Abstract
Oxalacetate acetylhydrolase (OAH), a member of the phosphoenolpyruvate mutase/isocitrate lyase superfamily, catalyzes the hydrolysis of oxalacetate to oxalic acid and acetate. This study shows that knock-out of the oah gene in Cryphonectria parasitica, the chestnut blight fungus, reduces the ability of the fungus to form cankers on chestnut trees, suggesting that OAH plays a key role in virulence. OAH was produced in Escherichia coli and purified, and its catalytic rates were determined. Oxalacetate is the main OAH substrate, but the enzyme also acts as a lyase of (2R,3S)-dimethyl malate with ∼1000-fold lower efficacy. The crystal structure of OAH was determined alone, in complex with a mechanism-based inhibitor, 3,3-difluorooxalacetate (DFOA), and in complex with the reaction product, oxalate, to a resolution limit of 1.30, 1.55, and 1.65 Å, respectively. OAH assembles into a dimer of dimers with each subunit exhibiting an (α/β)8 barrel fold and each pair swapping the 8th α-helix. An active site “gating loop” exhibits conformational disorder in the ligand-free structure. To obtain the structures of the OAH·ligand complexes, the ligand-free OAH crystals were soaked briefly with DFOA or oxalacetate. DFOA binding leads to ordering of the gating loop in a conformation that sequesters the ligand from the solvent. DFOA binds in a gem-diol form analogous to the oxalacetate intermediate/transition state. Oxalate binds in a planar conformation, but the gating loop is largely disordered. Comparison between the OAH structure and that of the closely related enzyme, 2,3-dimethylmalate lyase, suggests potential determinants of substrate preference.
Keywords: Enzyme Catalysis, Enzyme Structure, Fungi, Gene Knock-out, X-ray Crystallography, Chestnut Blight, Oxalic Acid, Oxaloacetate Hydrolase, Virulence
Introduction
Oxalacetate acetylhydrolase (OAH,2 EC 3.7.1.1) catalyzes the hydrolysis of oxalacetate to oxalic acid and acetate (Fig. 1) (1). In fungi causing human and plant diseases, oxalic acid production and secretion are closely associated with fungal pathogenesis and virulence (2–5). The virulence mechanism is believed to be due to acidification that triggers lignocellulose degradation, reduces viability of host tissue in favor of pathogen proliferation, and induces crystallization of calcium oxalate, which blocks vessels or bronchioles (4, 6, 7). Studies of the human pathogen Aspergillus fumigatus and the plant pathogen Botrytis cinerea showed that oxalic acid secretion is solely due to hydrolysis of oxalacetate catalyzed by OAH (1).
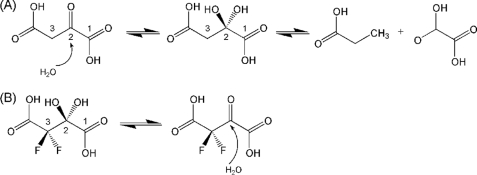
FIGURE 1.
A, OAH-catalyzed reaction involves a gem-diol intermediate/transition state. B, nonhydrolyzable inhibitor, 3,3-difluorooxalacetate, exists in two structural forms, a gem-diol (the major form in aqueous solution) and a ketone (minor form).
OAH is a member of the phosphoenolpyruvate mutase (PEPM)/isocitrate lyase (ICL) superfamily. Members of this superfamily act on α-oxycarboxylate substrates to cleave/form C–C or P–C bonds. All PEPM/ICL superfamily members are oligomeric proteins, mostly tetramers, and each subunit adopts an (α/β)8 barrel fold. The tetramer is composed of a dimer of dimers arranged such that the two molecules within a dimer swap the 8th helix of the α/β barrel. The active site is located at the C-terminal side of the β-barrel. In most PEPM/ICL superfamily members, although not all, a mobile loop controls access to the active site. Upon substrate binding, this gating loop closes to sequester the catalytic site from bulk solvent. All PEPM/ICL superfamily members require Mg2+ or Mn2+ for activity, and the metal mediates the interactions between the protein and the substrate (8–19).
Phylogeny analysis of the PEPM/ICL superfamily highlighted an intriguing clade that contains enzymes with different catalytic activities, in contrast to other phylogeny branches, which include enzymes that perform a single catalytic activity (13). This cluster includes the Petal Death Protein (PDP), shown to be a lyase that cleaves α-keto and α-hydroxycarboxylic acids with broad substrate specificity (20), carboxy-PEPM (21), 2,3-dimethylmalate lyase (DMML) (14), and OAH (1). The carboxy-PEPM exhibits an unusually low catalytic rate toward its substrate, and as of this writing no additional substrates have been discovered (21). The remaining three enzymes, PDP, DMML, and OAH, whose amino acid sequences are aligned in Fig. 2, exhibit varying levels of substrate promiscuity except that they do not act on carboxy-PEP. PDP possesses the broadest substrate profile and catalyzes both the cleavage of (2R)-ethyl-(3S)-methylmalate and the hydrolysis of oxalacetate with comparable efficacies (kcat/Km = 2 × 104 for both) (20). From the perspective of function evolution and amino acid sequence homology, DMML and OAH are striking examples of enzymes with overlapping catalytic activities and at the same time substrate specialization. The Aspergillus niger genome encodes both DMML and OAH, which share 60% sequence identity, yet the two enzymes perform distinct functions (14). The A. niger DMML catalyzes the cleavage of its primary substrate, (2R,3S)-dimethylmalate with kcat/Km = 9 × 104 m−1 s−1 and the hydrolysis of oxalacetate at nearly 50-fold lower efficacy of kcat/Km = 2 × 103 m−1s−1(14). The A. niger OAH was not produced in soluble form, but the closely related B. cinerea OAH catalyzes the hydrolysis of its primary substrate, oxalacetate, with kcat/Km = 1.5 × 105 m−1 s−1 and the cleavage of (2R,3S)-dimethylmalate with 750-fold lower efficacy of kcat/Km = 2 × 102 m−1 s−1 (1).
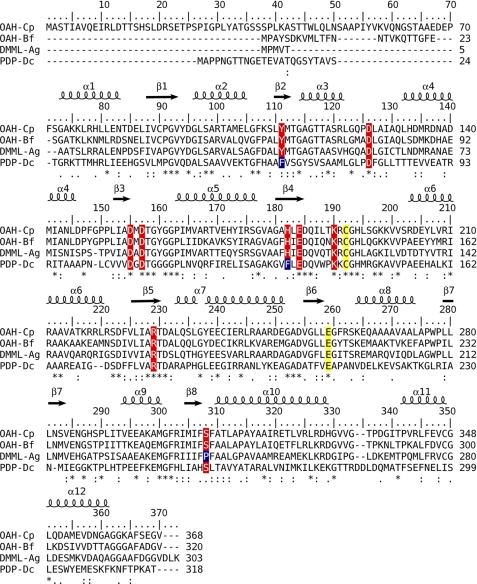
FIGURE 2.
Structure-based amino acid sequence alignment of enzymes belonging to the phylogeny clade of the PEPM/ICL superfamily whose members exhibit substrate promiscuity. Mg2+/Mn2+ coordinating residues are highlighted in red, as are several other key active site residues, except the catalytic Cys and Glu are highlighted in yellow. The α-helical and β-strand secondary structure units, as defined in the reported OAH structure, are depicted above the sequence blocks. Abbreviations uses are as follows: Cp, C. parasitica (GenBankTM accession number GU932672); Bf, B. cinerea; Ag, A. niger; Dc, Dianthus caryophyllus (carnation, clover pink).
Cryphonectria parasitica is a fungus-causing blight of chestnut trees (genus Castanea). The introduction of this fungus into North America and Europe from Asia resulted in the destruction of billions of mature American and European chestnut trees during the 20th century (22). Infection of C. parasitica by RNA hypoviruses results in virulence attenuation, providing the basis for biological control (23). Hypovirus infection also results in other phenotypic changes, including the loss of oxalic acid production (24). A strategy to reduce the pathogenicity of C. parasitica by transforming chestnut trees with the oxalate-degrading enzyme, oxalate oxidase, was proposed based on studies that showed that transgenic American chestnut callus tissue expressing oxalate oxidase protected the tissue from loss of lignin (25).
The C. parasitica genome contains only a single gene that is likely to encode OAH. The sequence exhibits 70% identity to the sequence of OAH from B. cinerea. Thus, we have undertaken to characterize the function of the enzyme in chestnut tree canker infection, its in vitro catalytic rates, and its crystal structure in the ligand-free state and bound to the oxalacetate hydrolysis product, oxalate, and to a mechanism-based inhibitor, 3,3-difluorooxalacetate (DFOA), an analog of the oxalacetate intermediate/transition state (Fig. 1B).
MATERIALS AND METHODS
Cloning, Production, and Purification of Recombinant C. parasitica OAH
The full-length oah gene from C. parasitica EP155 and several gene truncation versions were cloned to facilitate soluble production of active enzyme and crystal growth. The N-terminal truncated oah gene that was ultimately pursued (amino acid residues 66–368) was amplified by PCR with a forward primer ATGGTTCCCATATGGCGGAAGATGAGCCCTTCTCA containing an NdeI site and a reverse primer GGAAAGATCTTTAGACACCCTCCGAAAATGC containing a BglII restriction site. The template was a cDNA clone containing the full-length oah. The PCR product was digested by NdeI and BglII and ligated to the NdeI and BamHI sites of pET28a. The sequence was verified by DNA sequencing. The recombinant plasmid was used to transform Escherichia coli (Rosetta 2 DE3, Novagen), and the cells were grown overnight at 37 °C in Luria Broth (LB) containing 50 μm kanamycin and 50 μm chloramphenicol with vigorous agitations (250 rpm). Next, 50 ml of the culture, diluted into 1.0 liter of LB medium containing antibiotics, were grown until reaching A600 nm 0.6–1.0, when induction was initiated by adding isopropyl d-thiogalactopyranoside to a final concentration of 0.1 mm, and the temperature was decreased to 20 °C. After 20–24 h, the cells were harvested by centrifugation at 7000 rpm, and the pellet was resuspended in buffer containing 20 mm Tris-HCl (pH 8.0), 0.5 m NaCl, 5 mm MgCl2, and 5 mm imidazole. Protease inhibitor mixture (Sigma) was added to prevent degradation. The cells were disrupted by sonication, and the supernatant was harvested by ultracentrifugation at 48,000 rpm for 1 h.
The supernatant was applied onto a nickel-nitrilotriacetic acid column (Qiagen); the bound protein was washed with 20 mm imidazole-containing buffer and then eluted with 250 mm imidazole-containing buffer. The protein solution was incubated overnight at 4 °C with thrombin (∼1:1000 OAH/thrombin molar ratio) to cleave the His6 tag. Thrombin and uncleaved OAH were removed by passing the solution though a column containing nickel-nitrilotriacetic acid and benzamidine resin (GE Healthcare). The protein was further purified by size exclusion chromatography on a Superdex-200 10/300 GL column (GE Healthcare) with buffer A (10 mm Na+HEPES (pH 7.4), 50 mm NaCl, 5 mm MgCl2, and 1 mm DTT). The purified protein was concentrated to 120 mg/ml with a Centricon spin concentrator. The purity was judged to be better than 95% by SDS-PAGE showing an apparent molecular mass of ∼33 kDa (the calculated molecular mass is 33,063 Da). The protein concentration was determined by UV280 nm absorption (Δϵ = 15,930 m−1 cm−1) indicating a yield of 40 mg/liter of E. coli culture. The native molecular mass of the recombinant OAH was estimated to be ∼120 kDa based on the elution from the Superdex-200 column, consistent with the previous observation that OAH from B. cinerea forms tetramers (1).
Disruption of the C. parasitica oah Gene
Disruption of the C. parasitica oah gene was performed using the PCR-based strategy described by Davidson et al. (26) to produce a gene disruption cassette. The coding region extending from Ala-74 to Ala-179 was replaced with a 1.5-kb fragment containing the neomycin phosphotransferase gene regulated by the Cochliobolus heterostrophus gpd promoter and the Neurospora crassa β-tubulin terminator. The 5′- and 3′- terminal portions of the oah gene were individually amplified with primer pair 1 (OAH-KF1, TCAGGGATTATAGAGCAGGG; OAH-KR1, TTCGCCCTATCCAACATGGTGATCCTGAGAAGGGCTCATCTT) and primer pair 2 (OAH-KF2, AGTGTCTACTGCTGGCGTCGACGAGCACATCTCGAAGATCAA; OAH-KF2, AGCTATTACCAGACACATGG), respectively, using the C. parasitica strain EP155 genomic DNA as template. The 5′-terminal fragment extended from a position 1161 nucleotides upstream of the AUG initiator codon to a position in the coding region corresponding to Ala-74. The 3′-fragment extended from the codon for Ala-174 to a position 401 nucleotides downstream of the TAA termination codon. Primers Neo-F (5′-TCACCATGTTGGATAGGGCGAA-3′) and Neo-R (5′-GTCGACGCCAGCAGTAGACACT-3′) were used to amplify the G418 resistance gene cassette present in plasmid pSK666 (pSK666 was a kind gift from Dr. Seogchan Kang, Pennsylvania State University). The portions of the primers OAH-KR1 and OAH-KF2 corresponding to Neo-F and Neo-R are underlined. The amplified products were gel-purified with the aid of the QIAquick gel extraction kit (Qiagen) according to the manufacturer's instructions. After performing an intermediate PCR to anneal these three fragments, primers OAH-KF1 and OAH-KR2 were used to produce the full-length disruption cassette with a final round of PCR amplification. This full-length disruption cassette was gel-purified and used to transform spheroplasts of C. parasitica strain cpku80 (27) according to the method of Churchill et al. (28), followed by selection on regeneration medium containing 20 μg/ml G418 antibiotic. G418-resistant transformants were screened by PCR for oah gene disruption, and single conidial isolates of two positive candidates were further characterized.
Virulence Assay of C. parasitica
Virulence assays were performed with dormant American chestnut tree stems as described previously by Hillman et al. (29), with six duplicate inoculations per fungal strain. Inoculated stems were kept at room temperature in a glass tank to maintain moisture. Cankers were measured 3 weeks after inoculation. The growth characteristics and colony morphology under standard laboratory light conditions were evaluated on PDA plate (22–24 °C, 12:12-h light cycle with intensity at 1300–1600 lux).
Enzymatic Assays
OAH activity was assayed following a published procedure (1). The reactions were carried out in 96-well microplates at 25 °C. The 100-μl reaction mixtures contained 0.1–2.0 mm oxalacetate, 0.3 μm OAH, 5 mm MgCl2 (or 5 mm MnCl2 with 0.15 μm OAH and 0.2–5.0 mm oxalacetate or 5 mm CaCl2 with 0.15 μm OAH and 0.2–2.0 mm oxalacetate), and 50 mm Na+HEPES (pH 7.4). The reaction progress was monitored at 255 nm for the disappearance of the enol tautomer of oxalacetate (Δϵ = 1.1 mm−1 cm−1). The rate of oxalacetate consumption via spontaneous decarboxylation was measured prior to initiating the enzymatic reaction and subtracted from the reaction rate measured in the presence of OAH.
The OAH (20 μm) 2,3-dimethylmalate lyase activity was measured in 96-well microplates at 25 °C as reported previously (14). The 100-μl reaction mixtures contained 5 mm MgCl2 (or MnCl2 or CaCl2), 2 units of lactate dehydrogenase, 0.2 mm NADH, and 50 mm Na+HEPES (pH 7.4). The decrease in absorbance of NADH was monitored at 340 nm (Δϵ = 6.2 mm−1 cm−1). The kinetic constants were determined with assay solutions containing 0.2–4.0 mm (2R,3S)-dimethylmalate.
The steady-state kinetic parameters (Km and kcat) were determined from the initial velocity data measured as a function of substrate concentration. The initial velocity data were fitted to Equation 1 with GraFit (30),
where [S] is the substrate concentration; V0 is the initial velocity; Vmax is the maximum velocity, and Km is the Michaelis-Menten constant for the substrate. The kcat value was calculated from Vmax, and the enzyme concentration using the equation kcat = Vmax/[E], where [E] is the protein subunit molar concentration in the reaction calculated from the ratio of measured protein concentration and the protein molecular mass.
Crystallization and Structure Determination
Crystals of the apo-OAH (10 mg/ml in buffer A) were grown by the hanging-drop method at room temperature. Drops containing a 1:1 ratio of protein to reservoir solutions were equilibrated against the reservoir solution. The reservoir solution contained 0.2 m CaCl2, 0.1 m Na+HEPES (pH 7.5), and 30% v/v polyethylene glycol (PEG) 400. Crystals appeared within a week. The crystals were flash-cooled in their original mother liquor by immersing them into liquid propane reservoir cooled by liquid nitrogen.
Diffraction data of a ligand-free OAH crystal were acquired at the Advanced Photon Source on the SER-CAT 22BM beamline equipped with a MAR MX-225 CCD detector, and the data were processed with the computer program XDS (31). Data collection statistics are provided in Table 1. Structure factors were calculated with the program TRUNCATE (32) as implemented in the CCP4 package (33). The structure was determined by molecular replacement using the program PHASER (34) with the structure of DMML as the search model (Protein Data Bank code 3FA3) (14).The gating loop, the ligand, and water molecules were excluded from the search model. The initial OAH model was automatically rebuilt using ARP/WARP (35), manually adjusted using the interactive computer graphics program XTALVIEW (36), and refined with CNS (37) and PHENIX (38). Metal ions and water molecules were assigned gradually as the refinement progressed. Individual anisotropic temperature factors and riding hydrogen atoms were included toward the end of the refinement, which led to the reduction of both the free R and working R factors. No distance or geometry restraints were applied to the metal coordination. Table 1 provides the refinement statistics.
TABLE 1.
Data collection and refinement statistics
| OAH | OAH/DFOA/Mn2+ | OAH/OA/Mn2+ | |
|---|---|---|---|
| Space group | P42212 | P42212 | P42212 |
| Cell dimension | a = b = 82.7, c = 73.8 Å | a = b = 82.1, c = 72.7 Å | a = b = 82.1, c = 72.7 Å |
| No. of molecule/asymmetric units | 1 | 1 | 1 |
| Data collection | |||
| Wavelength | 1.0000 Å | 1.5418 Å | 1.5418 Å |
| Resolutiona | 45.9-1.30 Å (1.33-1.30 Å) | 20.0-1.55 Å (1.59-1.55 Å) | 20.0-1.65 Å (1.70-1.65 Å) |
| No. of observed reflections | 809,806 | 389,690 | 333,866 |
| No. of unique reflections | 58,937 | 36,608 | 30,472 |
| Completenessa | 92.8% (50.7%) | 99.9% (99.9%) | 99.8% (99.8%) |
| Rmergea | 0.044 (0.345) | 0.068 (0.492) | 0.076 (0.51) |
| 〈I/σ(I)〉a | 36.2 (2.7) | 21.1 (3.7) | 22.3 (3.0) |
| Refinement | |||
| Resolutiona | 45.9-1.30 Å (1.35-1.30 Å) | 20-1.55 Å (1.59-1.55 Å) | 20-1.65 Å (1.7-1.65 Å) |
| Rworka | 0.131 (0.249) | 0.173 (0.331) | 0.172 (0.295) |
| Rfreea | 0.166 (0.301) | 0.199 (0.360) | 0.201 (0.307) |
| r.m.s.d.b bond length | 0.010 Å | 0.006 Å | 0.005 Å |
| r.m.s.d.b bond angle | 1.3° | 0.9° | 0.9° |
| 〈B〉 Wilson statistic | 13.2 Å2 | 12.5 Å2 | 14.5 Å2 |
| Estimated coordinate error (maximum likelihood) | 0.14 Å | 0.18 Å | 0.21 Å |
| Ramachandran plot | |||
| Allowed | 99.6% | 99.6% | 99.6% |
| Disallowed | 0.4% | 0.4% | 0.4% |
| No. of non-hydrogen protein atoms | 2159 | 2236 | 2179 |
| No. of water molecules | 410 | 335 | 306 |
| No. of Ca2+/Mn2+/DFOA/OA | 3/0/0 | 1/1/1/0 | 1.0/0.7 + 0.3/0/0.7 |
a Rmerge = Σhkl((Σj|Ij − 〈I〉|)/Σj|Ij|). R = Σhkl‖Fo| − |Fc‖/Σhkl|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rwork corresponds to the reflections used in the refinement; Rfree refers to 5% randomly selected reflections omitted from the refinement (52). Values in parentheses are for the highest resolution shell.
b r.m.s.d. is root mean square deviation.
Efforts to co-crystallize OAH with the inhibitor DFOA failed. Instead, soaking of ligand-free OAH crystals in solution containing 0.2 m MnCl2 (or 0.2 m MgCl2), 10 mm CaCl2, 2 mm DFOA, 0.1 m Na+HEPES (pH 7.5), and 30% v/v PEG 400 for 5–10 min yielded the desired enzyme·inhibitor complex. The crystals soaked in MgCl2-containing solution tended to crack within 2 min and diffracted only to 2.5 Å resolution, whereas those soaked with MnCl2-containing solution for 15 min remained intact and diffracted to high resolution. Therefore, the structure of the OAH·DFOA complex was pursued with a Mn2+-containing crystal. Soaking of crystals with oxalacetate was performed following the same protocol as that used with DFOA. Diffraction data for both OAH·ligand complexes were collected on a Rigaku MicroMax 007HF rotating anode generator (CuKα radiation) equipped with a MAR345 imaging plate detector (Table 1). Molecular replacement, using the apo-OAH structure as the search model and the program PHASER (34), resulted in an electron density map that showed clearly the DFOA bound in the active site. For the oxalacetate-soaked crystal, the electron density accounted only for oxalate, consistent with hydrolysis of the oxalacetate substrate.
Structure refinements of the OAH·ligand complexes were carried out at the resolution limits of 1.55 Å (DFOA) and 1.65 Å (oxalate) using PHENIX (38). No geometrical or distance restraints were applied to the metal coordination. TLS parameters (39) were refined in the final refinement step, which improved both the free R and working R factors. Refinement statistics are provided in Table 1.
Structure quality was assessed with the program PROCHECK (40). Superposition of structures was carried out with XtalView or PyMOL (41). PyMOL was also used to generate figures.
RESULTS
Protein Production
To obtain recombinant protein, the full-length C. parasitica oah gene was cloned into pET28a, and the plasmid was used to transform E. coli as described under “Materials and Methods.” This expression system yielded high molecular weight soluble protein aggregates, evidenced by size exclusion chromatography. Native disorder analysis using the program DISOPRED2 with a 5% false-positive rate threshold (42) indicated that long stretches of the 74-amino acid N-terminal region might be intrinsically disordered. Thus, four oah truncation constructs were cloned beginning with residues 52, 59, 66, and 72. All but the last construct yielded soluble active enzymes, but only the construct beginning with residue 66 yielded well diffracting crystals. This fragment was chosen for subsequent enzyme kinetics and structure analyses.
Targeted Disruption of the oah Gene and Phenotypic Characterization in C. parasitica
To define physiological roles for OAH in C. parasitica, a PCR-based strategy was used to generate a disruption fragment consisting of a neomycin selectable marker flanked by oah gene-specific sequences that were used to transform C. parasitica strain cpku80 (27). The growth characteristics and colony morphology under standard laboratory light conditions of the two disruption mutants were very similar to that of strain cpku80, except that the mutants produced a little more orange pigment at late growth stage (data not shown). However, the disruption mutants showed dramatically reduced virulence when they were inoculated onto American chestnut stems. As illustrated in Fig. 3 and Table 2, strain cpku80 produced large cankers with densely packed orange spore-containing stromal pustules protruding through the bark surface. In contrast, the ability of the disrupted oah mutant to expand on chestnut tissue was severely impaired and yielded a comparable canker to that formed by the hypovirus CHV1-EP713-infected strain cpku80/CHV1-EP713 (27). This is consistent with an earlier observation that oxalic acid production is dramatically reduced in hypovirus-infected strains (24). These results establish a significant role for the C. parasitica OAH in virulence and reinforce previous correlative evidence that oxalic acid production contributes to canker formation (24).
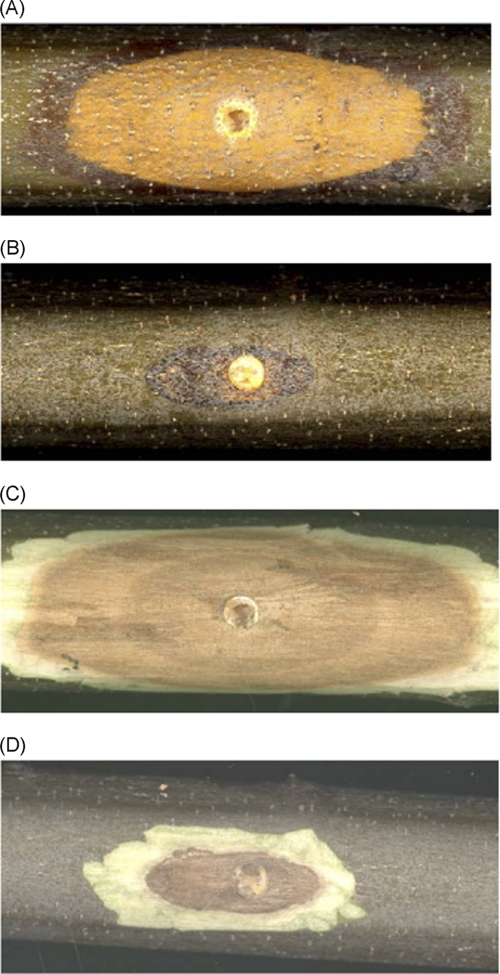
FIGURE 3.
Virulence assay on dormant American chestnut stems. Representative cankers were formed by C. parasitica strain cpku80 (A), and the oah deletion mutant was made in the cpku80 background (B). Photographs of cankers were taken 3 weeks post-inoculation. The Δoah mutant strain forms a much smaller canker with few orange-colored spores forming stromal pustules (termed pycnidia) that are shown in the early stage of development in the cpku80-incited canker. C and D, the bark was removed from stems shown in A and B to reveal the necrotic tissue caused by the infecting fungal strains.
TABLE 2.
Virulence assay
Size of cankers (cm2) produced on dormant American chestnut stems at 3 weeks post-inoculation.
a cpku80/CHV1-EP713, C. parasitica strain cpku80 infected with virulence-attenuating hypovirus CHV1-EP713; cpku80/Δoah, C. parasitica strain cpku80 in which the oah gene has been disrupted.
Enzyme Kinetics
C. parasitica OAH cleaves oxalacetate with high efficiency (kcat/Km = 7.3 × 104 s−1 m in the presence of 5 mm Mn2+ and kcat/Km = 2.7 × 104 s−1 m in the presence of 5 mm Mg2+; see Table 3). Although the kcat value is higher in the presence of Mn2+ than in the presence of Mg2+, the Km is lower for Mg2+. Enzyme catalysis is 20–50-fold less efficient in the presence of Ca2+, yet this is not an insignificant activity. We note that although the crystals were obtained in the presence of Ca2+ (in addition to binding in the active site, the calcium mediates crystal contacts), ligand binding required Mn2+.
TABLE 3.
Steady-state kinetic constants for divalent metal-activated OAH from C. parasitica-catalyzed hydrolysis of oxaloacetate and cleavage of (2R,3S)-dimethylmalate at 25 °C
| Cofactor | Substrate | kcat | Km OAH | kcat/Km |
|---|---|---|---|---|
| s−1 | μm | m−1s−1 | ||
| Mn2+ | Oxalacetatea | 35.2 ± 1.0 | 480 ± 7a | 7.3 × 104 |
| (2R,3S)-Dimethylmalateb | (6.1 ± 0.1) ×10−3 | 147 ± 26 | 4.1 × 101 | |
| Mg2+ | Oxalacetatec | 3.6 ± 0.5 | 134 ± 19 | 2.7 × 104 |
| (2R,3S)-Dimethylmalateb | (1.6 ± 0.1) ×10−2 | 377 ± 50 | 4.2 × 101 | |
| Ca2+ | Oxalacetated | 1.9 ± 0.05 | 1248 ± 94 | 1.5 × 103 |
| (2R,3S)-Dimethylmalateb | Undetectable |
a Reaction solutions contained 0.2–5.0 mm oxalacetate, 5 mm Mn2+, and 0.15 μm OAH.
b Reaction solutions contained 0.2–4.0 mm (2R,3S)-dimethylmalate, 5 mm metal2+, and 20 μm OAH.
c Reaction solutions contained 0.1–2.0 mm oxalacetate, 5 mm Mg2+, and 0.3 μm OAH.
d Reaction solutions contained 0.2–2.0 mm oxalacetate, 5 mm Ca2+, and 0.15 μm OAH.
OAH also exhibits low but measurable C–C bond cleavage of (2R,3S)-dimethylmalate. The hydrolysis of oxalacetate (Fig. 1) is 1780- and 643-fold more efficient than cleavage of (2R,3S)-dimethylmalate in the presence of Mn2+ and Mg2+, respectively. In contrast, the closely related family member, DMML, possesses the reverse kinetic properties; DMML cleaves (2R,3S)-dimethylmalate 750-fold more efficiently than it hydrolyzes oxalacetate (14). Yet the two enzymes exhibit high amino acid sequence identity (60%), and most of the active site residues are conserved in OAH and DMML. The same observation was made with the B. cinerea OAH (1).
Crystallization
The OAH crystals grew in the presence of 0.2 m CaCl2, and attempts to grow them in a wide range of concentrations of Mg2+ or Mn2+ failed. All attempts to obtain crystals of the OAH/DFOA by co-crystallization were unsuccessful. Thus, soaking experiments were explored. Diffraction data from crystals soaked with DFOA solution containing 0.2 m CaCl2 (used to obtain the apo-OAH crystals) for several hours yielded an electron density map that showed very low occupancy of the inhibitor. In contrast, a 15-min soak in DFOA solution containing 0.2 m MnCl2 or a 2-min soak in DFOA solution containing 0.2 m MgCl2 led to electron density maps that revealed full occupancy of both the bound inhibitor and an ordered gating loop in the closed conformation. The crystal quality and the diffraction resolution of the crystal soaked with Mn2+ solutions were far superior to those soaked in Mg2+-containing solutions, which rapidly cracked the crystals. Similarly, a 15-min soak with the substrate oxalacetate in 0.2 m MnCl2-containing solution yielded the best diffracting crystals (although the diffraction was lost after a 20-min soak). Consequently, the OAH/DFOA and OAH/oxalate structure refinements were carried out with diffraction data acquired from crystals soaked with Mn2+-containing ligand solutions.
Quality of the Structures
The apo-OAH structure was refined at 1.30 Å, yielding a high quality structure (Table 1). The model spans amino acid residues 68–189 and 198–361. There is no interpretable electron density associated with the terminal residues 66–67, 362–368, and with the active site gating loop residues 190–197. The crystal structure model contains three Ca2+ cations (one at the active site, one mediating dimer interaction within a tetramer, and one mediating tetramer-tetramer crystal contacts) and 410 water molecules. The backbone dihedral angle values of all residues except Asp-157 are within the allowed region of the Ramachandran plot (43). The backbone conformation of Asp-157, involved in metal cofactor coordination, is strained. Analogous steric strain occurs in the structures of other PEPM/ICL superfamily members. The association of steric and electrostatic strains with active centers is well documented in protein structures and attributed to more stringent requirements for function than folding and stability (44).
The DFOA-Mn2+-bound OAH crystal was isomorphous with the ligand-free OAH crystal and diffracted to 1.55 Å. The refined structure contains amino acid residues 68–364, whereas residues 66–67 and 365–368 are disordered. In contrast with the ligand-free structure, the entire active site gating loop is well defined in the OAH/DFOA structure. The model includes the DFOA, a Mn2+ cofactor bound in the active site, and the Ca2+ ion that mediates dimer interaction within a tetramer. The Ca2+ ion binds at the same position as observed in the ligand-free OAH structure. The third Ca2+ site, present in the structure of the ligand-free OAH, is missing in DFOA-bound OAH crystal structure. Finally, the model includes 335 water molecules.
The oxalate and Mn2+-bound OAH crystal was isomorphous with the ligand-free OAH crystal and diffracted to 1.65 Å. The refined structure contains amino acid residues 68–192 and 198–361. The gating loop residues 193–197, 66–67, and 362–368 are disordered. The model includes Mn2+ and oxalate with partial occupancy, complemented by a partial occupancy Mn2+ binding in the same site as the active site Ca2+ in the apo-OAH structure. The metal arrangement in the active site is described below. In addition, the model includes a Ca2+ ion that mediates dimer interaction and 306 water molecules.
Overall OAH Structure
The crystal asymmetric unit contains a single OAH molecule placed in the unit cell such that the crystallographic symmetry operators generate the physiologically relevant tetramer. The overall oligomeric structure of OAH is similar to that of other members of the PEPM/ICL superfamily (10–18), forming a dimer of dimers consisting of four (α/β)8 barrels (Fig. 4, A and B), with two subunits swapping their 8th helices (Fig. 4C). The bottom of each (α/β)8 barrel is flanked by an additional α-helix. Following a barrel 8th helix, the polypeptide chain meanders to form two more α-helices and traverses the top of the (α/β)8 barrel of the partner above its active site entrance (indicated as “partner subunit cap” in Fig. 4C). The C terminus reaches a third subunit, although this interaction is not extensive (interaction between the orange-colored C terminus of the yellow subunit with the molecule colored green in Fig. 4A).
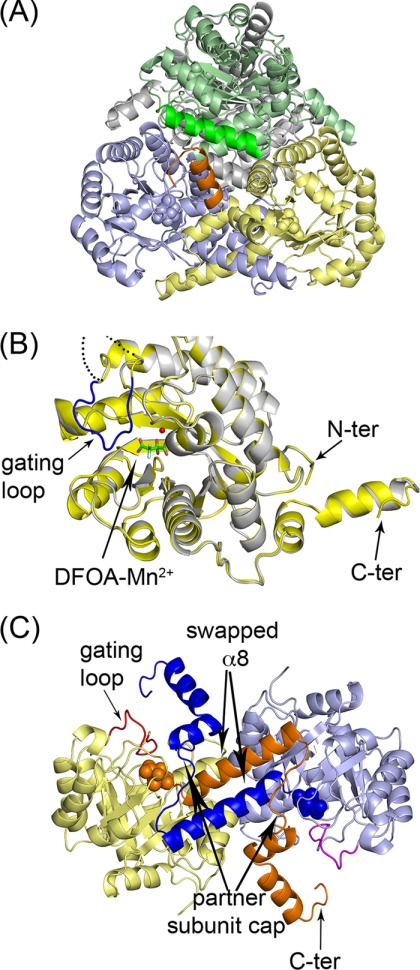
FIGURE 4.
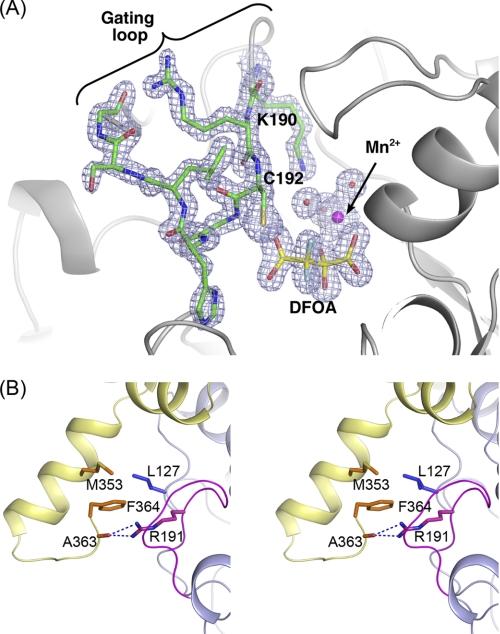
Ribbon representation of the overall fold and oligomeric association of OAH. A, dimer of dimers association that define the physiologically relevant tetramer. Bound DFOA molecules are shown as space-filling models. The helix colored dark orange depicts the C terminus of the yellow subunit that interacts with the green subunit. B, overall fold of a monomer. Superposition of the ligand-free and DFOA-bound OAH. The ordered gating loop corresponding to the ligand-bound state is colored in blue. The disordered gating loop in the ligand-free state is indicated by dashed line in an arbitrary trace because there is no associated electron density to enable chain tracing of this segment. The bound inhibitor, DFOA, is shown in stick model, and the Mn2+ cofactor is shown as a sphere. C, dimer formed by swapping α-helices. The swapped 8th α-helices and the ensuing chains are colored orange and dark blue. The inhibitor is shown as space-filling model. ter, terminal.
Much of the active site gating loop in the ligand-free OAH and OAH/oxalate structures is disordered. In contrast, the DFOA-bound OAH exhibits a well ordered gating loop in the closed conformation (illustrated in Fig. 4B and evidenced by the high quality electron density shown in Fig. 5A). The electron density associated with the C terminus of the DFOA-bound enzyme defines three additional residues because of interactions with the ordered gating loop. These residues turn toward the helix-swapping partner molecule and pack above its active site gating loop so that Phe-364 side chain is anchored in a hydrophobic pocket and Ala-363 carbonyl oxygen forms a hydrogen bond with the guanidinium group of Arg-191 located on the gating loop (Fig. 5B). In that manner, the C terminus participates in the closure of the gating loop of the partner subunit.
FIGURE 5.
Gating loop conformation. A, electron density map associated with the DFOA and the gating loop. The map was calculated omitting the DFOA, Mn2+, and the gating loop from the model. A SIGMAA weighted 2Fo − Fc map is shown contoured at 1.2σ. The electron density map shows clearly that the inhibitor binds as the gem-diol. B, interactions of the gating loop with the C terminus of a partner molecule.
Active Site Structure
As with all members of the PEPM/ICL superfamily, the OAH active site is located in the center of the α/β barrel at the C-terminal side of the β-strands. Among the residues that flank the substrate-binding cavity are residues located on loops connecting the β-strands with the α-helices, of which the gating loop is the longest. Upon DFOA binding, the protein undergoes localized conformational transition that organizes the gating loop in the so-called closed conformation (Figs. 4B and 5). As discussed below, the metal-ligand coordination geometry and distances are consistent with the presence of Ca2+ in the active site of the ligand-free OAH structure (present at concentration of 0.2 m) and with Mn2+ in the ligand-bound state (present at concentration of 0.2 m).
The location of the metal co-factor in the absence and presence of the ligand differs (Fig. 6A). In the apo-OAH structure, the Ca2+ coordinates directly to two aspartic acids (Asp-155 and Asp-157), whereas the Mn2+ in the DFOA-bound structure coordinates directly only to one aspartic acid (Asp-155). In the apo-OAH, the Ca2+ coordinates seven ligands, in contrast to Mn2+ or Mg2+, which a survey of high resolution well refined crystal structures shows to coordinate only six ligands (45). The OAH active site Ca2+ assumes a distorted pentagonal bipyramidal geometry (46), which was also described as a split vertex octahedron (47). The mean Ca2+-ligand distance is 2.32 Å, and it coordinates directly the Asp-155 and Asp-157 carboxylate groups (Fig. 6A). The remaining five Ca2+ ligands are water molecules, four of which bridge the metal to protein carboxylate groups, two to Asp-126 and two to Glu-184 (not shown in Fig. 6A). The fifth solvent molecule does not interact with any protein group and is located 2.0 Å away from the Ca2+, an unusual although not unprecedented short distance for calcium coordination in high resolution protein crystal structures (45, 48). Excluding this distance, the average Ca2+-ligand coordination distance is 2.38 Å, which fits well with other high resolution refined Ca2+-containing protein crystal structures (45). Interestingly, the unusual solvent molecule occupies the same site occupied by Mn2+ in the OAH·DFOA complex (Fig. 6A). We considered the possibility that this is an alternate calcium position and examined how such a model would refine. However, the resulting Fo − Fc difference Fourier map yielded residual positive electron density for both metal sites. For lack of a better description, a water molecule was assigned in this site.
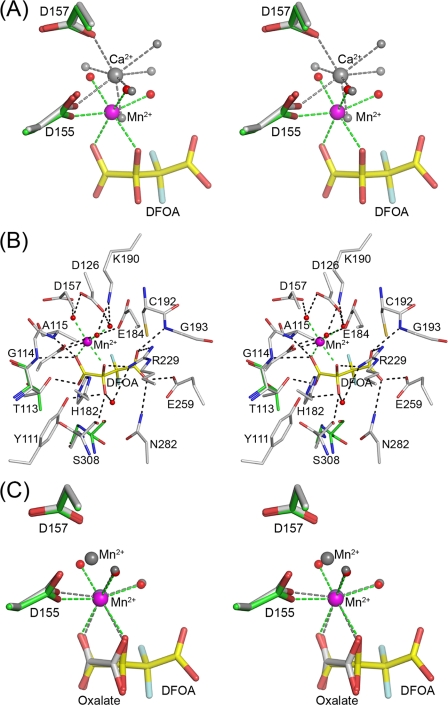
FIGURE 6.
Active site of OAH. A, stereoscopic representation of the metal coordination in the ligand-free and ligand-bound OAH structures. Atomic colors used are as follows: oxygen, red; fluorine, light blue; carbon, green (inhibitor-bound structure), gray (ligand-free structure; yellow, inhibitor. Mn2+ is depicted as a magenta sphere and Ca2+ as a gray sphere. Water molecules are represented by red spheres (the inhibitor-bounds structure) and gray spheres (ligand-free structure). Dashed lines show metal coordination. B, stereoscopic representation of the OAH active site containing the DFOA. Atomic colors used are as follows: carbon, gray for protein and yellow for inhibitor; oxygen, red; nitrogen, blue; fluorine, light blue; Mn2+, magenta. Also shown are two residues that adopt different conformations in the ligand-free structure; the backbone of Ser-308 is flipped, and the side chain of Thr-113 exhibits a different rotamer. The carbon atoms of these two residues are colored green. C, superposition of the metal ions and ligands in the OAH complexes bound with DFOA and oxalate. Color scheme as in A except that the gray colors are used to depict the Mn2+ and protein carbon atoms of the OAH·oxalate complex.
The Mn2+ binds in the presence of DFOA 1.7 Å away from the Ca2+ site seen in the apo-OAH structure. The Mn2+ forms an ideal octahedral coordination geometry (Fig. 6A) with an average metal-ligand distance of 2.19 Å, which is consistent with other high resolution Mn2+-containing protein structures (45). The Mn2+ engages the protein, the DFOA and three water molecules (Fig. 6, A and B). The DFOA binds as the C(2) gem-diol (two hydroxyl groups attached to C(2); Fig. 1B) such that the Mn2+ coordinates a DFOA C(1) carboxyl O and C(2) OH. The Asp-155 carboxylic group and three water molecules complete the octahedral coordination. Each of the coordinating water molecules forms hydrogen bonds with two carboxylate groups. One water molecule forms hydrogen bonds with Asp-126 and Asp-157, the second with Asp-155 and Glu-184, and the third with Asp-126 and the C(4) carboxylate of DFOA. The hydrogen bond geometry of the latter two water molecules enables their lone pair of electrons to form hydrogen bonds with Lys-190. In turn, Lys-190 interacts with Asp-157 and Glu-184. The exquisite interactions of Lys-190 balance the active site charge distribution and shield the amino group from Mn2+, two positively charged moieties that are placed only 3.7 Å apart. We previously proposed that this invariant lysine residue plays a role in regulating loop opening and closure in both the mutase and lyase branches of the PEPM/ICL superfamily (13, 14, 49).
In addition to providing two oxygen ligands to Mn2+, the DFOA interacts with the protein groups shown in Fig. 6B, many of which are conserved in other PEPM/ICL superfamily members. A C(1) carboxyl group oxygen atom is located in an oxyanion hole formed by the main chain amide groups of Gly-114 and Ala-115, and the second C(1) carboxyl oxygen forms a hydrogen bond with the side chain hydroxyl group of Thr-113. The pro-R C(2) O (coordinating to Mn2+) also interacts with Arg-221 guanidinium Nζ1, which in turn interacts with an internal water molecule. This water molecule interacts further with the pro-S C(2) O, and the pro-S C(2) O interacts with Ser-308 and Tyr-111 hydroxyl groups. Tyr-111 forms a hydrogen bond with His-182 Nϵ, an interaction that is conserved in the two lyases, DMML and 2-methylisocitrate lyase, but is absent in the family relative, the PDP, which contains two phenylalanine residues instead. Asn-282 Nδ anchors the Glu-259 carboxyl group, the group that shares a proton with the DFOA C(4) carboxyl oxygen. A shared proton between two carboxylate groups occurs in the lyase branch of the PEPM/ICL superfamily and is crucial for catalysis (13, 16). The DFOA C(4) carboxyl oxygen also interacts with Asn-282 Nδ. The second C(4) carboxyl oxygen atom interacts with the Arg-221 guanidinium Nζ2, as well as with a water molecule that coordinates to Mn2+. The intricate interactions extend to a second protein shell that supports the proper orientation of the catalytic groups with respect to the substrate.
DFOA binding induces closure of the gating loop, whereas the remaining residues are mostly unaltered except for two notable changes (Fig. 6B; the two residues in the unbound structure are highlighted in green). Thr-113 adopts an alternate side chain rotamer and forms a hydrogen bond with the C(1) carboxyl group. The main chain carbonyl of Ser-308 flips by 180° from a β-conformation in the apo structure that facilitates interaction with an adjacent β-strand to an α-helical conformation that eliminates this β-sheet interaction. The backbone flip shifts the position of the Ser-308 hydroxyl group by 0.6 Å and enhances its hydrogen bond interaction with the DFOA pro-S C(2) hydroxyl. Interestingly, the DMML and 2-methyl isocitrate lyase both contain a proline residue in place of the serine residue, which interacts with the respective substrate C(2) methyl groups (13, 14). In these structures, the proline backbone exists in the α-conformation in both ligand-free and ligand-bound structures. Thus, the energetic basis for the conformational switch in OAH is unknown.
The oxalacetate soaking experiments led to hydrolysis of the substrate and yielded bound oxalate in a planar conformation, in contrast to the equivalent moiety of DFOA, which is nonplanar (Fig. 6C). The gating loop is disordered. The structure represents a crystallographic average consistent with 70% of the crystal active sites occupied by the oxalate and Mn2+ and the remaining 30% corresponding to a ligand-free structure with Mn2+ bound in the site analogous to the Ca2+ site in the apo structure, 1.7 Å away from the major Mn2+. Although the electron density at the minor Mn2+ site may be accounted for by a water molecule, the short distances to the major Mn2+ and to Asp-155, Asp-157, and a water molecule (2.3, 2.2, and 2.1 Å, respectively) favor a bound metal rather than a water molecule.
Intermolecular Metal-binding Sites
In addition to the active site metal cofactors described above, two additional Ca2+-binding sites mediate intermolecular interactions. The first Ca2+ ion contributes to the interactions between the two subunits that swap their 8th α-helices. This Ca2+ ion is present in the ligand-free as well as ligand-bound structures. The Ca2+ exhibits an ideal octahedral coordination geometry involving the Glu-285 carboxyl oxygen and the Asn-286 amide oxygen of one OAH subunit (located on the C terminus of the 7th β-strand of the (α/β)8 barrel) and two coordinating ligands of the second subunits as follows: the Thr-334 backbone carbonyl oxygen and the Asp-336 carboxyl oxygen (located on the linker between the swapped 8th α-helix and the ensuing C-terminal α-helix). Interactions with two water molecules complete the octahedral coordination. The average Ca2+-ligand distance is 2.31, 2.35, and 2.35 Å in the apo-OAH, DFOA-bound, and oxalate-bound OAH structures, respectively. It is worth noting that although the crystal structures of the DFOA-bound and oxalate-bound OAH were obtained by soaking the ligand-free OAH crystal in solutions containing 20-fold excess MnCl2 over CaCl2, we model the metal as calcium ion rather than manganese ion because the 2.35 Å average coordination distance is closer to the coordination distance of the Ca2+ ion.
The second intermolecular Ca2+ site mediates crystal contacts across tetramers in the ligand-free OAH structure. This Ca2+ exhibits an ideal pentagonal bipyramidal coordination geometry involving the Glu-265 carboxyl group of one tetramer, the Phe-480 backbone carbonyl group of a second tetramer, and five water molecules. The mean Ca2+-ligand distance in this site is 2.45 Å, and it is not seen in the structures of the OAH·ligand complexes.
DISCUSSION
Structural Basis for Substrate Specificity
OAH, DMML, and PDP are distinguished from other members of the PEPM/ICL superfamily in that they exhibit substrate promiscuity. Yet each of these enzymes has a distinct substrate profile. OAH exhibits 1780-fold higher catalytic efficacy toward oxalacetate than toward (2R,3S)-dimethylmalate; DMML is 750-fold more efficient in cleaving (2R,3S)-dimethylmalate than oxalacetate, and PDP has equivalent catalytic efficacies toward these substrates. Despite the different substrate specificities, most of the groups involved in ligand binding, the likely determinants of substrate specificity, are conserved in these three enzymes (14, 18, 20). The few existing active site amino acid residue differences were therefore examined as potential specificity determinants.
Notably different in OAH and DMML is an active site residue that interacts with the pro-S C(2) hydroxyl of oxalacetate or with the equivalent methyl group of (2R,3S)-dimethylmalate. OAH contains a serine residue, Ser-308, and DMML contains a proline residue instead, Pro-240 (Figs. 2 and 7A). A serine side chain forms a favorable electrostatic interaction with the hydroxyl group of oxalacetate, whereas the hydrophobic proline interacts better with the methyl group of (2R,3S)-dimethylmalate, consistent with the catalytic rates of the two enzymes with oxalacetate and (2R,3S)-dimethylmalate substrates. Thus, one would expect that replacement of Ser for Pro-240 in DMML might improve its activity toward oxalacetate. Conversely, replacement of Pro for Ser-308 of OAH should improve cleavage of (2R,3S)-dimethylmalate. The reality proves more complex. Previously, we reported that the DMML P240S mutant lowered the catalytic efficiency of (2R,3S)-dimethylmalate cleavage by 45-fold and the catalytic efficiency toward oxalacetate by only 6-fold (14). The rate change indicates that although the trend follows the expectation based on the structural considerations, the enzyme structure is better optimized to accommodate the primary substrate ((2R,3S)-dimethylmalate), and a single residue replacement is insufficient to fully accomplish activity switch. For OAH from B. cinerea (for which a structure is unavailable), the mutant S260P (the counterpart of Ser-260 is Ser-308 in C. parasitica OAH) reduced its OAH activity 40-fold and increased the (2R,3S)-dimethylmalate lyase activity 15-fold (51), consistent with the structure-based prediction.
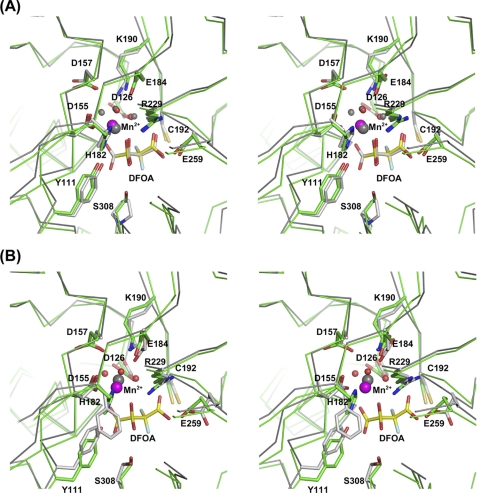
FIGURE 7.
Stereoscopic representations of superposed active sites of OAH and DMML (A) and OAH and PDP (B). The protein backbone Cα atoms are traced, and selected side chains are shown. OAH carbon atoms are colored green, and those of either DMML or PDP are colored gray. A, the DFOA-bound structures of both OAH and DMML are shown. B, the structure of PDP does not include DFOA. Instead, the closed conformation was obtained with glutaraldehyde that reacted with the catalytic cysteine (used to enable flash-cooling of the crystals). For clarity, the glutaraldehyde adduct is not depicted in the image. OAH Mn2+ is colored magenta and that of DMML in gray, as is the Mg2+ in the PDP structure.
The broad specificity lyase, PDP, exhibits similar oxalacetate hydrolase and (2R,3S)-dimethylmalate lyase activities (20). The sequence and structural differences between OAH and PDP, including some of the active site residues, are more substantial than those between OAH and DMML (Figs. 2 and 7B). The OAH Ser-308 counterpart in PDP is also a serine, Ser-257. The PDP substitution S257F affected both OAH and DMML activity similarly, reducing oxalacetate hydrolysis efficiency by 4-fold and the (2R,3S)-2-ethyl-3-methylmalate lyase activity by 2-fold (51). Moreover, PDP contains two phenylalanine residues (Figs. 2 and 7B) instead of the His-Tyr pair present in both DMML and OAH (His-182 and Tyr-111 in the OAH numbering scheme). The His-Tyr pair was shown to be crucial for the catalytic activities of DMML (14). The Tyr-His pair is also conserved in ICL and 2-methylisocitrate lyase, which are highly specialized enzymes. These residues participate in an extensive hydrogen bond network. Therefore, the broader substrate specificity in PDP may be linked to the replacement of the Tyr-His pair by the hydrophobic Phe-Phe pair that breaks the active site hydrogen bond network (Fig. 7B). This may increase flexibility in the active site cavity so that it accommodates productively a broader range of substrates with similar although lower catalytic efficiencies. The structural differences and their impact on substrate profile make the OAH/DMML/PDP branch of the PEPM/ICL superfamily an excellent model system to study and illuminate processes of function adaptation.
Catalytic Mechanism
The catalytic mechanism proposed for the lyase branch of the PEPM/ICL superfamily (13) is also operative in OAH. The proposed mechanism involves deprotonation of the pro-R C(2) hydroxyl triggered by the proximity to the Mg2+/Mn2+ cofactor and Arg-229, cleavage of the C(2)–C(3) bond to form an aci-carboxylate intermediate (generating acetate-leaving group in the case of OAH) assisted by a shared hydrogen bond between Glu-259 and the C(4) carboxyl group, and then proton transfer from the Cys-192 thiol group to the C(3) atom (13). Unique to OAH is the hydration of C(2) prior to cleavage of the C(2)–C(3) bond to form a gem-diol adduct. In the absence of the hydration step, the product would be glyoxylate rather than oxalate, as the product of isocitrate cleavage by ICL.
It worth noting that site-directed mutagenesis studies of DMML have shown that elimination of the catalytic thiol group abolishes (2R,3S)-dimethylmalate cleavage. In contrast, the mutant enzymes exhibited reduced low oxalacetate hydrolytic activity (which is very low from the outset) but remained significantly active with only a 5-fold reduction in kcat (14). The proposed explanation is that the substrate itself assists with catalysis; the pro-S C(2)OH (the binding OH) of the hydrated oxalacetate acts as a surrogate proton donor. This implies that equivalent catalytic cysteine mutants of OAH will also retain residual activity.
The DFOA gem-diol is an analog of the C(2) hydrated oxalacetate intermediate postulated along the reaction pathway (Fig. 1). The gem-diol is the major form of DFOA in solution (1), and the gem-diol is only a minor component (7%) of solvated oxalacetate (50). DFOA inhibits both OAH and DMML (1, 14) because the two fluoride atoms stabilize the C(2)–C(3) bond that is otherwise cleaved by the lyase branch of the PEPM/ICL superfamily members (13). Hence, the DFOA-protein interactions are expected to resemble closely those of the oxalacetate gem-diol intermediate.
It remains unknown whether an active site water molecule hydrates the oxalacetate or whether from the outset the hydrated gem-diol, the minor structural form in aqueous solution, binds in the active site. If oxalacetate is hydrated prior to binding, then formally OAH should be considered a lyase rather than hydrolase. The crystal structure of the ligand-free OAH contains a water molecule hydrogen-bonded to Ser-308 and Tyr-111, overlapping the site of the pro-S C(2) hydroxyl group of the DFOA gem-diol in the OAH·inhibitor complex. However, the presence of such a water molecule is not evidence that the oxalacetate becomes hydrated upon binding to OAH. An oxalacetate gem-diol may simply displace such a water molecule. Combined mutagenesis and crystallographic studies may shed light on this ambiguous aspect of the OAH catalytic mechanism.
A second intriguing observation that emerges from this study is the two alternative locations of the Ca2+ in the active site. Because of the requirement for exact substrate alignment with respect to the entire catalytic machinery, the Ca2+ site in the ligand-free structure probably represents an unproductive metal binding mode. The C. parasitica OAH exhibits significant activity in the presence of Ca2+, which implies that the Ca2+ must move upon substrate binding to the site where the Mn2+ binds in the structure of the OAH·DFOA complex. Perhaps the nonproductive binding site in the ligand-free state is preferred simply because direct interactions with two carboxylate groups (Asp-155 and Asp-157) are stronger than an interaction with a single carboxylate group. Once the negatively charged oxalacetate binds, the cation shifts away from Asp-157 to establish direct interaction with the oxalic group of the substrate, which in turn triggers catalysis.
Acknowledgments
We thank Debra Dunaway-Mariano for stimulating discussions and for the DFOA and the (2R,3S)-dimethyl malate samples. We thank Celia Chen and Kap Lim for helpful advice and Natalia Gorlatova for help with the kinetic assays. We also thank the APS SER-CAT staff for assistance in data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants P01-GM057890 (to O. H.) and GM55981 (to D. N.). This work was also supported by National Science Foundation Grant MCB9813271 (to O. H.).
We dedicate this paper to the memory of our colleague Qihong Sun.
The atomic coordinates and structure factors (codes 3LYE, 3M0J, and 3M0K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) GU932672.
- OAH
- oxalacetate acetylhydrolase
- DMML
- 2,3-dimethylmalate lyase
- ICL
- isocitrate lyase
- PEPM
- phosphoenolpyruvate mutase
- PDP
- petal death protein
- DFOA
- 3,3-difluorooxalacetate.
REFERENCES
- 1.Han Y., Joosten H. J., Niu W., Zhao Z., Mariano P. S., McCalman M., van Kan J., Schaap P. J., Dunaway-Mariano D. (2007) J. Biol. Chem. 282, 9581–9590 [DOI] [PubMed] [Google Scholar]
- 2.Guimarães R. L., Stotz H. U. (2004) Plant Physiol. 136, 3703–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutton M. V., Evans C. S. (1996) Can. J. Microbiol. 42, 881–895 [Google Scholar]
- 4.Nakagawa Y., Shimazu K., Ebihara M., Nakagawa K. (1999) J. Infect. Chemother. 5, 97–100 [DOI] [PubMed] [Google Scholar]
- 5.Kirkland B. H., Eisa A., Keyhani N. O. (2005) J. Med. Entomol. 42, 346–351 [DOI] [PubMed] [Google Scholar]
- 6.Jordan C. R., Dashek W. V., Highly T. L. (1996) Holzforschung 50, 312–318 [Google Scholar]
- 7.Lane B. G. (2002) IUBMB Life 53, 67–75 [DOI] [PubMed] [Google Scholar]
- 8.Britton K., Langridge S., Baker P. J., Weeradechapon K., Sedelnikova S. E., De Lucas J. R., Rice D. W., Turner G. (2000) Structure 8, 349–362 [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri B. N., Sawaya M. R., Kim C. Y., Waldo G. S., Park M. S., Terwilliger T. C., Yeates T. O. (2003) Structure 11, 753–764 [DOI] [PubMed] [Google Scholar]
- 10.Chen C. C., Han Y., Niu W., Kulakova A. N., Howard A., Quinn J. P., Dunaway-Mariano D., Herzberg O. (2006) Biochemistry 45, 11491–11504 [DOI] [PubMed] [Google Scholar]
- 11.Grimm C., Evers A., Brock M., Maerker C., Klebe G., Buckel W., Reuter K. (2003) J. Mol. Biol. 328, 609–621 [DOI] [PubMed] [Google Scholar]
- 12.Huang K., Li Z., Jia Y., Dunaway-Mariano D., Herzberg O. (1999) Structure 7, 539–548 [DOI] [PubMed] [Google Scholar]
- 13.Liu S., Lu Z., Han Y., Melamud E., Dunaway-Mariano D., Herzberg O. (2005) Biochemistry 44, 2949–2962 [DOI] [PubMed] [Google Scholar]
- 14.Narayanan B., Niu W., Joosten H. J., Li Z., Kuipers R. K., Schaap P. J., Dunaway-Mariano D., Herzberg O. (2009) J. Mol. Biol. 386, 486–503 [DOI] [PubMed] [Google Scholar]
- 15.Narayanan B. C., Niu W., Han Y., Zou J., Mariano P. S., Dunaway-Mariano D., Herzberg O. (2008) Biochemistry 47, 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma V., Sharma S., Hoener zu Bentrup K., McKinney J. D., Russell D. G., Jacobs W. R., Jr., Sacchettini J. C. (2000) Nat. Struct. Biol. 7, 663–668 [DOI] [PubMed] [Google Scholar]
- 17.Simanshu D. K., Satheshkumar P. S., Savithri H. S., Murthy M. R. (2003) Biochem. Biophys. Res. Commun. 311, 193–201 [DOI] [PubMed] [Google Scholar]
- 18.Teplyakov A., Liu S., Lu Z., Howard A., Dunaway-Mariano D., Herzberg O. (2005) Biochemistry 44, 16377–16384 [DOI] [PubMed] [Google Scholar]
- 19.von Delft F., Inoue T., Saldanha S. A., Ottenhof H. H., Schmitzberger F., Birch L. M., Dhanaraj V., Witty M., Smith A. G., Blundell T. L., Abell C. (2003) Structure 11, 985–996 [DOI] [PubMed] [Google Scholar]
- 20.Lu Z., Feng X., Song L., Han Y., Kim A., Herzberg O., Woodson W. R., Martin B. M., Mariano P. S., Dunaway-Mariano D. (2005) Biochemistry 44, 16365–16376 [DOI] [PubMed] [Google Scholar]
- 21.Pollack S. J., Freeman S., Pompliano D. L., Knowles J. R. (1992) Eur. J. Biochem. 209, 735–743 [DOI] [PubMed] [Google Scholar]
- 22.Anagnostakis S. L. (1982) Science 215, 466–471 [DOI] [PubMed] [Google Scholar]
- 23.Nuss D. L. (2005) Nat. Rev. Microbiol. 3, 632–642 [DOI] [PubMed] [Google Scholar]
- 24.Havir E. A., Anagnostakis S. L. (1983) Physiol. Plant Pathol. 23, 369–376 [Google Scholar]
- 25.Welch A. J., Stipanovic A. J., Maynard C. A., Powell W. A. (2007) Plant Sci. 172, 488–496 [Google Scholar]
- 26.Davidson R. C., Blankenship J. R., Kraus P. R., de Jesus Berrios M., Hull C. M., D'Souza C., Wang P., Heitman J. (2002) Microbiology 148, 2607–2615 [DOI] [PubMed] [Google Scholar]
- 27.Lan X., Yao Z., Zhou Y., Shang J., Lin H., Nuss D. L., Chen B. (2008) Curr. Genet. 53, 59–66 [DOI] [PubMed] [Google Scholar]
- 28.Churchill A. C., Ciufetti L. M., Hansen D. R., Van Etten H. D., Van Alfen N. K. (1990) Curr. Genet. 17, 25–31 [Google Scholar]
- 29.Hillman B. I., Shapira R., Nuss D. L. (1990) Phytopathology 80, 850–956 [Google Scholar]
- 30.Leatherbarrow R. J. (2007) GraFit, Version 6, 6.0 Ed., Erithacus Software Ltd., Horley, UK [Google Scholar]
- 31.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 32.French S., Wilson K. (1978) Acta Crystallogr. A 34, 517–525 [Google Scholar]
- 33.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 34.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamzin V. S., Perrakis A., Wilson K. S. (2001) in International Tables for Crystallography Volume F: Crystallography of Biological Macromolecules (Rossmann M. G. A., Arnold E. eds) pp. 720–722, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 36.McRee D. E. (1992) J. Mol. Graphics 10, 44–46 [Google Scholar]
- 37.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 38.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 39.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D. Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 41.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 42.Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. (2004) Bioinformatics 20, 2138–2139 [DOI] [PubMed] [Google Scholar]
- 43.Ramachandran G. N., Ramakrishnan C., Sasisekharan V. (1963) J. Mol. Biol. 7, 95–99 [DOI] [PubMed] [Google Scholar]
- 44.Herzberg O., Moult J. (1991) Proteins 11, 223–229 [DOI] [PubMed] [Google Scholar]
- 45.Harding M. M. (2001) Acta Crystallogr. D. Biol. Crystallogr. 57, 401–411 [DOI] [PubMed] [Google Scholar]
- 46.Herzberg O., James M. N. (1988) J. Mol. Biol. 203, 761–779 [DOI] [PubMed] [Google Scholar]
- 47.Swain A. L., Kretsinger R. H., Amma E. L. (1989) J. Biol. Chem. 264, 16620–16628 [PubMed] [Google Scholar]
- 48.Castagnetto J. M., Hennessy S. W., Roberts V. A., Getzoff E. D., Tainer J. A., Pique M. E. (2002) Nucleic Acids Res. 30, 379–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S., Lu Z., Han Y., Jia Y., Howard A., Dunaway-Mariano D., Herzberg O. (2004) Biochemistry 43, 4447–4453 [DOI] [PubMed] [Google Scholar]
- 50.Emly M., Leussing D. L. (1981) J. Am. Chem. Soc. 103, 628–634 [Google Scholar]
- 51.Joosten H. J., Han Y., Niu W., Vervoort J., Dunaway-Mariano D., Schaap P. J. (2008) Proteins 70, 157–166 [DOI] [PubMed] [Google Scholar]
- 52.Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]