Abstract
The bacterial infected mouse model is a powerful model system for studying areas such as infection, inflammation, immunology, signal transduction, and tumorigenesis. Many researchers have taken advantage of the colitis induced by Salmonella typhimurium for the studies on the early phase of inflammation and infection. However, only few reports are on the chronic infection in vivo. Mice with Salmonella persistent existence in the gastrointestinal tract allow us to explore the long-term host-bacterial interaction, signal transduction, and tumorigenesis. We have established a chronic bacterial infected mouse model with Salmonella typhimurium colonization in the mouse intestine over 6 months. To use this system, it is necessary for the researcher to learn how to prepare the bacterial culture and gavage the animals. We detail a methodology for prepare bacterial culture and gavage mice. We also show how to detect the Salmonella persistence in the gastrointestinal tract. Overall, this protocol will aid researchers using the bacterial infected mouse model to address fundamentally important biological and microbiological questions.
Protocol
This protocol includes three portions: bacterial culture, mouse gavage, and Salmonella detection.
1. Salmonella growth condition.
Prepare the Salmonella Luria-Bertani broth (LB) plate, incubate at 37° C overnight.
Pick a clone from the LB plate and put into 7 ml LB in a 12 ml tube, and shake at 37°C for about 5 hours.
Inoculate 50 ml LB with 0.05 ml of a stationary phase culture and incubate at 37°C without shaking for about 18 hours.
Spin overnight bacterial culture at room temperature with 6000 rpm for 10 minutes, suspend the bacteria in HBSS using ratio 100:3 (LB: HBSS). For example:Every 50 ml LB culture will be suspended in 1.5 ml HBSS.
For the animal gavage, further dilute the bacterial culture at 1:10 ratio. For example:Every 1.5 ml LB culture will be suspended in 15 ml HBSS.
2. Salmonella-infection model*.
Prepare the streptomycin solution. Each mouse will be given 7.5 mg of streptomycin in 100 μl HBSS. For example, prepare total 90 mg of streptomycin in 1.2 ml HBSS for 10 mice. Always have some extra volume when preparing the streptomycin solution.
Withdraw water and food 4 hours before oral gavage treatment.
Gavage mice with streptomycin with 7.5 mg of streptomycin (100 μl of HBSS for control mice). Grab the skin over the mouse shoulder firmly with the thumb and middle finger, stretch the head and neck with the index finger to make the esophagus straight. Direct the ball-tip of the feeding needle along the roof of the mouth and toward the right side of the back of the pharynx, then gently pass down into the esophagus and inject the 100 μl solution. No resistance should be felt.
At 20 hours after streptomycin treatment, withdrawn water and food again before the mice are infected with bacteria.
Gavage each mouse with 100 μl suspension in HBSS or treated with sterile HBSS (control) by oral gavage. The gavage procedure is the same as 2.3) gavage mice with streptomycin.
*Animal experiments were performed by using specific-pathogen-free female C57BL/6 mice (Taconic, Hudson, NY) that were 6-7 weeks old as previously described1. The protocol was approved by the University of Rochester University Committee on Animal Resources (UCAR).
3. Detection of Salmonella in intestine.
Collect mouse fecal (about 100 mg).
Transfer fecal sample to a 1.5 micro centrifuge tube with 1ml PBS and vortex vigorously.
Centrifuge for 10 min at 800 rpm. Transfer the supernatant into a clean microfuge tube.
Centrifugation at 6000 rpm for 5min. Supernatant is discarded and 200 μL PBS is added to the pellets and vortexed.
Streak the 200 μl PBS with a disposable cell spreader on a BBL CHROMagar plate to detect Salmonella. Salmonella species appear mauve (rose to purple, see Fig.1). If 200 μl yields too many colonies on the plate, use 100 μl or 50 μl for the streak.
4. Representative Results.
When the protocol is done correctly, Salmonella typhimurium colonization can be detected in the mouse intestine over 6 months. Salmonella could be detected by fecal culture over 6 months (Fig.1). As a typical out come of this model, body weigh loss and death occur within 4 weeks post infection. Dependent on the Salmonella strains used for infection, some mice may no survive over 6 months.
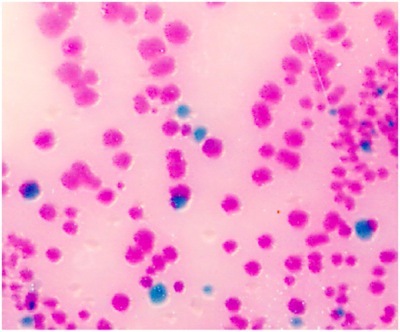 Figure 1. Intestinal Salmonella in the Salmonella species appear mauve (rose to purple) in color, due to metabolic differences in the presence of selected chromogens. Other bacteria are either inhibited or produce blue-green or colorless colonies.
Figure 1. Intestinal Salmonella in the Salmonella species appear mauve (rose to purple) in color, due to metabolic differences in the presence of selected chromogens. Other bacteria are either inhibited or produce blue-green or colorless colonies.
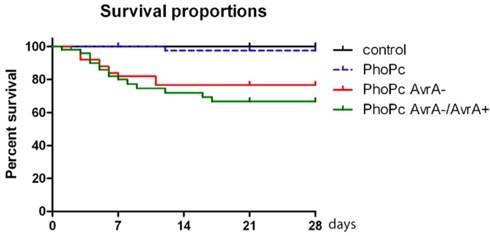 Figure 2. Survival proportions of mice infected with Salmonella mutant strain PhoPc, PhoPc AvrA-, and PhoPc AvrA-/AvrA+, PhoPc AvrA for 4 weeks (28 days).
Figure 2. Survival proportions of mice infected with Salmonella mutant strain PhoPc, PhoPc AvrA-, and PhoPc AvrA-/AvrA+, PhoPc AvrA for 4 weeks (28 days).
Table 1. Body weight of mice infected with Salmonella for 4 weeks.
| 0 | 1 week | 2 weeks | 3 weeks | 4 week | |
| control | 16.78 ± 1.05 | 16.91 ± 1.28 | 18.26 ± 1.23 | 19.31 ± 1.26 | 20.26 ± 1.15 |
| PhoPc | 16.89 ± 1.03 | 17.14 ± 1.19 | 17.43 ± 1.63* | 18.68 ± 1.78 | 20.05 ± 1.11 |
| PhoPcAvrA- | 16.91 ± 1.12 | 16.96 ± 1.39 | 17.06 ± 2.14** | 18.71 ± 2.18 | 20.15 ± 1.56 |
| PhoPc AvrA-/AvrA+ | 16.94 ± 0.96 | 17.17 ± 1.02 | 17.63 ± 1.42* | 18.44 ± 2.03 | 20.09 ± 1.17 |
*compared to control group p<0.05 ** compared to control group p<0.01
Table 2. Salmonella strains used in this study.
| Name | Description | Reference or source |
| Salmonella 14028s | Wild-type S. typhimurium | ATCC |
| PhoPc | Non-pathogenic complex regulator mutant | Miller et al., 1990 |
| PhoPc AvrA- | AvrA- mutation | Collier-Hyams et al., 2002 |
| PhoPcAvrA-/AvrA+ | PhoPcAvrA- with complemented plasmid encoding AvrA | Collier-Hyams et al., 2002 |
Discussion
To use this system, it is necessary for the researcher to learn how to gavage the animals. We detail a methodology for prepare bacterial culture and gavage the mice. We also show how to monitor the Salmonella persistence in the gastrointestinal (GI) tract. The critical steps in this protocol including:
Streptomycin-pretreatment: Streptomycin-pretreatment could get rid of some commensal gut flora and make the mice susceptible to the Salmonella infection2.
Salmonella gavage: for the beginner, the gavage could be challenging. Sometime, gavage fails because the solution is accidentally injected to the airway and causes mouse death.
Persistent bacterial colonization: need close monitoring bacteria in the GI tract. In addition to mouse fecal culture, contents of cecum can also be used for the Salmonella detection in BBL CHROMagar plates. Collect cecum contents when mice are sacrificed after Salmonella infection.
We have tested the Salmonella colonization in the mouse at different concentration: 1 x 103 colony-forming units to 1 x 108 colony-forming units (100 μl/mouse). At 1 x 106 to 10 x 108 colony-forming units, Salmonella was able to colonize the mice. Therefore, based on our previous publications1, 5 and unpublished data, the final concentration of Salmonella in this solution will not be measured before gavage.
In this experimental procedure, we used specific-pathogen-free female C57BL/6 mice (Taconic, Hudson, NY) that were 6-7 weeks old. Bacterial strains used include Salmonella typhimurium wild-type strain ATCC14028s (WT-SL), non-pathogenic Salmonella mutant strain PhoPc3, PhoPc AvrA-, and PhoPc AvrA-/AvrA+, PhoPc AvrA(table 2). Using this protocol, Salmonella typhimurium colonization can be detected in the mouse intestine over 6 months (Fig1). Salmonella infection and inflammation were measured by observing fecal matter and boy weight. When tissue samples and blood were collected, the length of intestine was measured as a feature of intestinal inflammation. The weights of spleens and livers were also examined. Mouse serum cytokine were tested by ELISA5. Further more, biochemical and molecular methods are further used to test the changes of inflammatory cytokine and pathology changes induced by Salmonella 5.
Typically, body weight loss and death occur within 4 weeks post infection (Table 1). Post Salmonella infection, there was significant body weight loss in the bacterial infected groups compared to the control group without bacterial treatment (*p< 0.05 or p<0.001 Table 1). The body weight was measured weekly. The survival proportions 4 weeks post infection were shown in Fig. 2. Overall, 92% of mice (n=50) infected with Salmonella strains PhoPc, 78% PhoPc AvrA-, or 70% PhoPc AvrA-/AvrA+ can survive with persistent Salmonella in intestine. After the acute infection, all survivors still carried Salmonella , which could be detected post infection 6 months using the method described in this proposal. Once mice survive 3 weeks post infection, mice gained body weight and less death occurred (Fig.2). Moreover, immunofluroscence staining data also show the invasion of Salmonella in intestinal mucosa 27 weeks post infection (Fig.3). These data showed that PhoPc, PhoPc AvrA-, or PhoPc AvrA-/AvrA+ can be used for the chronic infection model.
Mice with host resistance factor Nramp1+/+ are resistant to Salmonella infection 3-6. C57/BL6 mice with Nramp defective were use in the current studies. We found that 90% mice infected with pathogenic Salmonella strain 14028s cannot survive over 4 weeks (data not shown). To understand the long-term effects of wild-type S. Typhimurium infection, mice with Nampr1+/+ could be used.
This model is established in a BSL-2 laboratory. Our protocol is approved by the University of Rochester University Committee on Animal Resources (UCAR). Mice infected with Salmonella are likely to suffer discomfort and distress and become less active and move around slowly. Their fur may become ruffled and they may not feed or drink as normal. The animals will be observed closely and if any sign of discomfort such as unable to ambulate well enough to maintain hydration and caloric intake If a mouse showed indication that it had aspirated fluid or significant body weight loss (10% or more)7, and did not die immediately, the mouse was humanely euthanized. All laboratory personnel will be trained in observing the mice appropriately8, 9.
Many studies used the streptomycin-pretreatment have taken advantage of the colitis induced by Salmonella typhimurium for the studies on at the early phase of infection1,2,10. However, only few report on the chronic infection in mouse model in vivo11. Mice with Salmonella persistent existence in the GI tract allow us to explore the long-term host-bacterial interaction, signal transduction, and tumorigenesis. We have established a chronic bacterial infected mouse model with Salmonella typhimurium colonization in the mouse intestine over 6 months. This model can also be used for E.coli pathogenesis and probiotics studies. Overall, this protocol will aid researchers using the mouse model to address fundamentally important biological and microbiological questions.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by the NIDDK KO1 DK075386 grant and the American Cancer Society RSG-09-075-01-MBC to Jun Sun.
References
- Duan Y. beta-Catenin activity negatively regulates bacteria-induced inflammation. Lab Invest. 2007;87(6):613–624. doi: 10.1038/labinvest.3700545. [DOI] [PubMed] [Google Scholar]
- Barthel M. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71(5):2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Mekalanos JJ. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. Journal of bacteriology. 1990;172(5):2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez Y. Nramp1 expression by dendritic cells modulates inflammatory responses during Salmonella Typhimurium infection. Cell Microbiol. 2008;10(8):1646–1661. doi: 10.1111/j.1462-5822.2008.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H, Okamoto S, Guiney D, Gunn JS, Fierer J. A model of Salmonella colitis with features of diarrhea in SLC11A1 wild-type mice. PLoS ONE. 2008;3(2):e1603–e1603. doi: 10.1371/journal.pone.0001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL. Molecular call-and-response: how Salmonella learns the gospel from its host. Trends Microbiol. 2003;11(6):245–246. doi: 10.1016/s0966-842x(03)00127-6. [DOI] [PubMed] [Google Scholar]
- Zaharik ML, Vallance BA, Puente JL, Gros P, Finlay BB. Host-pathogen interactions: Host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15705–15710. doi: 10.1073/pnas.252415599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend P, Morton DB. Laboratory Animal Care Policies and Regulations: United Kingdom. ILAR J. 1995;37(2):68–74. doi: 10.1093/ilar.37.2.68. [DOI] [PubMed] [Google Scholar]
- Olfert ED, Godson DL. Humane endpoints for infectious disease animal models. ILAR J. 2000;41(2):99–104. doi: 10.1093/ilar.41.2.99. [DOI] [PubMed] [Google Scholar]
- Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171(3):882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassl GA, Valdez Y, Bergstrom KS, Vallance BA, Finlay BB. Chronic enteric salmonella infection in mice leads to severe and persiste nt intestinal fibrosis. Gastroenterology. 2008;134(3):768–780. doi: 10.1053/j.gastro.2007.12.043. [DOI] [PubMed] [Google Scholar]
- Sukupolvi S, Edelstein A, Rhen M, Normark SJ, Pfeifer JD. Development of a murine model of chronic Salmonella infection. Infection and immunity. 1997;65(2):838–842. doi: 10.1128/iai.65.2.838-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biosafety in Microbiological and Biomedical Laboratories (BMBL) Fifth Edition. US Department of Health and Human Services, Centers for Disease Control and Prevention and National Institutes of Health; 2007. Feb, [Google Scholar]
- Wu S, Lu R, Zhang Y, Sun J. Chronic effects of a Salmonella type III secretion effector protein AvrA in vivo. PLoS ONE. 2010 doi: 10.1371/journal.pone.0010505. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]


