Abstract
EmrE, a member of the small multidrug transporters superfamily, extrudes positively charged hydrophobic compounds out of Escherichia coli cytoplasm in exchange for inward movement of protons down their electrochemical gradient. Although its transport mechanism has been thoroughly characterized, the structural basis of energy coupling and the conformational cycle mediating transport have yet to be elucidated. In this study, EmrE structure in liposomes and the substrate-induced conformational changes were investigated by systematic spin labeling and EPR analysis. Spin label mobilities and accessibilities describe a highly dynamic ligand-free (apo) conformation. Dipolar coupling between spin labels across the dimer reveals at least two spin label populations arising from different packing interfaces of the EmrE dimer. One population is consistent with antiparallel arrangement of the monomers, although the EPR parameters suggest deviations from the crystal structure of substrate-bound EmrE. Resolving these discrepancies requires an unusual disposition of TM3 relative to the membrane-water interface and a kink in its backbone that enables bending of its C-terminal part. Binding of the substrate tetraphenylphosphonium changes the environment of spin labels and their proximity in three transmembrane helices. The underlying conformational transition involves repacking of TM1, tilting of TM2, and changes in the backbone configurations of TM3 and the adjacent loop connecting it to TM4. A dynamic apo conformation is necessary for the polyspecificity of EmrE allowing the binding of structurally diverse substrates. The flexibility of TM3 may play a critical role in movement of substrates across the membrane.
Keywords: Drug Transport, Electron Paramagnetic Resonance (EPR), Membrane Biophysics, Membrane Proteins, Multidrug Transporters, Multidrug Resistance, Spin Labeling
Introduction
One mechanism of multidrug resistance involves the active extrusion of toxic molecules out of the cell by dedicated membrane transporters. In prokaryotes, five superfamilies of transporters handle vectorial drug traffic moving energetically uphill against substrate concentration gradients (1). The thermodynamics of the problem are rendered favorable through coupling of substrate movement to the direct use of ATP energy or the discharge of electrochemical ion gradients. Small multidrug resistance transporters are the smallest bacterial transporters with four predicted transmembrane α-helices and no significant extramembrane domain (2, 3). They function as dimers coupling translocation of positively charged hydrophobic substrates out of the cell to the inward movement of protons.
Much of the current mechanistic understanding of small multidrug resistance transporters has emerged from seminal studies in the laboratory of Schuldiner and co-workers (4–7)defining fundamental steps in the transport cycle of Escherichia coli EmrE. Protons and substrates share a common binding site organized around the absolutely conserved, membrane-embedded, glutamate 14 in transmembrane (TM)3 helix 1 (8). Binding of positively charged hydrophobic substrates is coordinated by the Glu-14 side chain and stabilized by interactions with aromatic residues in TM segments 1–3 (9–11). Proton release at the cytoplasmic side, facilitated by the perturbed pKa of Glu-14, is concomitant with substrate binding (12). Mutually exclusive occupancy of the binding site by protons and substrates provides the basis for coupling of the two fluxes (13, 14).
The structural framework that mediates vectorial transport continues to be controversial. Specifically, the orientation of the two monomers in the dimer has emerged as a focal point of contention. EmrE structures, determined from two- and three-dimensional crystals (15–17), provide compelling evidence supporting antiparallel orientation of the monomers. In addition, the sequence determinants of membrane topology, the arginine and lysine contents of EmrE loops (i.e. the K+R bias), do not favor a unique orientation in the membrane implying that EmrE could insert with dual topology (18). However, experiments designed to verify or alter the relative orientation of the monomers were inconclusive (19).
In contrast, a body of biochemical data supports a parallel orientation of the dimer. These include the design and construction of functional EmrE chimeras where monomers are linked by short polar loops not favored energetically to cross the bilayer (20). Furthermore, residues predicted to be on opposite sides of the membrane in the antiparallel dimer model can be cross-linked without significant perturbation of transport (21). Finally, limited structural constraints derived from EPR analysis of spin-labeled EmrE in TM3 were interpreted as consistent with a parallel orientation (19).
Contributing to the structure mechanism divide is the absence of direct analysis of the structure and conformational dynamics of EmrE in lipid bilayers in various transport intermediates. Specifically, the structural changes induced by substrate/proton binding have not been described as both available structures were obtained in the presence of the substrate tetraphenylphosphonium (TPP+). In this study, we report results from a systematic analysis of EmrE structure in the (substrate-free) apo- and TPP+-bound conformations in liposomes. For this purpose, spin label probes were introduced one at a time along the EmrE sequence (residues 3–110). EPR spectroscopy was used to characterize spin label dynamics, accessibility to the lipid and water phases, as well as pairwise short range proximity across the dimer interface (22–25). Changes in the EPR constraints upon substrate binding reveal movements in TM1 and -2 and ordering of TM3 and the adjacent loop linking it to TM4. The compatibility between the EPR constraints and existing structures and models is evaluated to assess whether an antiparallel dimer is populated in liposomes.
EXPERIMENTAL PROCEDURES
Cloning and Site-directed Mutagenesis
The emrE gene is cloned between the restriction sites NdeI and BamHI of plasmid pET15b+. The construct has a His6 tag linked to the EmrE N terminus by a thrombin sequence. The three native cysteines were mutated to alanines (C39A, C41A, and C95A) to yield a cysteine-less EmrE hereafter referred to as WT*. Single cysteine mutants were generated for all amino acids spanning the first three transmembrane segments on pET15b+ WT* background using the QuikChange method (Stratagene). Residues in the fourth transmembrane segment were cloned in pET21a+ WT* between NdeI and HindIII. This construct has a spacer and a His6 tag added at the C terminus as described previously (26). All 110 cysteine mutants were also generated in pET20b+ WT* as described previously (27) for use in resistance phenotype activity assays. All the mutants were confirmed by sequencing.
EmrE Expression, Purification, and Reconstitution
The plasmids containing the EmrE single cysteine mutation were used to transform into BL21 (DE3) E. coli competent cells. The EmrE mutants were overexpressed in 1 liter of minimal medium A as described previously (27). Cells were grown at 37 °C until an absorbance of 1.4 was reached, and protein expression was induced at 28 °C for most mutants by the addition of 1 mm isopropyl β-d-1-thiogalactopyranoside. For mutants at positions 66 and 77, induction was carried out at 20 °C overnight. After 5 h of induction, the cells were harvested by centrifugation, and resuspended in lysis buffer (27). The cells were then sonicated and homogenized using an EmulsiFlex-C5 (Avestin). The membrane pellets were solubilized with 1.5% n-dodecyl β-d-maltoside in resuspension buffer (50 mm sodium phosphate monobasic, 300 mm sodium chloride, and 10 mm imidazole, pH 8). The solubilized membranes were ultracentrifuged at 388,000 × g for 1 h at 4 °C.
Solubilized EmrE was purified by nickel affinity chromatography (27). The eluted EmrE mutants were spin-labeled with 20-fold molar excess of methanethiosulfonate spin label (28) and incubated at room temperature for 2 h followed by a second and a third addition of 10-fold molar excess spin label every 2 h (29). After 6 h at room temperature, the samples were stored overnight at 4 °C. Spin-labeled EmrE mutants exhibiting dipolar broadening were underlabeled as follows. Mutants 14, 40, 44, 60, and 64 were spin-labeled with 0.5-fold molar excess of methanethiosulfonate spin label and incubated at room temperature for 1.5 h after which a 20-fold molar excess of diamagnetic spin label (1-acetyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methane thiosulfonate was added, and the samples were stored overnight at 4 °C. The labeled protein was purified by size exclusion chromatography (SEC) on a Superdex-200 column (Amersham Biosciences) in SEC buffer (50 mm sodium phosphate monobasic, 50 mm NaCl, 0.02% n-dodecyl β-d-maltoside, and 0.02% NaN3, pH 7.2).
Preparation of Unilamellar Liposomes
Asolectin (Avanti Polar Lipids) dried from chloroform solution was resuspended in reconstitution buffer (50 mm HEPES, 50 mm sodium chloride, 2 mm magnesium chloride, pH 7.5), and unilamellar vesicles were prepared by extrusion through 50-nm Nucleopore Track-Etch membrane filters (Schleicher & Schuell). The protein sample was mixed at 1:500 to 1:1000 molar excess of preformed unilamellar liposomes. The mixture was incubated with gentle agitation at 4 °C for 2 h and then was diluted to a total volume of 10 ml with reconstitution buffer. Bio-Beads SM2 (Bio-Rad) were added to the sample at the quantity of 80 mg/ml initial mixture and incubated for 2 h at 4 °C, and addition of fresh Bio-Beads was repeated twice at a 2-h interval followed by overnight incubation at 4 °C. The reconstituted proteoliposomes were collected by ultracentrifugation at 388,000 × g for 45 min at 4 °C. The pellets were then resuspended in reconstitution buffer.
EPR Spectroscopy
The EPR spectra of all reconstituted spin-labeled EmrE mutants were collected on a Varian EPR spectrometer equipped with a loop-gap resonator at room temperature. Substrate binding to reconstituted EmrE was initiated by the addition of 16 mm of TPP+ to ∼100 μm EmrE sample. The mixture was incubated at room temperature for 20 min. The samples were then drawn into round capillaries (VitroCom). The incident microwave power was 2 milliwatts, and the modulation amplitude was 1.6 G. Power saturation experiments were carried out on a Bruker ELEXSYS spectrometer equipped with a dielectric resonator (Bruker BioSpin). Samples were loaded in gas-permeable methylpentene polymer TPX® capillaries, and the measurements were carried out under nitrogen and in the presence of 20% oxygen or 50 mm nickel diaminediacetic acid (NiEDDA). The data were analyzed to obtain the parameter P½. The EPR accessibility parameter Π was calculated as described previously.
Ethidium Resistance Assay
The ability of mutants to confer resistance to ethidium bromide was tested as described previously (26). Overnight cultures of E. coli expressing mutant EmrE were grown to saturation. The cultures were diluted 10-, 103-, and 106-fold in fresh media, and 5 μl of each dilution was plated on LB-amp plates containing 200 μg/ml ethidium bromide. Growth was examined after incubation at 37 °C for 24 h. The empty vector showed no growth at any dilutions and was used as a negative control. EmrE WT* exhibited growth at all dilutions and was used as a positive control. For all mutants, growth at 103-fold dilution and above was considered active. Some mutants with growth at 0- and 10-fold dilutions were considered to have compromised activity. Finally, mutants with no growth at any dilution were considered inactive.
TPP+ Binding Assay
TPP+ binding experiments were carried out on an L-format fluorometer from Photon Technology International. The extent of binding of TPP+ was estimated by quenching of native intrinsic tryptophan (Trp) fluorescence emission (10). Samples containing 3 μm of spin-labeled EmrE mutants were mixed with 0, 6, and 12 μm TPP+ and incubated for 20 min at room temperature. The samples were loaded in a 1-cm path length quartz cuvette. Tryptophan emission spectra of the samples were recorded in the 310–350 nm range after excitation at 295 nm. For the high concentration experiments, the spin-labeled EmrE samples were diluted to a final concentration of 20 μm, and TPP+ was added to a final concentration of 80 and 120 μm.
RESULTS
Biochemical Analysis of EmrE Mutants
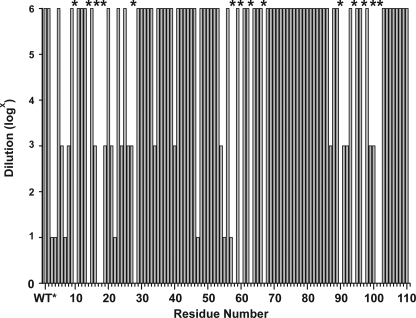
To assess the transport activity of the EmrE cysteine mutants, we determined whether they conferred drug resistance on transformed E. coli cells. For this purpose, overnight cultures were diluted, spotted on agar plates containing 200 μg/ml EtBr, and allowed to grow for 24 h at 37 °C (26). Bacterial growth was qualitatively characterized as described under “Experimental Procedures.” Fig. 1 shows that most of the mutants are active at the highest dilution tested reflecting efficient EtBr clearance out of the cytoplasm. In contrast, a number of substitutions did not confer resistance at any dilution, including at residues in TM1 (e.g. residues 10, 14, 17, and 18) and TM3 (e.g. residues 58, 60, 63, and 67) previously identified as important for substrate binding (10, 11, 30–32). Similarly, substitutions of glycine residues in TM4 (residues 90 and 97) led to inactive transporters. For a few sites, growth was observed at only one of the three dilutions suggesting compromised EmrE transport activity. Resistance reflects the balance between multiple factors, including levels of EmrE expression and the competition between passive inward leaks of EtBr and its extrusion by the transporter. Thus, lack of resistance in some mutants may reflect changes in binding affinity as well as the kinetics of the transport cycle.
FIGURE 1.
Growth phenotypes of cells expressing EmrE single cysteine mutants. Saturated cultures of each mutant were diluted by a factor of 106, 103, and 10 and spotted on agar plates containing ethidium bromide as described under “Experimental Procedures.” The height of the bar indicates the maximal dilution at which cell growth was observed. * indicates mutants that did not support a resistance phenotype.
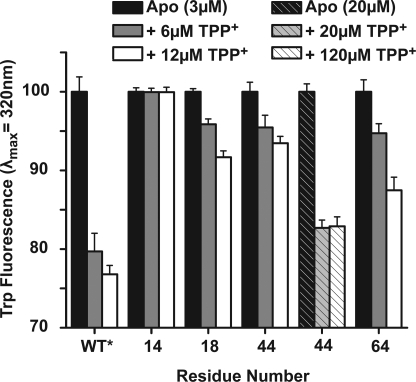
Therefore, to complement the coarse phenotypic screen, we analyzed the substrate affinity of the purified, spin-labeled proteins in dodecyl maltoside micelles. EmrE Trp fluorescence, specifically of Trp-63, is quenched by binding of TPP+ (10). For each spin-labeled mutant, the change in Trp fluorescence upon addition of TPP+ was compared with that of the WT* (supplemental Fig. 1). Lack of change in Trp fluorescence implies a reduction in affinity due to the cysteine substitution and/or spin labeling. The binding assay identified multiple residues in TM1–3 but not in TM4 with reduced affinity indicating that the latter is not directly involved in substrate binding and coordination. Mutations that reduce TPP+ affinity tend to cluster on the same face of each helix spaced by 3–4 residues and facing away from the lipid phase (see below). For a subset of sites such as 40, 44, and 60, Trp quenching was restored at higher concentrations of EmrE and TPP+ suggesting that these mutants are capable of binding TPP+ albeit with lower affinity (Fig. 2). The lower affinity is presumably responsible for the loss of resistance in the cell assay of Fig. 1.
FIGURE 2.
Changes in TPP+ binding affinity of selected spin-labeled EmrE detected by the level of Trp quenching. For a number of residues (e.g. 44), binding required high concentration of EmrE and TPP+ indicating reduction in affinity due to the cysteine substitution and/or attachment of the spin label.
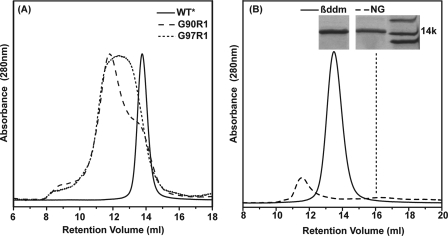
Most spin-labeled mutants assembled into dimers with similar profiles to the WT* on size exclusion chromatography (data not shown). Where detected, changes in retention times reflect formation of larger oligomers rather than dissociation into a monomer as illustrated in Fig. 3A for sites 90 and 97. Remarkably, the structural destabilization due to the substitution of these residues does not lead to loss of substrate affinity (supplemental Fig. 1).
FIGURE 3.
Analysis of EmrE dimeric assembly by size exclusion chromatography. A, spin labeling of selected residues in TM4 leads to changes in the retention time indicative of aggregation. B, detergent nonyl glucoside (NG) destabilizes the apo EmrE resulting in dissociation to a monomer. The inset is a cut out from an SDS-PAGE confirming the protein identity in each SEC peak. βddm, β-dodecyl maltoside.
The choice of detergent has a profound effect on the integrity of the EmrE dimer. Substitution of dodecyl maltoside with nonyl glucoside, the detergent used in the crystal structure determination, alters apo EmrE SEC profile particularly in pH 8 buffered solutions. Overall, there is a significant loss in protein yield accompanied by a shift of the peak to retention times indicative of dissociation to a monomer (dashed line in Fig. 3B). These results are in agreement with cross-linking analysis in this detergent suggesting destabilization of the EmrE dimer (21). They are also consistent with the conclusion of Chen et al. (17) that the crystal structure of apo EmrE in this detergent is likely to be misfolded.
Accessibility Profiles Identify Transmembrane Segments and Water-accessible Regions
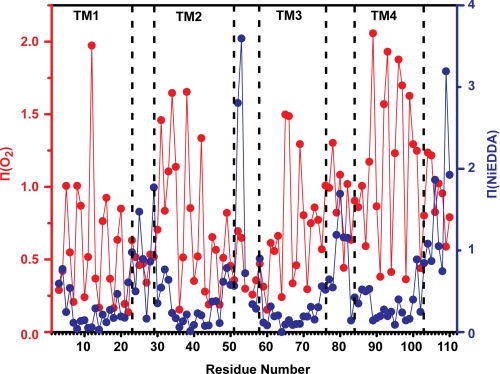
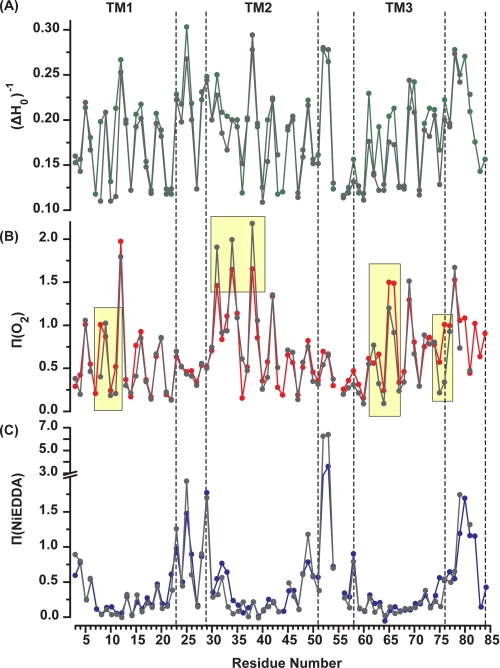
The orientation and boundaries of EmrE transmembrane helices were deduced from measurement of collision frequency of each spin-labeled site with paramagnetic probes that are either lipid-soluble (molecular oxygen) or water-soluble (NiEDDA) (24, 33). For a TM helix, the O2 concentration gradient in the bilayer leads to a signature accessibility pattern consisting of 3.6 periodicity with increasing values at maxima near the middle of the membrane. This pattern is observed along TM1, although its modifications at the N termini of TM2 and TM3 suggest specific structural features (Fig. 4). Large Π(O2) in the 30–35-amino acid stretch of residues along with an offset in the values at the minimum at residue 32 indicate loose packing of the N terminus of TM2 and/or a tilt relative to the membrane normal. In contrast, low O2 accessibilities and nonhelical periodicity at the N terminus of TM3 (residues 58–64) reflect a sterically packed environment. TM3 emerges from these contacts at residues 65 and 66 where exposure to O2 continues through the loop connecting TM3 and -4 and coincides with increased NiEDDA accessibility following residue 75.
FIGURE 4.
Accessibility profiles of apo EmrE. Red, Π(O2); blue, Π(NiEDDA). Both parameters show periodic variation as a function of residue number in the regions of the TM helices. The dashed lines indicate helix and loop boundaries on the basis of the crystal structure assignment.
The O2 accessibility profile along helix 4 deviates significantly from that of a TM helix parallel to the membrane normal. Although the values at the maxima indicate direct contacts with the lipid phase, the pattern is asymmetric, becoming successively smaller toward the C terminus. The changes coincide with attenuated contrast between the two faces of the helix and an increase in the accessibility to the water-soluble NiEDDA.
As expected, large NiEDDA accessibilities segregate predominantly to loops connecting the various TM segments. Compared with absolute values obtained in other membrane proteins such as MsbA (34), the EmrE accessibilities localize the loops to the membrane/water interface except for the amino acids 52 and 53. Measurable collisions with NiEDDA are observed at the termini of TM1 and -2 at sites of minimal oxygen exposure. They are 180° out of phase with O2 indicating water penetration to the transmembrane segment presumably near the substrate-binding site.
Spin Label Mobility Identifies Sites of Tertiary Contacts and Residues at Dimer Interfaces
EPR spectra for spin-labeled EmrE reconstituted in liposomes are shown in supplemental Figs. 2–5. The shapes of these spectra report the dynamics of the spin label side chain, i.e. its mobility, relative to the protein (22). The determinants of the rate and/or amplitude of spin label dynamics are the local steric crowding in the immediate vicinity of the spin label and the flexibility of the backbone to which it is attached. The spectra can also have contributions from broadening by short range dipolar coupling between symmetry-related spin labels in the dimer.
Line shapes characteristic of motionally restricted spin labels recur at sites within the TM helices with a characteristic 3.6 periodicity (supplemental Figs. 2–5). Together they define the surface of each TM that packs against other TM helices. On the opposite surface, the line shapes reflect highly mobile spin labels as expected at lipid-exposed sites. This motif is observed along the entire lengths of TM1 and -4, although TM2 and -3 show distinct evidence of asymmetric packing. The N-terminal two turns of TM2 (residues 30–36) have uniformly mobile EPR spectra suggesting that this segment is lipid-exposed with little tertiary contacts. In contrast, spin labels introduced at residues 56–60 of TM3 have restricted motion consistent with a sterically packed environment.
To quantitatively compare the mobility of spin labels, the inverse of the central linewidth (ΔH0)−1 was calculated from the EPR spectrum (Fig. 5A). (ΔH0)−1 is a phenomenological descriptor of mobility whose value was shown to correlate with the structural environment of the label (22). The 3.6 periodic variation in (ΔH0)−1 (Fig. 5A) reflecting alternation between buried, exposed, and tertiary contact environments is consistent with four α-helical segments. The overall mobility pattern reflects the packing characteristic of each helix discussed above.
FIGURE 5.
Sequence-specific environmental parameters in the apo (color) and TPP+ bound intermediates (gray). A, mobility parameter (ΔH0)−1. B, accessibility to molecular O2, and C, accessibility to NiEDDA. The regions of accessibility changes are highlighted in yellow.
The EPR line shapes at sites 14, 18, 40, 44, 60, and 64 have extensive broadening characteristic of strong dipolar coupling between spin labels separated by less than 15 Å. As noted previously in detergent micelles, the spectra reveal the presence of two populations, one in close proximity and one separated by more than 20 Å (19). Remarkably, these sites are located on the buried surfaces of TM1 (residues 14 and 18), TM2 (residues 40 and 44), and TM3 (residues 60 and 64) but not TM4. In TM1 and -2, they occur near the middle of the helix. Analysis of labeling efficiency using an experimentally determined extinction coefficient demonstrates that the second population does not arise from incomplete labeling (27).
Effects of TPP+ Binding
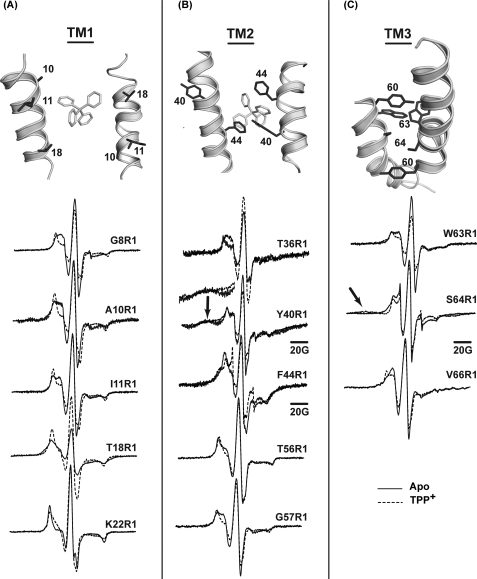
TPP+ binding leads to conformational changes that alter the environments of spin labels in TM1–3 as shown in Fig. 5 and supplemental Figs. 2–5. In TM1, line shape changes at residues N-terminal to Glu-14 (residues 8, 10, and 11) reveal an increase in motional restriction and order although those C-terminal to Glu-14 such as residues 19, 21, and 22 near the C terminus of TM1 report a decrease in steric restriction of the spin labels (Fig. 6A and supplemental Fig. 2). In parallel, TPP+ binding eliminates dipolar coupling at residue 18 arising from the packing of the two spin labels at the dimer interface. Finally, oxygen accessibilities at residues 8 and 11 are dramatically reduced indicating transition of these residues from lipid exposed to buried environments (Fig. 5B).
FIGURE 6.
EPR line shape changes upon TPP+ binding reveal changes in packing and proximity along the interfaces of TM1–3. For A–C, a close up view of the structure highlighting the spin-labeled residues is shown. A and B, the bound TPP molecule is also visible. All spectra were normalized to the same number of spins and then scaled to reveal the details of the line shapes.
To establish the specificity of TPP+-induced conformational change, we took advantage of the absolute requirement of a glutamate at position 14 for substrate binding. For this purpose, the spectral changes at residues 10 and 18 were examined in a background where Glu-14 was substituted with an alanine. The supplemental Fig. 6 shows that the mutation eliminates the spectral sensitivity to the addition of TPP+ demonstrating the lack of bulk effects on the EPR line shapes. In addition, the E14A mutation alters the EPR spectral line shape eliminating dipolar coupling at site 18. This may reflect the transition of the transporter to a different conformational state as a result of the Glu-14 substitution.
TPP+ binding induces opposite changes in O2 and NiEDDA accessibilities at the two N-terminal turns of TM2 (Fig. 5, A and B). Spin labels at residues 31, 34, and 38, already in a lipid-exposed environment, report an increase in O2 and a decrease in NiEDDA accessibility. This movement also affects the dipolar coupling between symmetry-related spin labels at sites 40 and 44, although the magnitude of the change is rather small (Fig. 6B). The width of the broad component at site 40 is reduced (arrow in Fig. 6B), although the dipolar splittings at site 44 become more prominent.
Starting at residue 56 and continuing through the N-terminal part of TM3, TPP+ binding uniformly reduces the mobility of spin labels (Fig. 6, B and C). This is consistent with the observation of a rigid loop connecting TM2 and -3 in the EM structure (16, 35). At site 64, the strength of the dipolar coupling increases resulting in a spectrum similar to that of residues 14 and 40 indicating spin labels separated by less than 10 Å. Changes in O2 accessibility are observed at a number of sites in TM3 and in the loop connecting TM3 and -4 (residues 75 and 76).
In contrast, we did not observe significant TPP+-induced changes in the EPR line shapes in TM4. Given previous evidence, this helix does not participate in substrate binding (30, 36, 37), and line shape changes were screened in detergent micelles where EmrE has been shown to be functional (supplemental Fig. 5). The lack of spectral changes upon TPP+ binding was also confirmed in liposomes for a number of residues (data not shown).
DISCUSSION
An alternating access model of vectorial transport by EmrE posits transitions between at least four intermediates. The cycle begins by binding of protons (substrate) to an outward (inward)-facing conformation and ends at either of the two putative apo intermediates. Although direct information regarding the conformational changes that mediate transition between these intermediates is lacking, most prevalent models invoke reorientation of the binding site to shuttle the protons and substrates between the two sides of the membrane (35, 38).
Structure of Apo EmrE
Given a pKa of around 8.0 for the two Glu-14 residues (12), the substrate-free (apo) intermediate investigated here corresponds to the protonated state. The EPR parameters provide constraints on the local environment, the orientation and tilt of the TM segments, and the packing of the two monomers in the dimer. TPP+-induced changes in these parameters reflect structural rearrangements induced by substrate binding.
The accessibility data clearly delineates four helices as predicted by sequence analysis and verified by the EM and crystal structures (15, 17, 39). Each of TM1–3 is directly packed against its counterpart in the dimer as deduced from dipolar coupling between spin labels in these segments. A recurrent theme in the corresponding EPR line shapes is their two-component nature reflecting at least two spin label populations only one of which is in close proximity. Because all these mutants have similar SEC profiles to the WT* with no evidence of dissociation, the second population is likely to reflect a different packing arrangement of the EmrE dimer. Remarkably, weak dipolar coupling observed in TM4 (residues 96 and 97) is not consistent with either its packing in the structures or its role as the dimerization helix of the antiparallel dimer. Although the EPR line shapes along TM4 reveal a peripheral location in direct contact with the lipid phase, the O2 accessibilities pattern suggests that TM4 is on average tilted relative to the membrane normal at a steeper angle compared with the TPP+-bound EmrE crystal structure.
TPP+-bound EmrE Intermediate
The simplest interpretation of the changes in EPR parameters along TM1 would involve a kink in the helix near residue 14 allowing tighter packing of the turns N-terminal to the critical Glu-14 but reducing steric contacts C-terminal to this residue along the lines of an alternating access mechanism. This interpretation, however, does not exclude the possibility of more complex movements. For instance, the increased steric restrictions at residues 8 and 10 and the reduced dipolar coupling at residue 18 caused by TPP+ binding may reflect limited local helix rotation. The EPR data also suggest tilting of TM2 at its two highly dynamic N-terminal turns (residues 30–40), which are already in direct contact with the lipid bilayer. TPP+ binding changes the backbone structure of TM3 and the two adjacent loops. O2 accessibility changes along TM3 indicate ordering of the helical backbone in the TPP+-bound state (Fig. 5B). Finally, the loop linking TM3 and -4 becomes more lipid-exposed, a rearrangement likely to require a dynamic and flexible TM4.
Comparison of the EPR Parameters with the Structure of the TPP+-bound EmrE
As noted above, there are multiple EmrE regions where the EPR parameters of the apo intermediate deviate from the expected environments in the crystal structure (17) and a model based on the EM structure (35). These deviations may reflect the conformational changes upon TPP+ binding or arise from differences between the structure in liposomes and in the crystal.
Overall, the spectral broadening at sites in TM1 and -2 can be reconciled with the packing geometry of these segments in the crystal structure. Although the buried environments of these sites hinder a quantitative comparison, we evaluated whether the pattern of proximity agrees with the pairwise packing of helices in the crystal structure by modeling the spin label in cones that project along the Cα–Cβ bond (40). At residue 14 in TM1, the spin labels are expected to directly point toward each other thus rationalizing the broadened population. The projection of the side chains at residue 18 is in agreement with the absence of dipolar coupling in the TPP+-bound state (Fig. 6A). Thus, the observed spectral broadening at this residue in the apo intermediate reveals structural changes in the C-terminal region of TM1 reducing the separation between the spin labels and increasing spin label mobility at residues 19, 21, and 22.
Residues 41 and 44 in TM2 are within the range of dipolar coupling, although at residue 40 one of the labels is predicted to project away from the interface (Fig. 6B). Therefore, comparison of the EPR data in the apo state with the TPP+-bound structure suggests structural changes in this part of TM2. A tilting motion away from the interface following TPP+ binding, predicted by the accessibility data, would reduce dipolar coupling at 40 as is experimentally observed.
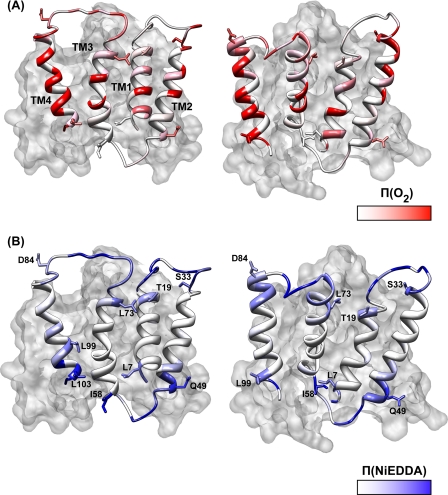
Mapping the accessibility parameters onto the crystal structure provides a local perspective on the correspondence between the crystal structure and the EPR data (Fig. 7). In general, the O2 accessibility profile along TM1 and -2 is in reasonable agreement with the disposition of the two helices in the structure. Although the comparison is complicated by the structural asymmetry between the two monomers (see Fig. 7), residues with large exposure to O2 map to the lipid-facing surfaces of TM1 and -2. Because the EPR parameters represent an average from the two spin labels, deviations from the crystal structures are not unexpected.
FIGURE 7.
Π(O2) (A) and Π(NiEDDA) (B) were mapped onto a ribbon representation of the two EmrE monomers. The second monomer is shown in a surface rendering. Selected residues from each TM are shown to provide markers and highlight the structural asymmetry between the two monomers.
The most pronounced disagreements between the EPR constraints and the crystal structure are in TM3 and -4. Previously, dipolar coupling along TM3 (residues 60 and 75) was interpreted as indicative of parallel packing of the two TM3s at the dimer interface (19). With the finding that spin labels at residue 64 have strong dipolar coupling, the data could be brought into closer agreement with an antiparallel model if the N terminus of TM3 is located well below the membrane/water interface, placing residues 60 and 64 in close proximity. We note, however, that the strong dipolar coupling predicted by the crystal structure at residue 63 was not observed experimentally. The proximities at residues 60 and 64 are not consistent with the EM-based model (35) where the N terminus of TM3 is located at opposite end of the bilayer.
Moreover, there are striking discrepancies in the orientation of the TM3 C terminus relative to the lipid phase. Residues of maximum O2 accessibility (e.g. residue 74) are buried in the crystal structure, and residues of minimal O2 accessibility are lipid-facing (Figs. 4 and 7). Although NiEDDA accessibility is observed in the 75–80-residue loop (Fig. 7B), relatively high O2 accessibility in this region also suggests exposure to the hydrocarbon phase of the bilayer. Concurrent accessibility to O2 and NiEDDA can only be rationalized by a highly dynamic backbone allowing large amplitude excursion of the loop between the aqueous and lipid phases.
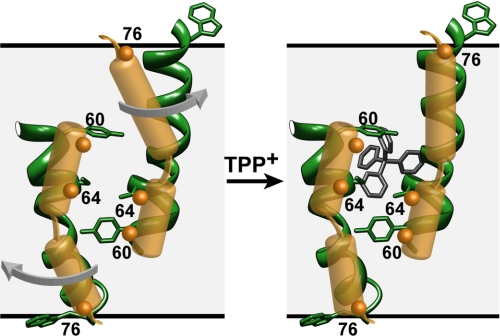
As noted by Fleishman et al. (35), TM3 contains sequences, including a GXG motif (residues 65–67), that are likely to impart a degree of flexibility and account for a kink in the EM-based model. The lack of continuous periodicity in the O2 accessibility may reflect a break in the helix that allows a dynamic C-terminal region to bend back toward the bilayer without perturbation of the tight packing at the N terminus. The EPR data along the loop connecting TM3 and -4 hints at large amplitude motion resulting in simultaneous accessibility to the bilayer and aqueous phases (Fig. 8). An ensemble of conformations in this region may have an average EPR accessibility that deviates from the unique conformation trapped in the crystal.
FIGURE 8.
Model of TM3 based on the EPR data (yellow) superimposed on a ribbon representation of the helix in the crystal structure. It assumes a 2-fold symmetry axis near the middle of the bilayer. This configuration satisfies the constraints of proximity between residues 60 and 64. The arrows indicate movement of the C-terminal tail that allows residues at the C terminus and in the loop linking TM3 to -4 to sample the membrane and aqueous environments.
Although confirming that TM4 is in direct contact with the lipid bilayers, the EPR data imply a substantial tilt of its axis relative to the bilayer normal. The decrease in the values at successive maxima and the loss of helical periodicity indicate that after residue 103 the helix does not undergo stable tertiary contacts. This latter result is not inconsistent with the TPP+-bound crystal structure where the segments following residues 99 and 104 in each monomer are not resolved. The concomitant increase in NiEDDA accessibility at this turn suggests that this part of the helix is emerging out of the membrane (Fig. 7B).
Concluding Remarks
How does the EPR data weigh in on the dual topology controversy? As noted above, we find substantial deviations between the average liposome conformation and the x-ray and EM structures. Furthermore, the persistence of second populations in the dipolar-coupled spectra reveals conformations of EmrE with different packing interfaces. Nevertheless, the pattern of proximity along TM1 and -2 suggests that one EmrE conformation is similar to the TPP+-bound crystal structure. Flexibility and dynamics are invoked to rationalize the discrepancy between the EPR data along TM3 and its crystallographic conformation. The comparison is complicated by the low resolution of the latter, which at 4 Å implies that residue assignment can be off by as much as a full helical turn. While noting this agreement, the definite test of the veracity of the crystal structure as a model of the antiparallel dimer requires verification of the helical packing in the monomer as well as long range distance measurements in the dimer (41). These experiments are currently in progress.
Vectorial transport by EmrE requires a re-orientation of the binding site from inward- to outward-facing. The structural changes that underlie the switch in accessibility have not been described experimentally, but a model has been proposed on the basis of the available structures of TPP+-bound EmrE (35). Starting from the asymmetric dimer, changes in substrate-binding site exposure require a simple interchange of the conformation of the two monomers. Consequently, the inward- and outward-facing conformations are linked by 180° rotation around the in-plane axis of symmetry. This conceptualization of alternating access implies no net change in residue environment, accessibility, or proximity across the dimer interface during the transition. Consequently, it does not predict changes in the EPR parameters as these are not affected by the symmetric exchange of spin label environments.
The results presented here demonstrate that transition from the apo- to the TPP+-bound intermediate does not follow the putative symmetric interchange described above. When mapped onto the crystal structure, the segments reporting changes in the EPR parameters appear to be distributed on both sides of the asymmetric dimer tracing a pathway through which the substrate may permeate. Specifically, the tilt at the N terminus of TM2 provides direct access to the bilayer from which hydrophobic substrates may partition. A detailed global model of this conformational transition will also benefit from long range distance measurements (42).
Supplementary Material
Acknowledgments
We thank Derek Claxton for critical reading of the manuscript and Nathan Alexander for generating the model of EmrE with side chains.
This work was supported, in whole or in part, by National Institutes of Health Grant GM077659 from the NIGMS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
- TM
- transmembrane
- TPP+
- tetraphenylphosphonium
- NiEDDA
- nickel(II) ethylenediamine diacetate
- SEC
- size exclusion chromatography
- WT*
- cysteine-less wild type.
REFERENCES
- 1.Higgins C. F. (2007) Nature 446, 749–757 [DOI] [PubMed] [Google Scholar]
- 2.Chung Y. J., Saier M. H., Jr. (2001) Curr. Opin. Drug Discov. Dev. 4, 237–245 [PubMed] [Google Scholar]
- 3.Paulsen I. T., Brown M. H., Skurray R. A. (1996) Microbiol. Rev. 60, 575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuldiner S. (2009) Biochim. Biophys. Acta 1794, 748–762 [DOI] [PubMed] [Google Scholar]
- 5.Schuldiner S. (2007) Trends Biochem. Sci. 32, 252–258 [DOI] [PubMed] [Google Scholar]
- 6.Schuldiner S., Granot D., Mordoch S. S., Ninio S., Rotem D., Soskin M., Tate C. G., Yerushalmi H. (2001) News Physiol. Sci. 16, 130–134 [DOI] [PubMed] [Google Scholar]
- 7.Schuldiner S., Lebendiker M., Yerushalmi H. (1997) J. Exp. Biol. 200, 335–341 [DOI] [PubMed] [Google Scholar]
- 8.Yerushalmi H., Mordoch S. S., Schuldiner S. (2001) J. Biol. Chem. 276, 12744–12748 [DOI] [PubMed] [Google Scholar]
- 9.Sharoni M., Steiner-Mordoch S., Schuldiner S. (2005) J. Biol. Chem. 280, 32849–32855 [DOI] [PubMed] [Google Scholar]
- 10.Elbaz Y., Tayer N., Steinfels E., Steiner-Mordoch S., Schuldiner S. (2005) Biochemistry 44, 7369–7377 [DOI] [PubMed] [Google Scholar]
- 11.Rotem D., Steiner-Mordoch S., Schuldiner S. (2006) J. Biol. Chem. 281, 18715–18722 [DOI] [PubMed] [Google Scholar]
- 12.Soskine M., Adam Y., Schuldiner S. (2004) J. Biol. Chem. 279, 9951–9955 [DOI] [PubMed] [Google Scholar]
- 13.Yerushalmi H., Schuldiner S. (2000) FEBS Lett. 476, 93–97 [DOI] [PubMed] [Google Scholar]
- 14.Muth T. R., Schuldiner S. (2000) EMBO J. 19, 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tate C. G., Kunji E. R., Lebendiker M., Schuldiner S. (2001) EMBO J. 20, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ubarretxena-Belandia I., Baldwin J. M., Schuldiner S., Tate C. G. (2003) EMBO J. 22, 6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y. J., Pornillos O., Lieu S., Ma C., Chen A. P., Chang G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18999–19004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapp M., Seppälä S., Granseth E., von Heijne G. (2007) Science 315, 1282–1284 [DOI] [PubMed] [Google Scholar]
- 19.McHaourab H. S., Mishra S., Koteiche H. A., Amadi S. H. (2008) Biochemistry 47, 7980–7982 [DOI] [PubMed] [Google Scholar]
- 20.Steiner-Mordoch S., Soskine M., Solomon D., Rotem D., Gold A., Yechieli M., Adam Y., Schuldiner S. (2008) EMBO J. 27, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soskine M., Mark S., Tayer N., Mizrachi R., Schuldiner S. (2006) J. Biol. Chem. 281, 36205–36212 [DOI] [PubMed] [Google Scholar]
- 22.Mchaourab H. S., Lietzow M. A., Hideg K., Hubbell W. L. (1996) Biochemistry 35, 7692–7704 [DOI] [PubMed] [Google Scholar]
- 23.Mchaourab H. S., Kálai T., Hideg K., Hubbell W. L. (1999) Biochemistry 38, 2947–2955 [DOI] [PubMed] [Google Scholar]
- 24.Hubbell W. L., Gross A., Langen R., Lietzow M. A. (1998) Curr. Opin. Struct. Biol. 8, 649–656 [DOI] [PubMed] [Google Scholar]
- 25.Dong J., Yang G., McHaourab H. S. (2005) Science 308, 1023–1028 [DOI] [PubMed] [Google Scholar]
- 26.Mordoch S. S., Granot D., Lebendiker M., Schuldiner S. (1999) J. Biol. Chem. 274, 19480–19486 [DOI] [PubMed] [Google Scholar]
- 27.Koteiche H. A., Reeves M. D., McHaourab H. S. (2003) Biochemistry 42, 6099–6105 [DOI] [PubMed] [Google Scholar]
- 28.Berliner L. J., Grunwald J., Hankovszky H. O., Hideg K. (1982) Anal. Biochem. 119, 450–455 [DOI] [PubMed] [Google Scholar]
- 29.Koteiche H. A., Mchaourab H. S. (2002) FEBS Lett. 519, 16–22 [DOI] [PubMed] [Google Scholar]
- 30.Elbaz Y., Salomon T., Schuldiner S. (2008) J. Biol. Chem. 283, 12276–12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yerushalmi H., Schuldiner S. (2000) J. Biol. Chem. 275, 5264–5269 [DOI] [PubMed] [Google Scholar]
- 32.Gutman N., Steiner-Mordoch S., Schuldiner S. (2003) J. Biol. Chem. 278, 16082–16087 [DOI] [PubMed] [Google Scholar]
- 33.Altenbach C., Froncisz W., Hemker R., McHaourab H., Hubbell W. L. (2005) Biophys. J. 89, 2103–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou P., McHaourab H. S. (2009) J. Mol. Biol. 393, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleishman S. J., Harrington S. E., Enosh A., Halperin D., Tate C. G., Ben-Tal N. (2006) J. Mol. Biol. 364, 54–67 [DOI] [PubMed] [Google Scholar]
- 36.Gottschalk K. E., Soskine M., Schuldiner S., Kessler H. (2004) Biophys. J. 86, 3335–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soskine M., Steiner-Mordoch S., Schuldiner S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12043–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yerushalmi H., Schuldiner S. (2000) Biochemistry 39, 14711–14719 [DOI] [PubMed] [Google Scholar]
- 39.Edwards R. A., Turner R. J. (1998) Biochem. Cell. Biol. 76, 791–797 [DOI] [PubMed] [Google Scholar]
- 40.Alexander N., Bortolus M., Al-Mestarihi A., Mchaourab H., Meiler J. (2008) Structure 16, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borbat P. P., Surendhran K., Bortolus M., Zou P., Freed J. H., McHaourab H. S. (2007) PLoS Biol. 5, 2211–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou P., Bortolus M., McHaourab H. S. (2009) J. Mol. Biol. 393, 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.