAD formation. The isonicotinyl-NAD•+ radical is identified as a reaction intermediate, and its reduction by superoxide is proposed. Binding sites for INH and its co-substrate, NAD+, are identified for the first time in crystal complexes of Burkholderia pseudomallei catalase-peroxidase with INH and NAD+ grown by co-crystallization. The best defined INH binding sites were identified, one in each subunit, on the opposite side of the protein from the entrance to the heme cavity in a funnel-shaped channel. The NAD+ binding site is ∼20 Å from the entrance to the heme cavity and involves interactions primarily with the AMP portion of the molecule in agreement with the NMR saturation transfer difference results.
Keywords: Crystal Structure, Enzyme Mechanisms, Peroxidase, Spectroscopy, Superoxide Ion
Introduction
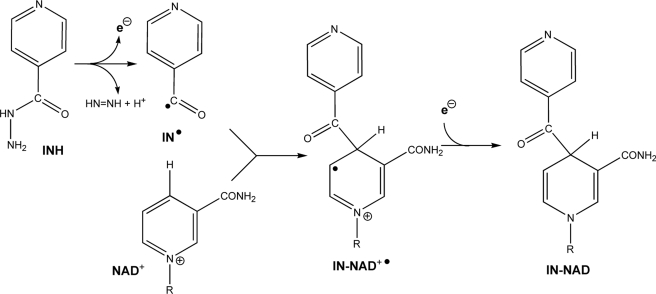
Isonicotinic acid hydrazide (isoniazid or INH)3 is a widely used anti-tubercular pro-drug that requires activation in a reaction involving the catalase-peroxidase KatG of Mycobacterium tuberculosis (MtKatG) (1) whereby the hydrazine group is removed and the isonicotinyl portion is added to NAD+ to generate isonicotinyl-NAD or IN·NAD (see Fig. 1). Once formed, IN·NAD inhibits the synthesis of mycolic acids, and therefore, the growth of M. tuberculosis, by binding to the long-chain enoyl acyl carrier protein reductase (InhA) (2). Despite understanding the role of IN·NAD in the inhibition of mycolic acid synthesis and knowing that KatG is required for INH activation in vivo, uncertainties about the mechanism of its formation remain.
FIGURE 1.
Scheme describing the conversion of INH and NAD+ into IN·NAD, including the key radical intermediates.
The involvement of MtKatG in INH activation suggested that the peroxidatic process had a role, and this provided a focus for several studies employing external oxidants such as peroxyacetic acid (3), t-butyl hydroperoxide (4–6) and low levels of H2O2 (7, 6) to activate the peroxidatic pathway for INH oxidation and activation. In addition, EPR studies have demonstrated that INH can serve as an electron source to reduce peroxidatic intermediates, including specific Trp• radical species following peroxyacetic acid oxidation of MtKatG (3, 8).
A multiplicity of methods has been employed to directly and indirectly assay INH activation, including the determination of INH oxidation to isonicotinic acid (9, 10), the HPLC assay of INH disappearance (11), the inactivation of InhA in a mixture of InhA and KatG (7, 12, 13, 14), the HPLC detection of IN·NAD (4, 15), and the direct measurement of IN·NAD using its characteristic absorbance at 326 nm (6, 7, 12, 15, 16). Reports of INH activation in mixtures lacking an external oxidant (4, 5, 6, 9, 12, 14, 15) initially suggested that the peroxidatic process may not be required, but the mixtures of INH, NADH, and KatG would have supported NADH reduction of molecular oxygen to superoxide and low levels of H2O2 (15) to activate the peroxidase reaction.
Despite the considerable evidence that a peroxidatic process is involved in IN·NAD synthesis, attempts to rationalize the reaction entirely in terms of the peroxidatic pathway generally produced models that were incomplete from the standpoint of electron balance (5, 7, 10) or that involved hypothetical intermediates not characterized in any other system (5, 6). For example, a common scheme shows the isonicotinyl radical, generated in a peroxidatic reaction, reacting with NAD+ to yield IN·NAD, when such a reaction would actually yield the IN·NAD+• radical (Fig. 1). The need to reduce this radical in an oxidizing environment suggests that more than a simple peroxidatic pathway is involved in the INH activation process. This rationale leads to superoxide, O2˙̄, a known reducing agent with a documented role in INH activation (4–6, 11, 17, 18). Peroxidases, including KatGs, can generate O2˙̄ from O2 and an electron donor such as INH or NADH (11, 15, 19–21), and O2 has a role, as yet not fully defined, in INH activation (4, 5, 11, 14, 17). Finally, three reports have described IN·NAD synthesis in mixtures of KatG, INH and NAD+ with no oxidant other than O2, conditions that also give rise to O2˙̄ (6, 15, 22). There exists, therefore, a dichotomy between peroxidatic and superoxide-driven pathways of IN·NAD synthesis that has not been clearly resolved.
The crystal structures of KatG from four different organisms, including Haloarcula morismortui (23, 24), Synechococcus PCC7492 (25), Burkholderia pseudomallei (BpKatG) (26, 27), and M. tuberculosis (28), along with the structures of the predominant INH-resistant variant S315T of MtKatG (7) and its homologue in BpKatG, S324T (22), have been reported. A covalent modification unique to all KatGs involves the side chains of Met264–Tyr238–Trp111. This adduct, in combination with a nearby mobile Arg426, is essential for catalase activity, but not peroxidase activity. These structures have provided many insights into KatG structure and function and confirmed the considerable similarity in structure between BpKatG and MtKatG consistent with the similarity in enzymatic properties (16). Unfortunately, they have not led to an identification of the binding sites for the two substrates, INH and NAD+, which would potentially enhance our understanding of the INH activation process.
The work described in this report examines in more detail the role of KatG in IN·NAD synthesis. In particular, the apparent dichotomy between the peroxidase- and superoxide-driven paths is clarified in a confirmation that superoxide and its in situ generation by KatG is essential for IN·NAD synthesis, whereas the peroxidatic path enhances the reaction. In addition, while non-enzymatic aspects of the reaction are found to have a significant role, binding sites for both INH and NAD+ on BpKatG are identified in enzyme·ligand complexes generated by co-crystallization.
EXPERIMENTAL PROCEDURES
Variant Protein Construction, Purification, and Characterization
Standard chemicals and biochemicals were obtained from Sigma. The oligonucleotides GCCGACGGCGCCGGCGGCGCG (R123A), GGCGCGGGCGCAGGGCAGCAG (E128A), TGGGAGCCCGCGGACGTCTAC (E198A), TATTCGGGCGCCCGCCAGCTC (D222A), GGCAATCCCGCTCCGGTCGCC (D249A), GCCGCGGCGGCCGACATTCGT (R255A), and GACAAGGCGGCACTGCTGACG (Q622A) were purchased from Invitrogen and used to mutate a fragment of pBpKatG (29) following the Kunkel procedure (30). The mutated sequences were confirmed (31) and used to generate the plasmids pR123A, pE128A, pE198A, pD222A, pD249A, pD255A, and pQ622A. The native and variant proteins were expressed and purified as described (29). The variants R123A, E128A, E198A, D222A, D249A, R255A, and Q622A were unchanged compared with wild-type BpKatG in both catalase and peroxidase activities. The release of radicals from INH was assayed by the increase in absorbance at 560 nm caused by nitro blue tetrazolium reduction to formazan (ϵ = 18,500 m−1 cm−1) in a solution containing 1.0 mm INH, 2 μm MnCl2 and 0.2 mm nitro blue tetrazolium in 50 mm Tris-HCl, pH 8.0, at 37 °C (32). The synthesis of IN·NAD was assayed by the increase in absorbance at 326 nm (ϵ = 6,900 m−1 cm−1) (33) of a solution containing 0.4 mm NAD+, 1.0 mm INH, 2 μm MnCl2, and 50 mm Tris-HCl, pH 8.0, at 37 °C. Protein was estimated according to the methods outlined by Layne (34).
NMR-STD Analysis
NMR saturation transfer difference (STD) experiments (35, 36) were run at 500 MHz in a Varian INOVA-500 instrument. Saturation was achieved by a train of 50-ms Gaussian-shaped pulses with 5-ms intervals, and a total saturation time of ∼2.5 s. The on-resonance protein irradiation was performed at a chemical shift of −0.4 ppm, and off-resonance at 34.5 ppm, where no protein signals are present. The free induction decays with on-resonance and off-resonance protein irradiation were acquired in an interleaved mode using 16 transient blocs. The protein signal was partially suppressed by the application of a 20-ms spin lock after the observation pulse. The residual water signal was suppressed using an excitation sculpting module null at the water. The STD spectra were acquired with 1000 or 500 transients. The relative STD values were calculated by dividing the intensities of signals in the STD spectrum by the intensities of corresponding signals from the off-resonance control experiment. The protein concentration was 10 μm in 50 mm sodium acetate, pH 5. Ligand concentration was 40 mm. All NMR buffers were prepared in D2O.
MS Analysis
Samples were analyzed on a MALDI Qq Time-of-Flight mass spectrometer built and maintained in the Department of Physics and Astronomy at the University of Manitoba (37). No buffer exchange was done. For each sample, a 0.5-μl aliquot was mixed with an equal volume of saturated 2,5-dinitrobenzoic acid matrix. Tandem mass spectrometry was done on selected ions using collision voltages appropriate for similar sized peptides. Some samples were also analyzed on a Varian 500-MS ion trap mass spectrometer after dilution with an equal volume of 2% formic acid in methanol before electrospray ionization.
Crystallization and Structure Determination
INH, NAD+, or AMP in 50 mm potassium phosphate, pH 7.0, were added separately to a solution of 20 mg/ml BpKatG also in 50 mm potassium phosphate, pH 7.0, to a final concentration of 1 mm. This solution was mixed 2:1 (v/v) with the reservoir solution of 0.1 m sodium citrate, pH 5.6, 25 mm NaCl, 16–20% PEG 4000, 20% (v/v) 2-methyl-2,4-pentanediol. Brownish crystals were obtained at 20 °C by the vapor-diffusion, hanging drop method, which belonged to the orthorhombic space group P212121 and contained a dimer in the asymmetric unit. Data sets were collected using synchrotron beam line CMCF 08ID-1 at the Canadian Light Source in Saskatoon from crystals flash-cooled in the reservoir buffer. Diffraction data were processed and scaled using programs MOSFLM and SCALA (38), respectively (Table 2). Structure refinement was completed using program REFMAC (39) and manual modeling with the molecular graphics program COOT (40). As determined by SFCHECK (38), the Ramachandran distribution of residues for all structures was 90.9–91.6% in favored regions and 8.4–9.1% in allowed regions. The figures were generated using PyMOL (The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA). Structure factors and coordinates have been deposited in the Protein Data Bank with the accession numbers 3N3N, 3N3O, 3N3P, 3N3Q, 3N3R, and 3N3S for BpKatG·INH, BpKatG-NAD+, BpKatG-ADP, S324T·INH, E198A, and E198A-INH, respectively.
TABLE 2.
Mass spectrometry analysis of a reaction mixture containing INH, NAD+, Mn2+, BpKatG, and TEMPO (as described in Fig. 4), and MS-MS analysis of the “923/925” ion
| Observed ions |
Assumed structurea | Change | Formula | Expected ion | |
|---|---|---|---|---|---|
| MS | MS-MS of 923.17 | ||||
| “parent” | |||||
| 925.18 | IN·NAD·T | C36H49N9O16P2 | 925.28 | ||
| 923.17 | 923.17 | IN·NAD·T(-2H) | C36H47N9O16P2 | 923.26 | |
| 907.16 | 907.16 | IN·NAD·T-H2O | -H2O | C36H47N9O15P2 | 905.25 |
| 905.17 | 905.17 | IN·NAD·T(-2H)-H2O | -H2O | C36H45N9O15P2 | |
| 816.12 | 816.12 | NAD·T | -IN | C30H42N8O15P2 | 816.22 |
| 771.08 | IN·NAD | -T | C27H33N8O15P2 | 771.15 | |
| 753.04 | -H2O | C27H31N8O14P2 | 753.14 | ||
| 664.12 | NAD | C21H28N7O14P2 | 664.12 | ||
| “y” ions | |||||
| 657.06 | PPRN(IN)T | -A | C26H35N4O12P2 | 657.17 | |
| 576.10 | PRN(IN)T | -P | C26H33N4O9P | 576.20 | |
| 558.10 | PRN(IN)T-H2O | -H2O | C26H31N4O8P | 558.19 | |
| 319.07 | PRN | -IN -T | C11H16N2O7P | 319.07 | |
| “b” ions | |||||
| 524.06 | APPR | -N | C15H20N5O12P2 | 524.06 | |
| 428.04 | APP | -R | C10H16N5O10P2 | 428.04 | |
| 364.10 | AP | -P | C10H15N5O8P | 364.07 | |
| 348.07 | AP (-O) | -O | C10H15N5O7P | 348.07 | |
| 250.09 | A | -P | C10H12N5O3 | 250.09 | |
| 136.06 | Adenine | -ribose | C5H6N5 | 136.06 | |
a A, adenosine; P, phosphate; R, ribose; N, nicotinamide; IN, isonicotinyl portion of INH; T, TEMPO; dash, position of break.
RESULTS
Definition of the Minimal Components for IN·NAD Synthesis
INH activation by conversion to IN·NAD in M. tuberculosis requires MtKatG. BpKatG has catalytic properties almost identical to those of MtKatG and has the advantages of a large number of accessible variants and facile, reproducible crystallization. In addition, the properties of the widely studied Ser to Thr variants (MtKatG S315T, responsible for a majority of INH resistance and its homologue BpKatG S324T) (7, 22) are indistinguishable, a fact confirmed in the current work.
To systematically consider the impact of component changes and supplements on IN·NAD synthesis, a minimal or basal reaction mixture was first defined, and refined by the determination of saturating substrate concentrations. This basal mixture contained INH, NAD+, BpKatG, Mn2+ and O2 dissolved in the buffers (15, 16, 22) and supported IN·NAD synthesis with a relatively slow (compared with the catalase and peroxidase reactions) Vmax of 3.54 nmol of IN·NAD/min-nmol heme at 37 °C. Km values of 25 μm for NAD+ and 550 μm for INH were determined at an optimum pH of 8.25. Replacing NAD+ with NADH reduced the rate of reaction by >80%. Removal of either INH or NAD+ stopped the reaction completely, and the catalytic role of BpKatG is evident in the 7-times increase from the non-enzymatic rate of IN·NAD synthesis upon the addition of enzyme (0.31 nmol/min to 2.21 nmol/min with 0.625 μm BpKatG) (Table 1). All subsequent work utilizes this basal mixture unless modified in a specified way.
TABLE 1.
Rates of IN·NAD synthesis and radical generation assayed with NBT
| Enzyme | Additiona | IN·NAD | Radical |
|---|---|---|---|
| nmol/min | mol/min | ||
| Controlsa | |||
| 0.31 ± 0.02 | 0.29 ± 0.12 | ||
| SOD | NDb | 0.05 ± 0.01 | |
| EDTA | ND | <0.01 | |
| NaCN | ND | 0.40 ± 0.05 | |
| NaCN + SOD | ND | 0.12 ± 0.01 | |
| GOx | ND | 0.16 ± 0.04 | |
| GOx + SOD | ND | 0.04 ± 0.01 | |
| PAA | 0.09 ± 0.02 | 0.38 ± 0.05 | |
| PAA + SOD | ND | 0.05 ± 0.01 | |
| MtKatG and variantc | |||
| MtKatG | 2.17 ± 0.02 | 1.42 ± 0.02 | |
| MtKatG | No Mn2+ | 0.08 ± 0.01 | 0.17 ± 0.03 |
| MtKatG | SOD | 0.24 ± 0.04 | 1.10 ± 0.09 |
| MtKatG | GOx | 2.51 ± 0.19 | 1.71 ± 0.05 |
| MtKatG | GOx + SOD | 1.78 ± 0.22 | 1.60 ± 0.04 |
| MtKatG | tBHP | 3.45 ± 0.29 | 1.35 ± 0.02 |
| MtKatG | tBHP + SOD | 2.60 ± 0.30 | 1.04 ± 0.04 |
| MtKatG | PAA | 2.24 ± 0.14 | 1.31 ± 0.01 |
| MtKatG | PAA + SOD | 0.17 ± 0.02 | 1.21 ± 0.02 |
| S315T | 1.06 ± 0.12 | 0.53 ± 0.02 | |
| S315T | SOD | 0.11 ± 0.01 | 0.31 ± 0.02 |
| S315T | GOx | 1.23 ± 0.10 | 0.63 ± 0.01 |
| S315T | GOX + SOD | 0.33 ± 0.01 | 0.43 ± 0.02 |
| BpKatG and variantsc | |||
| BpKatG | 2.21 ± 0.17 | 1.12 ± 0.04 | |
| BpKatG | No Mn2+ | 0.04 ± 0.01 | 0.16 ± 0.01 |
| BpKatG | SOD | 0.07 ± 0.01 | 0.68 ± 0.07 |
| BpKatG | HPC | 1.82 ± 0.05 | 0.77 ± 0.18 |
| BpKatG | EDTA | 0.10 ± 0.02 | 0.09 ± 0.01 |
| BpKatG | NaCN | 0.55 ± 0.02 | 0.25 ± 0.02 |
| BpKatG | NaCN + SOD | 0.43 ± 0.11 | 0.23 ± 0.02 |
| BpKatG | GOx | 2.70 ± 0.29 | 1.47 ± 0.07 |
| BpKatG | GOx + SOD | 1.47 ± 0.24 | 1.09 ± 0.03 |
| BpKatG | GOx + HPC | 2.18 ± 0.01 | 1.32 ± 0.06 |
| BpKatG | tBHP | 2.84 ± 0.05 | 1.15 ± 0.11 |
| BpKatG | tBHP + SOD | 1.45 ± 0.05 | 1.04 ± 0.07 |
| BpKatG | PAA | 2.11 ± 0.17 | 1.01 ± 0.07 |
| BpKatG | PAA + SOD | 0.07 ± 0.02 | 0.86 ± 0.14 |
| BpKatG | XOx | 1.87 ± 0.31 | 2.23 ± 0.29 |
| H112A | 0.65 ± 0.05 | 0.40 ± 0.02 | |
| H112A | SOD | 0.10 ± 0.02 | 0.11 ± 0.02 |
| H112A | GOx | 0.36 ± 0.07 | 0.32 ± 0.02 |
| H112A | GOx + SOD | 0.26 ± 0.05 | 0.23 ± 0.01 |
| R108A | 1.05 ± 0.04 | 0.65 ± 0.07 | |
| R108A | SOD | 0.12 ± 0.02 | 0.20 ± 0.03 |
| W111F | 0.92 ± 0.07 | 0.72 ± 0.10 | |
| W111F | SOD | 0.27 ± 0.02 | 0.45 ± 0.07 |
| Y238F | 1.95 ± 0.48 | 1.98 ± 0.36 | |
| Y238F | SOD | 1.06 ± 0.17 | 1.51 ± 0.22 |
| R426A | 3.20 ± 0.65 | 1.12 ± 0.02 | |
| R426A | SOD | 0.76 ± 0.21 | 0.95 ± 0.04 |
| M264A | 1.59 ± 0.19 | 1.40 ± 0.11 | |
| M264A | SOD | 0.55 ± 0.12 | 1.04 ± 0.09 |
| E198A | 2.01 ± 0.10 | 1.04 ± 0.02 | |
| E198A | SOD | 0.10 ± 0.02 | 0.67 ± 0.02 |
| S324T | 0.92 ± 0.17 | 0.59 ± 0.09 | |
| S324T | SOD | 0.14 ± 0.04 | 0.29 ± 0.08 |
| S324T | GOx | 1.16 ± 0.10 | 0.86 ± 0.02 |
| S324T | GOx + SOD | 0.43 ± 0.10 | 0.68 ± 0.01 |
| W139F | 2.29 ± 0.12 | 1.18 ± 0.04 | |
| W139F | SOD | 0.67 ± 0.12 | 0.83 ± 0.07 |
| W153F | 1.76 ± 0.19 | 1.18 ± 0.06 | |
| W153F | SOD | 0.29 ± 0.09 | 0.70 ± 0.06 |
| W202F | 2.00 ± 0.11 | 1.31 ± 0.04 | |
| W202F | SOD | 0.51 ± 0.14 | 0.77 ± 0.09 |
| W330F | 2.07 ± 0.17 | 0.95 ± 0.04 | |
| W330F | SOD | 0.22 ± 0.07 | 0.45 ± 0.04 |
| W139/153F | 1.86 ± 0.17 | 1.44 ± 0.25 | |
| W139/153F | SOD | 0.43 ± 0.05 | 0.76 ± 0.09 |
| W139/330F | 1.32 ± 0.02 | 1.01 ± 0.02 | |
| W139/330F | SOD | 0.29 ± 0.03 | 0.58 ± 0.05 |
| W139/153/330F | 0.96 ± 0.12 | 1.16 ± 0.14 | |
| W139/153/330F | SOD | 0.19 ± 0.04 | 0.94 ± 0.36 |
a The following supplements were included as indicted: SOD (5 μg/ml), EDTA (1 mm), NaCN (5 mm), GOx (3 μg of glucose oxidase + 5 mm glucose), PAA (100 μm), tBHP (0.4 mm), XOx (0.012 units/ml xanthine oxidase + 0.5 mm xanthine). Except where explicitly removed, 2 μm MnCl2 was included.
b ND, not detectable.
c 0.625 μm MtKatG, BpKatG, or the variants was used for the IN·NAD assays, and 1.25 μm was used for the radical assays.
Superoxide Is Essential for IN·NAD Synthesis and Is Produced by KatG
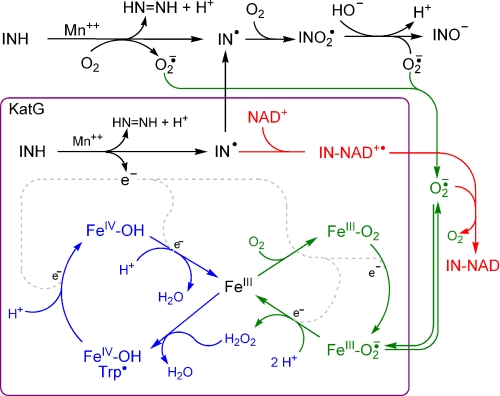
The requirement for O2 in INH activation has been linked to the involvement of O2˙̄ in IN·NAD synthesis (6, 11, 17), but without a clear picture for how it is used in IN·NAD synthesis or how O2 is reduced to O2˙̄. The essential role of O2˙̄ in IN·NAD synthesis was confirmed by 0.1 μm superoxide dismutase causing >95% inhibition (Table 1), whereas much larger amounts (>2 μm) of catalase or bovine serum albumin had no effect. Repeated flushing of the buffers with N2 gas reduced the rate of reaction by >50% confirming that O2 is required for O2˙̄ synthesis. Somewhat surprisingly, inclusion of the xanthine/xanthine oxidase system, to supplement O2˙̄ formation, caused only a small enhancement in the rate of IN·NAD synthesis suggesting that O2˙̄ generation is not the rate-limiting step in the reaction.
These observations led to the question: how is O2 reduced to O2˙̄? This was addressed using the radical sensor nitro blue tetrazolium with and without SOD present to quantify the rate of radical formation and the proportion of radicals that are O2˙̄ (32). Two sources of O2˙̄ were identified, a non-enzymatic Mn2+-catalyzed reaction and a KatG-catalyzed reaction. The Mn2+-catalyzed oxidation of INH (11) produces radicals at a basal rate of 0.31 nmol/min-ml of which >90% are O2˙̄ (Table 1). The predominance of O2˙̄ can be rationalized in terms of two one-electron transfer steps (Fig. 2, top); O2˙̄ and IN• radicals are generated in the first step, followed by IN• conversion to an IN-peroxy radical (41) to yield O2˙̄ and isonicotinic acid in the second. The KatG-catalyzed production of O2˙̄ differs in two key respects from the non-enzymatic process. First, the rate of radical formation is 4-times faster at 1.12 nmol/min-ml (in the presence of 1.25 μm KatG) (Table 1), and second, only ∼50% of the radicals are O2˙̄ (captured by SOD). Thus, while INH oxidation is faster in the presence of KatG, it involves a single one-electron transfer step producing O2˙̄ and the IN• radical required for IN·NAD synthesis. Despite the different reaction pathway, the rate of O2˙̄ formation by KatG is faster than from the Mn2+-catalyzed reaction and inhibition by NaCN confirms that O2 reduction occurs in the heme cavity.
FIGURE 2.
Scheme summarizing the enzymatic and non-enzymatic reactions involved in the synthesis of IN·NAD. The Mn2+-catalyzed oxidation of INH is shown in black at the top. The KatG-catalyzed oxidation of INH is also drawn in black but inside the maroon box representing KatG. Electrons from INH can be used to reduce peroxidatic intermediates (in blue) or O2 and O2˙̄ in the superoxide cycle (in green). Although a Cpd I* species (FeIV-OH, Trp•) is shown, a classic Cpd I species (implying a porphyrin cation radical) would initially be formed, and electron transfer from specific residues in the protein would rapidly give rise to the FeIV-OH Trp• species. The location of the reaction of IN• with NAD+ to form IN·NAD•+ (bound to the enzyme) and reduction of IN·NAD•+ (not bound to KatG) are inferred from the existence of a NAD+ binding site, the capture of IN·NAD•+ by TEMPO, and the involvement free O2˙̄ in IN·NAD•+ reduction, but is open to other interpretations.
The Peroxidase Cycle Enhances, but Is Not Essential for IN·NAD Synthesis
Several reports in which IN·NAD synthesis was assayed in the basal reaction mixture (INH, NAD+, Mn2+, and KatG) supplemented with H2O2, PAA, or tBHP suggested that the peroxidatic pathway had a role in INH activation (5–7). These studies did not determine the incremental effect of the oxidants on the basal O2˙̄-dependent rate of IN·NAD synthesis, or whether the peroxidatic process replaced, supplemented, competed with, or had no effect on the basal O2˙̄-dependent reaction. These questions are at the heart of the dichotomy between the superoxide and peroxidase roles in IN·NAD synthesis and are addressed here in two ways. The first was to investigate BpKatG variants with reduced peroxidase and catalase activities, including H112A, which lacks peroxidase (<4% of native) and catalase activity (<0.1% of native), and R108A, which has peroxidase and catalase rates that are 25% of native (15). Despite a significant reduction in the peroxidatic process in these two variants, the rates IN·NAD synthesis remained at 30% (H112A) and 50% (R108A) of the native enzyme rate, whereas the rate of radical formation was either not affected (R108A) or reduced to 30% (H112A) of native, respectively. This clearly demonstrates that the peroxidatic pathway is not essential for, but does enhance IN·NAD synthesis. Changes to other distal side residues, including those in the Met264–Tyr238–Trp111 adduct and the associated R426, which are essential for the catalase reaction but not the peroxidase reaction, elicit only small changes in the rates of IN·NAD synthesis and radical production (Table 1), albeit with subtle changes in the O2˙̄ dependence of IN·NAD synthesis and in the O2˙̄ proportion of radicals. In summary, none of the residues that significantly influence the catalase and peroxidase reactions have a similar impact on IN·NAD synthesis or INH breakdown.
The absence of a substantive impact of the peroxidatic process on IN·NAD synthesis is also evident in the small increases in rates of IN·NAD synthesis and radical formation caused by oxidant supplements to the basal mixture (28%, 22, and 0%, respectively, for H2O2, tBHP, and PAA). H2O2 and tBHP did reduce the requirement for O2˙̄ in IN·NAD synthesis such that SOD inhibited the reaction only 55 and 49%, respectively, but again, PAA had no effect (Table 1). This was also true for the H112A variant. The reduction in O2˙̄ dependence and decreased O2˙̄ production despite a normal reaction rate suggests that other radical species may be substituting for O2˙̄ in some aspect of the reaction.
UV-visible absorbance spectra of BpKatG, determined at steady state 30 s after starting the reaction under the same conditions as used for IN·NAD synthesis and INH oxidation, were obtained to determine if the spectral changes might be consistent with the formation of any previously characterized reaction intermediates. INH, alone or in combination with NAD+, caused a 15% decrease in intensity of the Soret and charge transfer bands, but with no shift in wavelength (Fig. 3). The addition of H2O2, tBHP, or PAA after INH did not change the INH-induced spectrum, whereas the characteristic oxoferryl spectrum caused by PAA rapidly changed to the INH-induced spectrum upon addition of INH. In conclusion, INH induces changes in the heme pocket that, at steady state, are not consistent with the accumulation of oxoferryl or oxoferrous intermediates.
FIGURE 3.
Effect of INH on the UV-visible absorption spectrum of BpKatG. The spectrum of 1.25 μm BpKatG before (black) and 30 s after (red) the addition of 400 μm INH is shown. The spectrum of BpKatG treated with 100 μm PAA is shown before (blue) and after (cyan) the addition of 400 μm INH.
Identification of the IN·NAD•+ Species
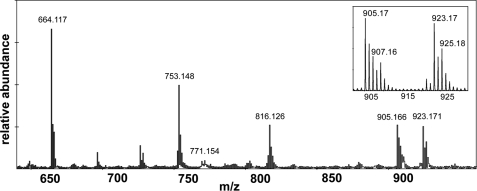
The first step in INH activation common to most schemes is the formation of an IN• radical along with diimide and a proton (Fig. 1). The formation of IN• in solutions lacking NAD+ has been confirmed both using the radical trap TEMPO (42) and in EPR spin trapping experiments, with the latter also identifying Pyr• and INOO• formed from decarbonylation of IN• and from reaction of IN• with O2, respectively (41). To identify radicals formed in the presence of INH and NAD+, TEMPO was included and the reaction mixture analyzed by MALDI MS (Fig. 4). Ions corresponding to NAD+ and IN·NAD, masses of 654 and 771 Da, respectively, have been observed and characterized previously (15), but two larger ions or groups of ions were evident at 816 and 925 Da. The larger ion was identified by MS-MS analysis as the IN·NAD·TEMPO adduct from the series of degradation products (Table 2). The 816-Da ion was identified as NAD·TEMPO, which along with the MS-MS breakdown pattern suggest that the TEMPO is attached to the nicotinamide ring of NAD, not to the isonicotinyl ring. As an aside, the inefficiency of radical trapping is evident in the significant amount of IN·NAD formed in the presence of TEMPO.
FIGURE 4.
MALDI spectrum of a sample from a typical experiment with 10 mm TEMPO included in a mixture of 1.25 μm BpKatG, 1.0 mm INH, 0.4 mm NAD+, and 2 μm MnCl2. The reaction was stopped by freezing after 8-h incubation. The most abundant monoisotopic ions unique to the experiment are identified, and details of the structures assigned to the different ions are in Table 2.
Identification of INH Binding Sites in BpKatG
A clear understanding of the INH activation process has suffered from uncertainty about the sites of INH and NAD+ binding. They have not been identified in any of the four reported KatG structures, and extrapolation from binding sites in plant peroxidases must be done with the caveat that access to the KatG heme cavity is more restricted in the larger protein. Attempts to generate a BpKatG·INH complex by crystal soaking proved unsuccessful, but ultimately co-crystallization conditions that included chloride were found to produce crystals of the complex from which diffraction data sets were obtained and refined to between 1.9 and 2.1 Å (Table 3).
TABLE 3.
Data collection and structural refinement statistics for BpKatG and its S324T and E198A variants with INH, NAD+, and AMP bound
| Variant | BpKatG·INH | BpKatG·NAD+ | BpKatG·AMP+INH | S324T·INH | E198A | E198A·INH |
|---|---|---|---|---|---|---|
| A) Data collection statistics | ||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| PDB | 3N3N | 3N3O | 3N3P | 3N3Q | 3N3R | 3N3S |
| Unit cell parameters | ||||||
| a (Å) | 100.1 | 100.6 | 100.4 | 100.2 | 100.3 | 100.6 |
| b (Å) | 113.3 | 114.9 | 114.2 | 112.6 | 116.0 | 115.9 |
| c (Å) | 174.6 | 174.5 | 173.2 | 173.7 | 174.7 | 174.7 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)a | 33.3-2.09 (2.21-2.09)a | 38.0-1.69 (1.78-1.69) | 35.1-1.9 (2.00-1.90) | 33.2-1.9 (2.00-1.90) | 29.1-1.60 (1.69-1.60) | 29.1-1.70 (1.79-1.70) |
| Unique reflections | 115,829 | 225,404 | 145,296 | 154,832 | 267,486 | 204,869 |
| Completeness (%) | 99.4 (96.9) | 99.9 (100.0) | 91.6 (87.0) | 100.0 (100.0) | 100.0 (100.0) | 91.8 (79.1) |
| Rsymb | 0.094 (0.372) | 0.084 (0.37) | 0.11 (0.72) | 0.242 (0.90) | 0.084 (0.55) | 0.102 (0.45) |
| 〈I/σI〉 (%) | 9.9 (3.0) | 5.7 (2.1) | 9.2 (1.9) | 4.8 (1.8) | 11.9 (2.8) | 8.9 (2.3) |
| Multiplicity | 4.2 (3.8) | 5.1 (5.0) | 3.6 (2.9) | 6.6 (6.5) | 5.9 (5.5) | 5.3 (4.6) |
| B) Model refinement statistics | ||||||
| No. reflections | 109.936 | 214,061 | 137,960 | 147,103 | 253,932 | 194,460 |
| Rcryst (%)c | 16.2 | 15.1 | 16.5 | 18.5 | 15.8 | 15.3 |
| Rfree (%)d | 20.2 | 17.8 | 20.6 | 22.6 | 18.5 | 18.4 |
| Non-H atoms | 12,374 | 12,772 | 12,634 | 12,489 | 12,983 | 12,676 |
| Water molecules | 1,198 | 1,546 | 1,422 | 1,292 | 1,809 | 1,519 |
| Average B-factor (Å2) | ||||||
| Protein | 24.4 | 21.7 | 22.9 | 23.4 | 18.9 | 19.4 |
| Heme group | 18.1 | 15.3 | 16.9 | 16.2 | 13.4 | 12.8 |
| Waters | 32.6 | 32.7 | 31.9 | 32.6 | 31.1 | 30.8 |
| Other | ||||||
| Coord. err. (Å)e | 0.103 | 0.048 | 0.092 | 0.093 | 0.042 | 0.051 |
| in bonds (Å) | 0.025 | 0.032 | 0.025 | 0.026 | 0.032 | 0.032 |
| in angles (°) | 1.80 | 2.36 | 1.81 | 1.94 | 2.51 | 2.27 |
a Values in parentheses correspond to the highest resolution shell.
b Rsym = Σhkl Σj|Ihkl,j − 〈Ihkl〉|/Σhkl〈Ihkl〉.
c Rcryst = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|.
d Rfree is as for Rcryst but calculated for a test set comprising reflections not used in the refinement.
e Based on maximum likelihood.
There were two concerns with regards the co-crystallization process. The first was that INH breakdown is accelerated significantly in the presence of KatG, and this would progressively lower the effective concentration of INH during the prolonged crystallization period. The second was that O2˙̄ and other radicals generated by INH breakdown might cause damage to the protein, which would interfere with the crystallization process. In the end, crystals grown for 3 or 4 weeks did exhibit poorer diffraction, lower resolution, and lower INH occupancy, whereas restricting crystal growth to 2 weeks or less produced BpKatG·INH crystals with good INH occupancy and reasonable diffraction patterns. However, even these crystals did not diffract to as high a resolution as crystals of BpKatG alone.
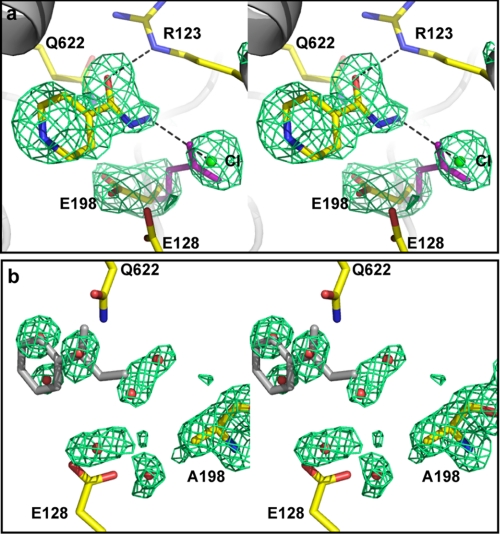
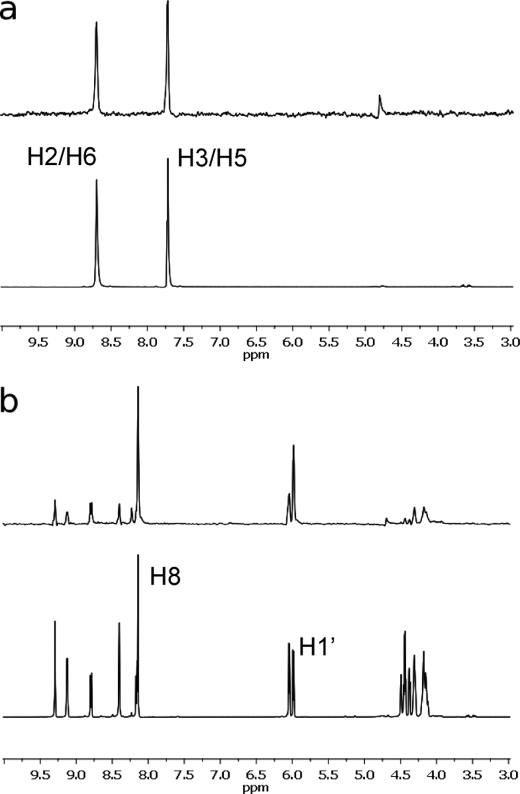
Two regions of electron density, not present in native BpKatG maps, one in each subunit adjacent to Glu198, were found that corresponded well to a molecule of INH. This is evident in the Fo − Fc maps calculated without INH in the model (Fig. 5a). For reasons which are not clear, the occupancy of INH, based on the electron density and B-factors of the INH atoms relative to nearby protein segments and overlapping waters, was invariably higher in the B subunit compared with the A subunit. These binding sites are not in the heme cavity as predicted by work with eukaryotic peroxidases (43, 44), but rather they are found at the end of a funnel shaped channel (27) on the opposite side of the subunit from the heme entrance channel. A change in the protein side chain location of Glu198, relative to the native structure, creates a cavity that is occupied by a chloride ion adjacent to which INH binds (Fig. 5a). STD spectra of INH-KatG mixtures confirmed that the protons on C2/C6 and C3/C5 make interaction with the protein (Fig. 6a), consistent with the x-ray structures, which show that these carbons are within 3.3 and 3.6 Å of the protein.
FIGURE 5.
Stereo view of an INH binding site on BpKatG in panel a and the equivalent site in its variant E198A in panel b. The Fo − Fc omit electron density maps are drawn at σ = 3.0 in green. In panel a, the map, calculated without INH, the chloride ion, or the Glu198 side chain in the model, is shown. Models of INH, the chloride ion, and the Glu198 side chain are superimposed in the figure. The solid purple side chain shows the location of Glu198 in the native structure that has not been exposed to INH or chloride ion. In panel b, the map, calculated without INH, the chloride ion, Ala198, or cavity waters in the model, is shown. INH, as bound in native BpKatG, is shown in gray. The Ala198 residue and a number of waters in red that satisfy the electron density reasonably well are superimposed. The orientations of the binding sites in the two panels is slightly different to provide the best view of the electron density maps in each case.
FIGURE 6.
STD NMR spectra of INH (a) and NAD (b) in the presence of KatG. STD NMR spectra were measured at pH 5. The corresponding reference one-dimensional NMR spectra are shown at the bottom in each section.
INH occupancy at these locations was also investigated in two variants of BpKatG. The E198A variant was constructed to investigate the effect of removing the side chain that must move for INH binding to occur. The variant crystallized well both in the presence and absence of INH, and the resulting crystals diffracted to 1.6 and 1.7 Å, without and with INH, respectively (Table 3). The resulting maps clearly show the presence of Ala at position 198, but lack any electron density that could be assigned as INH in the vicinity (Fig. 5b). Clearly, the E198A mutation prevents stable binding of INH at this location, although this is accompanied by only small decreases in the rates of radical formation and IN·NAD synthesis (Table 1) and no significant change in the Km for INH.
The crystal structure of the S324T variant has been published (22) and subsequently confirmed to be similar in most respects to the structure of the S315T variant of MtKatG (7). Crystallization of the S324T variant in the presence of INH produced crystals that diffracted to 2.0 Å, a somewhat lower resolution than crystals grown in the absence of INH, but there were regions of strong electron density near Glu198 at the same location and in the same orientation as in the native crystals that refined very well as INH. Surprisingly, the occupancy was 100% in both subunits, even subunit A. This is consistent with the slower rates of IN·NAD synthesis and INH oxidation in S324T, which would preserve a higher INH concentration during crystallization. Significantly, the Km for INH was unchanged in the variant compared with the native enzyme suggesting that there was no change in affinity for INH.
The small effect of the E198A variant on both IN·NAD synthesis and radical formation despite the significant change in binding led to a search for possible additional sites of INH binding. No evidence for binding as strong as at the Glu198 site was found, but there were changes in a region of electron density, compared with the native protein, near Arg730. The occupancy and shape of the density varied among different crystals making it difficult to fit INH into the density, and interpretation is further complicated by the side chain of Arg730 occupying the putative INH site prior to INH addition. Although this putative binding site is clearly weaker than the Glu198 site, it does provide some evidence that there may be more than one site where INH interacts with KatG.
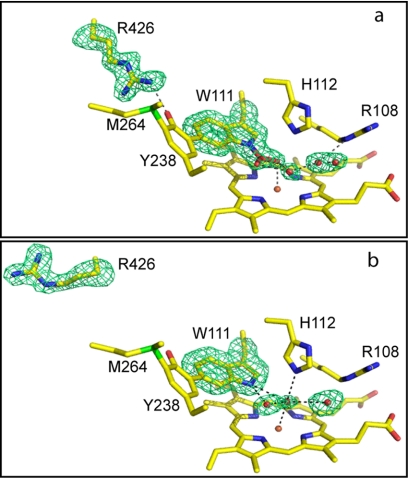
In reviewing the remainder of the protein structure in crystals grown in the presence of INH, two very striking changes in electron density were found compared with crystals grown in the absence of INH: one in the heme cavity and one involving the mobile Arg426. In the heme cavity, the perhydroxy modification on the distal Trp111, first observed in the S324T variant (22) and present in all crystals grown in the current work without INH present, is missing. Associated with the loss of the perhydroxy modification is a change in location of the mobile side chain of Arg426 from being >80% associated with Tyr238 of the M-Y-W adduct (Y conformation) in the absence of INH to >90% in the R conformation away from Tyr238 in the presence of INH (Fig. 7). The two modifications are consistent with a change in the electronic environment of the heme and the adduct consistent with the UV-visible absorbance spectra, and they have been associated with NADH oxidation that produces O2˙̄ (the perhydroxy modification) (45) and with a molecular switch modulating the catalytic process (Arg426 movement) (46).
FIGURE 7.
Comparison of the heme environment in crystals grown in the absence (a) and presence (b) of INH. The Fo − Fc electron density maps calculated using a model lacking Trp111 without (a) and with (b) the perhydroxy modification, Arg426 (both a and b), and the indicated waters are drawn at σ = 3.0 in green. A model containing the omitted residues and waters is superimposed.
The INH binding site near Glu198 is remote from the heme cavity creating the necessity for an electron transfer route to the heme where O2˙̄ is produced. Insight is provided by three earlier observations. First, Trp139, Trp153, and Trp330 have been identified as sites where stable oxoferryl-Trp• intermediates are formed in the peroxidase reaction of BpKatG (29). Second, two separate electron transfer pathways linking Trp139 (via Trp111) and Trp153 (via Trp94 and Trp95), respectively, with the heme and Trp330 have been defined (29). Third, INH reduces Trp radicals remote from the heme cavity generated by PAA treatment of MtKatG (8). Exchange of Trp111 in the W111F variant removes a residue that is part of one of the putative electron transfer paths and also an important residue in the heme cavity, and causes a reduction in the rate of IN·NAD synthesis to 50% of the native rate (Table 1). Therefore, single, double, and triple combinations of variants at the other key positions, including W139F, W153F, W202F (5 Å from INH bound near Glu198), and W330F, were examined. None of these variants exhibited changed catalase or peroxidase activities compared with wild-type BpKatG. However, although the single variants exhibited only small, if any, reduction in rates, all of the multiple variants exhibited rates of IN·NAD synthesis up to 55% slower than the native rate. This implies the existence of multiple routes for electron transfer, all of which must be blocked before an effect is seen. In fact, the situation is more complex, because only small reductions in the rate of O2˙̄ generation were evident in these variants, whereas the rate of production of other radicals increased and the dependence of IN·NAD synthesis on O2˙̄ decreased. In other words, changing residues that would be expected to support electron transfer from INH to the heme for O2˙̄ formation affect the reaction in ways that can be only partially explained.
Identification of a Binding Site for NAD+ on BpKatG
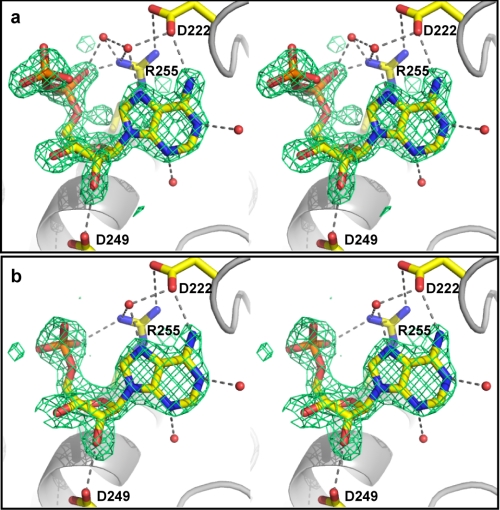
Co-crystallization was also successful in producing crystals of a BpKatG·NAD+ complex that diffracted to between 1.7 and 1.9 Å (Table 3). Electron density maps revealed a region of density not present in the native BpKatG maps ∼20 Å from the entrance to the heme channel (Fig. 8). The strongest region of this density refined very well as ADP, whereas a nearby weaker region could not be reliably refined, but is in a location consistent with a more weakly bound and disordered nicotinamide portion. The region of the Fo − Fc omit map calculated without ADP in the model satisfies the structure of ADP when superimposed (Fig. 8a). Strong interaction with the adenosine moiety was confirmed in a BpKatG·AMP crystal (Table 3) where the Fo − Fc omit maps calculated without AMP overlapped perfectly with the ADP portion of NAD+, differing only in a 60° rotation of the oxygens on the α-phosphate (Fig. 8b). The location of this site relative to the heme cavity and the Glu198 INH binding site is illustrated in Fig. 9. A matrix of interactions define the binding site of the AMP-derived nucleotides, including the carboxylates of Asp222 and Asp249, the guanidinium group of Arg255 and waters associated with each of N1, N3, and N7 of the adenine ring. Surprisingly, variants of these residues, either singly or in combination, did not affect the rate of IN·NAD synthesis or change the Km for NAD+, and AMP did not act as a competitive inhibitor for IN·NAD synthesis.
FIGURE 8.
Stereo view of the NAD+ (a) and AMP (b) binding sites on BpKatG. The Fo − Fc omit electron density maps calculated using a model lacking NAD+ or AMP are drawn at σ = 3.0 in green. A model of ADP is superimposed in panel a, and a model of AMP is superimposed in panel b. The nicotinamide portion of NAD+ in panel a appears to be disordered and has not been included. However, the presence of the second phosphate group of the NAD+ pyrophosphate is clearly visible.
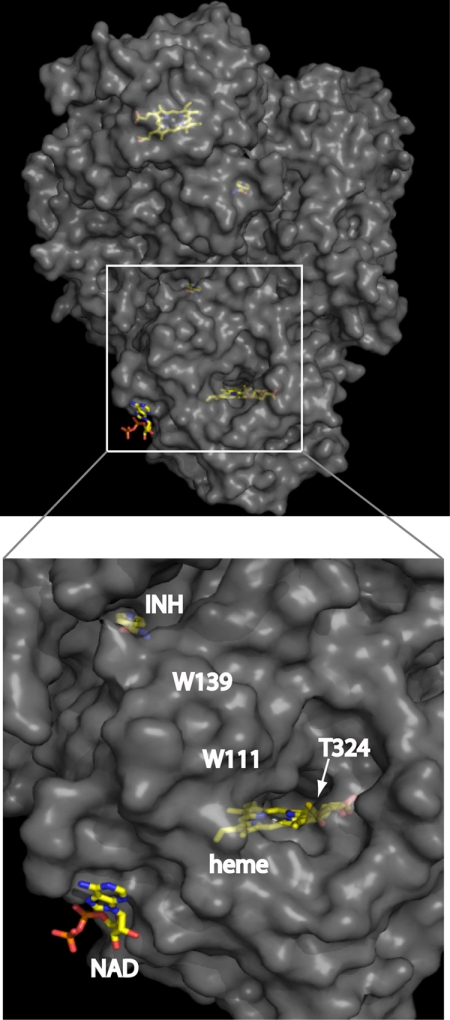
FIGURE 9.
View of KatG showing the INH and NAD+ binding sites in relation to the heme cavity. The complete KatG dimer in the upper panel contains the locations of the hemes, NAD+, and INH and is oriented so as to view the lower heme through the main entrance channel. The portion in the box is enlarged in the lower panel to reveal the orientation of the binding sites in more detail. The surface is rendered with partial transparency to reveal the buried heme and the INH on the opposite side of the protein. The approximate locations of Trp111 (W111) and Trp139 (W139) involved in electron transfer are indicated.
Interactions in solution, similar to those seen in the crystal structure, were suggested by STD spectra of KatG/NAD+ and KatG/AMP mixtures, which reveal weak interactions involving mainly two hydrogen atoms, one on C8 of the adenine ring and one on C1′ of the adenosine ribose, that exhibit stronger STD signals than the rest (Fig. 6b). This is consistent with C1′ being situated 3.5 Å from the methyl group of Ala252 and with C8 being situated 3.5 Å from the Arg255 NH1 and 3.7 Å from Cδ of Arg255. The high occupancy observed for this NAD+ binding site might have been enhanced by the proximity in the crystal of a neighbor molecule, which forms an extra hydrogen bond with the adenine ribose and reduces protein flexibility (as suggested by the reduced temperature factors).
DISCUSSION
Understanding the process of IN·NAD synthesis has been complicated by difficulties in rationalizing the role of superoxide within a peroxidatic reaction cycle. Identification of binding sites for INH and NAD+ far from the heme cavity of KatG introduces further complexity into the picture and has prompted the more detailed analysis of the reaction parameters described here. The study concludes that O2˙̄ is normally an essential component in the KatG-catalyzed synthesis of IN·NAD and that the peroxidase reaction cycle enhances the reaction. Furthermore, a clear definition of the roles of the superoxide and peroxidase systems within a complex medley of enzymatic and non-enzymatic reactions is presented, and two roles for O2˙̄ are defined. The first lies in the initiation and feeding of the peroxidase process that enhances INH cleavage into IN•, and the second lies in the reduction of the final intermediate, IN·NAD•+.
The fact that SOD is so efficient in stopping the reaction implies that the O2˙̄ required in the reaction is, at some point, free in the medium prior to its participation. This is probably a reflection of the relatively slow turnover rate (∼3 per min) that provides ample opportunity for dissociation and re-association. The proposed scheme in Fig. 2 details the roles of O2˙̄ and outlines how the peroxidase cycle enhances the processes of INH breakdown and IN·NAD synthesis. Significantly, it rationalizes the data in terms of known reaction intermediates.
KatG production of O2˙̄ using electrons from INH to reduce O2 may involve a transient Fe2+ species (17), but the INH-induced absorbance spectrum shows that such a species does not accumulate. Indeed the spectral changes do not suggest the accumulation of a new intermediate species and are more consistent with a simple change in the electronic environment of the heme. This is consistent with the structural changes observed in the heme cavity of the BpKatG·INH complex.
The inhibition by EDTA of all radical formation from INH breakdown and of IN·NAD synthesis shows that Mn2+ plays a key role both with and without KatG present. KatG does enhance the rate of radical production compared with the purely Mn2+-catalyzed reaction, but it also changes the process such that the proportion of radicals that are O2˙̄ decreases from >90% with Mn2+ to <40% with KatG, suggesting an increase in IN• accumulation. That is, there are two different reaction paths depending on whether or not KatG is present. In the absence of KatG, most of the radical is O2˙̄, but in the presence of KatG more IN• accumulates. This may be explained by the stabilization of IN• when bound to KatG and also by INH interaction with KatG creating a second pathway for IN• generation.
The reduction of O2˙̄ to H2O2 is favorable (E′° = +0.89 v) and occurs as part of the coupled oxidase-peroxidase reaction of peroxidases (47). The formation of H2O2 then provides the substrate to activate the peroxidase cycle creating two one-electron reduction opportunities with a much faster turnover rate, thereby enhancing the rate of INH breakdown. In this way, IN• is produced as part of both the superoxide cycle and the peroxidase cycle. The enhancement of INH breakdown provided by the peroxidase cycle is evident in the 60% slower rate of radical production by variant H112A where the peroxidase cycle does not function. It should also be noted that the S315T variant exhibits a similar reduction in the rate of IN·NAD synthesis, which is sufficient to impart the INH resistance phenotype in vivo. It follows therefore that absence of the peroxidase capability would also impart INH resistance even if it is not essential for IN·NAD formation.
The fate of IN• radicals depends on their location of formation relative to the NAD+ binding site. If remote from the NAD+ site, as suggested by the strongest INH and NAD+ binding sites identified (Fig. 9), it would have to dissociate and diffuse to the NAD+ site. Alternatively, if IN• is formed in close proximity to the NAD+, such as at the putative site near Arg730, it would be immediately adjacent to NAD+ and may react rapidly on formation precluding any need for dissociation.
Identification of the IN·NAD·TEMPO complex confirms that IN·NAD+• is an intermediate in the synthesis of IN·NAD. It is not possible to say whether it was formed in solution or on the protein followed by dissociation before capture by TEMPO. Most significantly, however, its presence confirms the need for a reduction step to convert IN·NAD•+ to IN·NAD, and this provides a second role for O2˙̄. The reduction of IN·NAD+• by O2˙̄ is a favorable reaction based on the standard reduction potentials of −0.33 and +0.3 V for the [O2/O2˙̄] and [H+, NAD•/NADH] half-cell reactions, respectively (48). When the basal system is perturbed to allow some IN·NAD synthesis in the presence of SOD, other radical species must replace O2˙̄ for this final reduction step.
Because the two INH binding sites identified in this work are remote from the heme and yet both the superoxide and peroxidase cycles occur at the heme, there must be paths in the protein for electron passage to the heme. This conclusion is consistent with EPR results that revealed INH reduction of Trp radicals remote from the heme cavity in MtKatG (8) and with the identification of two separate electron transfer pathways involving stable Trp• radicals on Trp139 (via Trp111 to the heme) and Trp153 (via Trp95 to the heme) (29). In fact, electron transfer must involve more routes than just these two, because eliminating both paths by mutating the key residues reduced KatG-catalyzed IN·NAD synthesis by only 55%. This may also imply the existence of more INH binding sites than just those identified in the crystal complexes.
The absence of any change in IN·NAD synthesis caused by mutations in the INH and NAD+ binding sites identified in the crystal structure may have several origins. The most obvious is that these sites do not have any physiological significance and are simply an artifact of crystal growth. Alternatively, the existence of multiple INH binding sites, as suggested by the electron density maps and by the partial impairment of IN·NAD synthesis upon mutation of internal tryptophan residues, would require the mutation of multiple sites to generate a significant physiological effect. These alternative sites must be weaker than those found in the crystal structure, and such an example lies in the putative site near Arg730. The binding of INH in the heme cavity with the hydrazine -NH2 within 4.5 Å of the heme iron (49) must be similarly weak, because the Km for INH in the S324T variant, in which access to the heme cavity is hampered, is unchanged from the native enzyme.
A major challenge in rationalizing the role of KatG in INH activation is to provide an explanation for INH resistance caused by the mutation of Ser315 to Thr in MtKatG. The only structural change in the variant is a narrowing of the channel leading to the heme cavity that even reduces water occupancy (7, 22). However, the remoteness of INH binding from the heme cavity and absence of a change in Km for INH in the S324T variant seems to preclude the single explanation that the narrower channel restricts entry of INH. An alternative explanation lies in the need for O2 and O2˙̄ to diffuse in and out of the heme pocket. In addition to physical hindrance to movement presented by the narrower channel, the O2˙̄ would be forced to pass closer to the negatively charged carboxylate of Asp141 situated in the heme channel (50). Evidence of 60% slower radical generation, including O2˙̄ formation in the S324T variant of BpKatG compared with the native enzyme (22), has been confirmed for the S315T variant of MtKatG. In addition, INH occupancy in the S324T·INH complex crystal is higher in both sites compared with the native complex, consistent with the slower turnover of INH.
One caveat that must be placed on this study is that it has been carried out in vitro, and it could be argued that the medley of reactions leading to IN·NAD might be different inside M. tuberculosis. However, evidence suggests that there may not be significant differences. The fundamental features, including the requirement for O2 and O2˙̄ (12) and the involvement of the peroxidase cycle (7) are unchanged, and the slower rate of IN·NAD synthesis by the S315T variant in vitro correlates well with the INH resistance phenotype of M. tuberculosis katG mutants harboring this change. The S315T variant is also instructive in revealing that it is not necessary to completely inhibit IN·NAD synthesis to impart the INH resistance phenotype; slowing the reaction by 50% is sufficient.
In summary, INH breakdown is coupled to the reduction of O2 to O2˙̄ in both Mn2+- and KatG-catalyzed reactions. Most of the O2˙̄ is either created in or diffuses into the medium at some point and is required in two key roles. The first is as a source of H2O2 to activate the peroxidase cycle and enhance the rate of INH oxidation, and the second is as a reductant of the final intermediate IN·NAD•+. Existence of an INH binding site near Glu198 necessitates the transfer of electrons to the heme cavity, which occurs, in part, via previously identified paths involving Trp139 and Trp153, but also other undefined routes. The modified electronic environment of the heme suggested by the UV-visible spectra of KatG in the presence of INH and by the movement of the mobile Arg426 and loss of the perhydroxy modification on Trp111 in BpKatG·INH complexes is consistent with the superoxide cycle involving O2 and O2˙̄ associated with, but not actually covalently linked to the heme iron.
Acknowledgments
We thank V. Spicer and V. Collado for technical assistance, and K. Standing and W. Ens for continued unrestricted access to the instruments in the Time-of-Flight Laboratory, Dept. of Physics and Astronomy, University of Manitoba.
This work was supported by a Discovery Grant 9600 from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to P. C. L.) and by the Canada Research Chair Program (to P. C. L.). The diffraction data were collected at the Canadian Light Source, which is supported by NSERC, National Research Council, Canadian Institutes for Health Research, and the University of Saskatchewan. This work was supported by Grants BFU2005-08686-C02-01 (to I. F.), BIO2007-63458 (to M. P.), both from the Spanish Ministry of Science and Innovation, and SGR2009-1352 (to M. P.) from the Generalitat de Catalunya.
- INH
- isonicotinic acid hydrazide or isoniazid
- PAA
- peroxyacetic acid
- tBHP
- t-butyl hydroperoxide
- SOD
- superoxide dismutase
- TEMPO
- 2,2,6,6-tetramethylpiperidine-1-oxyl
- MtKatG
- catalase-peroxidase KatG of M. tuberculosis
- STD
- saturation transfer difference
- BpKatG
- KatG from B. pseudomallei
- InhA
- long-chain enoyl acyl carrier protein reductase
- IN·NAD
- isonicotinyl-NAD.
REFERENCES
- 1.Zhang Y., Heym B., Allen B., Young D., Cole S. (1992) Nature 358, 591–593 [DOI] [PubMed] [Google Scholar]
- 2.Rozwarski D. A., Grant G. A., Bartom D. H., Jacobs W. R., Jr., Sacchettini J. C. (1998) Science 279, 98–102 [DOI] [PubMed] [Google Scholar]
- 3.Chouchane S., Lippai I., Magliozzo R. S. (2000) Biochemistry 39, 9975–9983 [DOI] [PubMed] [Google Scholar]
- 4.Ghiladi R. A., Cabelli D. E., Ortiz de Montellano P. R. (2004) J. Am. Chem. Soc. 126, 4772–4773 [DOI] [PubMed] [Google Scholar]
- 5.Ghiladi R. A., Medzihradszky K. F., Rusnak F. M., Ortiz de Montellano P. R. (2005) J. Am. Chem. Soc. 127, 13428–13442 [DOI] [PubMed] [Google Scholar]
- 6.Cade C. E., Dlouhy A. C., Medzihradszky K. F., Salas-Castillo S. P., Ghiladi R. A. (2010) Prot. Sci. 19, 458–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X., Yu H., Yu S., Wang F., Sacchettini J. C., Magliozzo R. S. (2006) Biochemistry 45, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 8.Singh R., Switala J., Loewen P. C., Ivancich A. (2007) J. Am. Chem. Soc. 129, 15954–15963 [DOI] [PubMed] [Google Scholar]
- 9.Wengenack N. L., Uhl J. R., Amand A. L., Tomlinson A. J., Benson L. M., Naylor S., Kline B. C., Cockerill F. R., III, Rusnak F. (1997) J. Infect. Dis. 176, 722–727 [DOI] [PubMed] [Google Scholar]
- 10.Pierattelli R., Banci L., Eady N. A., Bodiquel J., Jones J. N., Moody P. C., Raven E. L., Jamart-Grégoire B., Brown K. A. (2004) J. Biol. Chem. 279, 39000–39009 [DOI] [PubMed] [Google Scholar]
- 11.Wengenack N. L., Hoard H. M., Rusnak F. (1999) J. Am. Chem. Soc. 121, 9748–9749 [Google Scholar]
- 12.Lei B., Wei C. J., Tu S. C. (2000) J. Biol. Chem. 275, 2520–2526 [DOI] [PubMed] [Google Scholar]
- 13.Vilchèze C., Morbidoni H. R., Weisbrod T. R., Iwamoto H., Kuo M., Sacchettini J. C., Jacobs W. R., Jr. (2000) J. Bacteriol. 182, 4059–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zabinski R. F., Blanchard J. S. (1997) J. Am. Chem. Soc. 119, 2331–2332 [Google Scholar]
- 15.Singh R., Wiseman B., Deemagarn T., Donald L. J., Duckworth H. W., Carpena X., Fita I., Loewen P. C. (2004) J. Biol. Chem. 279, 43098–43106 [DOI] [PubMed] [Google Scholar]
- 16.Singh R., Wiseman B., Deemagarn T., Jha V., Switala J., Loewen P. C. (2008) Arch. Biochem. Biophys. 471, 207–214 [DOI] [PubMed] [Google Scholar]
- 17.Magliozzo R. S., Marcinkeviciene J. A. (1996) J. Am. Chem. Soc. 118, 11303–11304 [Google Scholar]
- 18.Wang J. Y., Burger R. M., Drlica K. (1998) Antimicrob. Agents Chemother. 42, 709–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoeb H. A., Bowman B. U., Jr., Ottolenghi A. C., Merola A. J. (1985) Antimicrob. Agents Chemother. 27, 404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoeb H. A., Bowman B. U., Jr., Ottolenghi A. C., Merola A. J. (1985) Antimicrob. Agents Chemother. 27, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoeb H. A., Bowman B. U., Jr., Ottolenghi A. C., Merola A. J. (1985) Antimicrob. Agents Chemother. 27, 408–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deemagarn T., Carpena X., Singh R., Wiseman B., Fita I., Loewen P. C. (2005) J. Mol. Biol. 345, 21–28 [DOI] [PubMed] [Google Scholar]
- 23.Yamada Y., Saijo S., Sato T., Igarashi N., Usui H., Fujiwara T., Tanaka N. (2001) Acta Crystallog. D57, 1157–1158 [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y., Fujiwara T., Sato T., Igarashi N., Tanaka N. (2002) Nature Struct. Biol. 9, 691–695 [DOI] [PubMed] [Google Scholar]
- 25.Wada K., Tada T., Nakamura Y., Kinoshita T., Tamoi M., Shigeoka S., Nishimura K. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 157–159 [DOI] [PubMed] [Google Scholar]
- 26.Carpena X., Switala J., Loprasert S., Mongkolsuk S., Fita I., Loewen P. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 2184–2186 [DOI] [PubMed] [Google Scholar]
- 27.Carpena X., Loprasert S., Mongkolsuk S., Switala J., Loewen P. C., Fita I. (2003) J. Mol. Biol. 327, 475–489 [DOI] [PubMed] [Google Scholar]
- 28.Bertrand T., Eady N. A., Jones J. N., Jesmin Nagy J. M., Jamart-Grégoire B., Raven E. L., Brown K. A. (2004) J. Biol. Chem. 279, 38991–38999 [DOI] [PubMed] [Google Scholar]
- 29.Colin J., Wiseman B., Switala J., Loewen P. C., Ivancich A. (2009) J. Am. Chem. Soc. 131, 8557–8563 [DOI] [PubMed] [Google Scholar]
- 30.Kunkel T. A., Roberts J. D., Zakour R. A. (1987) Methods Enzymol. 154, 367–382 [DOI] [PubMed] [Google Scholar]
- 31.Sanger F., Nicklen S., Coulson A. R. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auclair C., Torres M., Hakim J. (1978) FEBS Lett. 89, 26–28 [DOI] [PubMed] [Google Scholar]
- 33.Rawat R., Whitty A., Tonge P. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13881–13886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Layne E. (1957) Methods Enzymol. 3, 447–454 [Google Scholar]
- 35.Mayer M., Meyer B. (1999) Angew. Chem. Int. Ed. 38, 1784–1788 [DOI] [PubMed] [Google Scholar]
- 36.Klein J., Meinecke R., Mayer M., Meyer B. (1999) J. Am. Chem. Soc. 121, 5336–5337 [Google Scholar]
- 37.Loboda A. V., Krutchinsky A. N., Bromirski M., Ens W., Standing K. G. (2000) Rapid Commun. Mass Spectrom. 14, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 39.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 41.Wengenack N., Rusnak F. (2001) Biochemistry 40, 8990–8996 [DOI] [PubMed] [Google Scholar]
- 42.Amos R. I., Gourlay B. S., Schiesser C. H., Smith J. A., Yates B. F. (2008) Chem. Commun. 1695–1697 [DOI] [PubMed] [Google Scholar]
- 43.Metcalfe C., Macdonald I. K., Murphy E. J., Brown K. A., Raven E. L., Moody P. C. E. (2008) J. Biol. Chem. 283, 6193–6200 [DOI] [PubMed] [Google Scholar]
- 44.Singh A. K., Kumar R. P., Pandey N., Singh N., Sinha M., Bhushan A., Kaur P., Sharma S., Singh T. P. (2010) J. Biol. Chem. 285, 1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carpena X., Wiseman B., Deemagarn T., Herguedas B., Ivancich A., Singh R., Loewen P. C., Fita I. (2006) Biochemistry 45, 5171–5179 [DOI] [PubMed] [Google Scholar]
- 46.Carpena X., Wiseman B., Deemagarn T., Singh R., Switala J., Ivancich A., Fita I., Loewen P. C. (2005) EMBO Rep. 6, 1156–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunford H. B. (1999) Heme Peroxidases, pp 97–101, Wiley-VCH, New York [Google Scholar]
- 48.Farrington J. A., Land E. J., Swallow A. J. (1980) Biochim. Biophys. Acta 590, 273–276 [DOI] [PubMed] [Google Scholar]
- 49.Todorović S., Juranić N., Macura S., Rusnak F. (1999) J. Am. Chem. Soc. 121, 10962–10966 [Google Scholar]
- 50.Deemagarn T., Wiseman B., Carpena X., Ivancich A., Fita I., Loewen P. C. (2007) Proteins 66, 219–228 [DOI] [PubMed] [Google Scholar]