Abstract
A new approach is described for imaging mass spectrometry analysis of drugs and metabolites in tissue using matrix-assisted laser desorption ionization-Fourier transform ion cyclotron resonance (MALDI-FTICR). The technique utilizes the high resolving power to produce images from thousands of ions measured during a single mass spectrometry (MS)-mode experiment. Accurate mass measurement provides molecular specificity for the ion images on the basis of elemental composition. Final structural confirmation of the targeted compound is made from accurate mass fragment ions generated in an external quadrupole-collision cell. The ability to image many small molecules in a single measurement with high specificity is a significant improvement over existing MS/MS based technologies. Example images are shown for olanzapine in kidney and liver and imatinib in glioma.
An important phase of drug discovery is determining how a potentially therapeutic compound is distributed and metabolized within the body. Traditional imaging techniques like positron emission tomography, magnetic resonance imaging, and whole-body radiography share a key advantage in their compatibility with living systems but have limited spatial resolution and require molecular or radioisotope tags that do not distinguish molecular forms having the tag. The wide applicability of imaging mass spectrometry (IMS) has been demonstrated in recent studies investigating molecular classes that include peptides,1–4 proteins,5–8 and phospholipids.9–11 IMS also permits the analysis of drugs and metabolites direct from tissue without target specific labels or reagents.12–17
IMS analyses of pharmaceutical compounds present unique analytical challenges. The low-m/z region of a matrix-assisted laser desorption ionization (MALDI) spectrum (less than m/z 1000) contains a large population of ions from compounds endogenous to the tissue sample as well as matrix-related adduct clusters and fragments. Given such a high density of ions, it is inevitable that other species will share the same nominal mass as the target compound and often, as in the case of matrix ions, at substantially different intensities. The mass analyzer must therefore be capable of distinguishing target compounds of interest from this large chemical background. The traditional strategy for MALDI IMS analyses of pharmaceutical compounds is to perform the imaging experiment in MS/MS mode. Essentially, all ions in the range of the targeted precursor are fragmented and the abundance of the target compound is determined from the measured intensity of one or more of its structurally significant fragment ions. This measurement is repeated for all pixels as well as for each precursor of interest.
High-performance instruments like the Fourier transform ion cyclotron resonance (FTICR) offer new strategies for imaging small molecules that do not rely on the imaging of fragment ions. High mass-resolution makes it possible to measure many isobaric peptide ions from tissue.3,18–20 Similarly, for small molecules, a single imaging experiment in the MS-mode can yield ion images for many ions within a narrow range of only a few hundred m/z. Further, other FTICR studies have demonstrated the use of high measurement accuracy and resolving power to determine the elemental composition of small molecules.21–25 For many analytes of interest in a small-molecule imaging experiment, the specificity of having an elemental composition will be sufficient to confirm identity. For others, subsequent MS/MS measurements can be made at discrete pixel locations to confirm the presence of a molecular structure and to determine if multiple isomers contribute to the image. In this report we describe the use of high-performance FTICR for the image analysis of drugs and metabolites from dosed tissue. Unlike traditional MS/MS imaging, this new strategy can provide ion images for the hundreds of peaks measured in a single-scan experiment.
EXPERIMENTAL SECTION
Olanzapine and imatinib tablets were obtained from the Vanderbilt University Hospital Pharmacy. All animal studies were approved by the Institutional Animal Care and Use Committee at Vanderbilt University.
Olanzapine Animal Model
OLZ was administered (p.o.) at pharmacologically relevant doses (8 mg/kg) to 10 week-old male Fischer 344 rats. Animals were euthanized at 2 h postdose by isoflurane anesthesia followed by exsanguinations via decapitation. Liver and kidney were collected and frozen on powdered dry ice. The abundance of OLZ in the two tissues was quantified at 1778 ng/g for liver and 41123 ng/g for kidney.26
Intracranial Tumor Model
Six-week-old, male, C57BL6 mice were anesthetized with 100 mg/kg ketamine and 5 mg/kg xylazine. A 0.5 mm burr hole was drilled in the right forebrain, approximately 4 mm lateral from the midline and 1 mm anterior to bregma. Cancer cells, suspended in 0.1 mL of medium, were injected 3 mm deep (from the skull surface) into the burr hole. Tumors were allowed to grow over a period of 14 days. The mice received 3 mg of Imatinib in 100 mL of PBS administered by esophageal gavage. Two hours after treatment, the mice were euthanized via ketamine/xylazine anesthesia followed by decapitation. Brains were excised and frozen on powdered dry ice.
Tissue sections (12 μm thick) were cut on a cryostat, thaw-mounted onto gold-plated stainless-steel plates, and stored in a vacuum desiccator until analyzed. In order to avoid extracting the dosed analyte, no washing or fixation of the tissue was done. α-Cyano-4-hydroxycinnamic acid (HCCA) matrix was dissolved in water/acetonitrile (1:1) to a concentration of 10 mg/mL. Arrays of matrix microdroplets were applied using a Portrait 630 (Labcyte Inc., Sunnyvale CA). The arrays were deposited in a pattern of 1 drop/spot repeated 40 times at a pitch of 350 μm.
Digital images of the prepared sections were acquired using a flatbed scanner prior to MALDI analysis. Mass spectra were acquired using a 9.4 T Apex Qe (Bruker Daltonics, Billerica MA) equipped with an Apollo II dual ion source and a 355 nm solid-state laser focused to a diameter of 250 μm. Data were acquired and analyzed using the FlexImaging application. Mass spectra were accumulated from 3 scans of 20 laser shots acquired from each matrix spot without rastering, thereby establishing the spatial resolution of the ion images at 350 μm. Ion fragmentation for MS/MS analysis was done in the external quadrupole prior to their measurement in the ICR cell.
RESULTS AND DISCUSSION
Olanzapine (OLZ) is prescribed for the treatment of mood disorders and is absorbed rapidly and transported via plasma.27 Pharmacokinetic studies have shown OLZ accumulates in a number of organs including brain, kidney, liver, and spleen, with the latter two organs containing highest amounts.28 A recent IMS study of OLZ in whole, dosed rat sections15 measured similar accumulation patterns from a series of MS/MS ion images and served as reference for the present study. OLZ abundance in liver is quite high, and from previous imaging studies of proteins, this tissue is known to exhibit a high degree of molecular homogeneity. Kidney contains a lower abundance of the drug but offers distinct internal differentiation of medulla and cortex. Both tissues were collected from a dosed rat and analyzed by FTICR imaging to determine the presence and distribution of the drug and the two metabolites. The composition and exact mass of these compounds are listed in Table 1.
Table 1.
| compound | elemental formula (M + H) | exact mass | prominent fragment ions, m/z |
|---|---|---|---|
| olanzapine (OLZ) | C17H21N4S | 313.1481 | 256.0903, 282.1060 |
| N-desmethyl OLZ | C16H19N4S | 299.1325 | 256.0903, 282.1060 |
| 2-hydroxymethyl OLZ | C17H21N4OS | 329.1431 | 272.0852, 298.1009 |
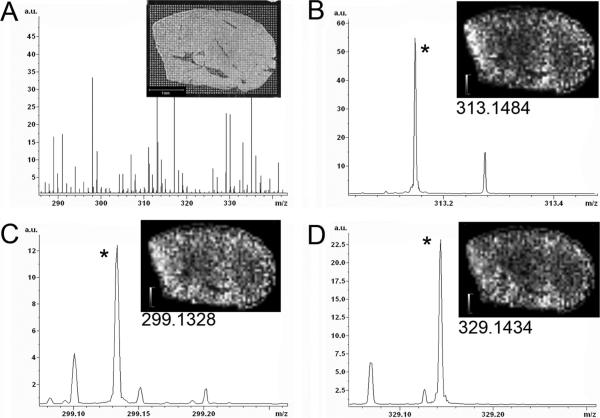
Data acquired from dosed rat liver are presented in Figure 1. The average of all spectra acquired from the section is shown in Figure 1a. A digital image of the liver section with microspotted matrix array is shown as an inset. MALDI spectra in this m/z range are quite complex and this is evident by the 8–10 isobaric peaks detected at each nominal m/z. Many of the more intense ions are related to the matrix rather than the tissue, and without the high resolving power these peaks would obscure neighboring peaks of lesser intensity at the same nominal m/z. Expanded views about the nominal m/z of the three OLZ compounds are shown in parts b–d of Figure 1. Each region contains multiple isobaric ions. The base peak of each region has a measured m/z that is within 1 ppm of the formula mass of the targeted OLZ compounds. Respective high resolution m/z ion images for each of these peaks are shown as insets to the spectra. Images were generated by integrating each peak using a 20 ppm wide window which is twice the measured peak resolution of 10 ppm (full width at half-maximum). The distribution of each compound was nearly uniform over the section, consistent with the homogeneous nature of liver.
Figure 1.
FTICR images from a liver of a rat dosed with 8 mg/kg olanzapine (2 h postdose). (A) Average of all spectra collected from matrix spots on the tissue (inset image). The m/z region of the average spectrum in the regions of protonated molecule of (B) olanzapine, (C) desmethyl metabolite, and (D) hydroxymethyl metabolite. Ion images of each marked peak is inset to each spectrum.
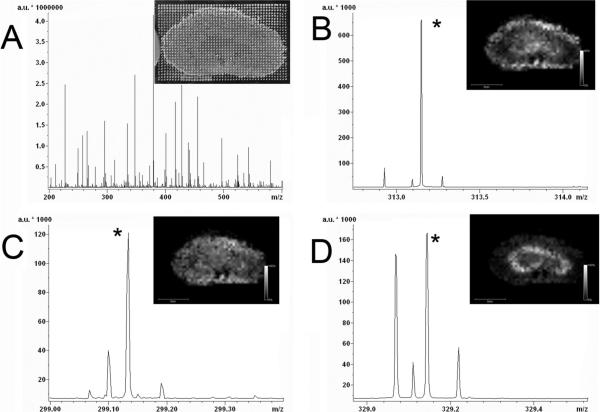
Images acquired from dosed kidney also contained intense ions within 1 ppm of the theoretical m/z of the three OLZ compounds. Ion images and spectral data from this section are shown in Figure 2. As before, ion images were extracted from the spectra using a 20 ppm integration window. Unlike liver, kidney exhibits a higher degree of differentiation for the drug and metabolites. Images indicate that these compounds accumulate in different regions of the kidney. For example, the intact drug, OLZ, appears most abundant along the outer cortex while the oxidized metabolite appears most abundant along the outer medulla. Similar OLZ distributions have been observed from this sample using traditional MS/MS imaging techniques, but such molecular-specific distribution information is not achievable with autoradiography imaging.29
Figure 2.
FTICR images from the kidney of a rat dosed with 8 mg/kg olanzapine (2 h postdose). (A) Average of all spectra collected from matrix spots on the tissue (inset image). The m/z region of the average spectrum in the regions of protonated molecule of olanzapine (B), metabolite (C), and hydroxymethyl metabolite (D). Ion images of each marked peak is inset to each spectrum.
The high-mass measurement accuracy in conjunction with isotopic profiles allows the elemental composition of these particular ions to be determined. The current software for image analysis is restricted to an external mass calibration. Over the course of several imaging experiments, the typical measurement error for externally calibrated images within the range of 300–500 m/z is ~1.5 ppm. However with custom scripting, spectra from individual pixels can be calibrated internally. The matrix ions, [M + Na]+, [M2 + H]+, and [M2 + H − H2O]+ were used to internally calibrate each spectrum using a two-factor linear function. The average and standard deviations for the measured m/z of the three OLZ target ions and matrix dimer calibrant are presented in Table 2. Combinations of the elements C, N, H, O, and S were identified that had formula weights within the standard deviation of the ion images. Only one combination for each image, matching the formulas in Table 1, was identified within tolerance of the calibration.
Table 2.
| ion | average m/z | std deviation |
|---|---|---|
| OLZ | 313.1475 | 0.000 271(0.87ppm) |
| N-desmethyl OLZ | 299.1325 | 0.000 195(0.65ppm) |
| 2-hydroxymethyl OLZ | 329.1419 | 0.000 209(0.63ppm) |
| HCCA dimer, [M2 + H]+ | 379.0934 | 0.000 261(0.69ppm) |
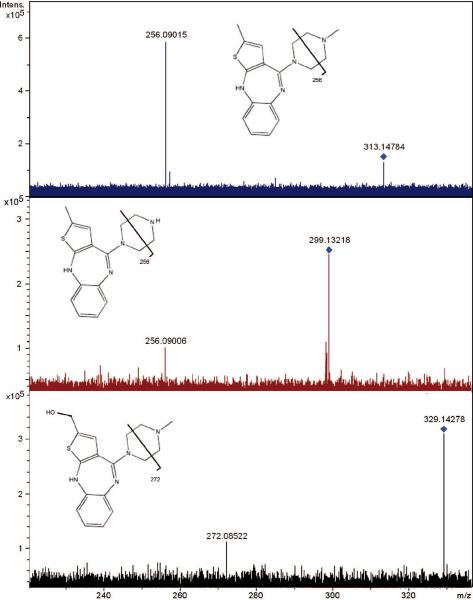
Establishment of an ion's identity requires confirmation of molecular structure using MS/MS analysis. Although an elemental composition can be established, the complex nature of biological samples requires some consideration that the ion images are representative of structural isomers of the OLZ compounds. A survey of the CAS Registry (Chemical Abstracts Service) revealed more than 100 compounds that share each formula listed in Table 1. An undetermined number of these are naturally occurring, and their presence should be considered. It is common practice in MALDI analysis of small molecules to infer the abundance of a specific compound from the measurement of one or more structurally specific fragment ions. Traditional IMS analysis of drugs relies on such measurements, made for each precursor ion at each pixel location. Similarly, MS/MS measurements can also be used to identify ion images from FTICR. However, it could be argued that the elemental composition is sufficiently specific that fragment ions need not be acquired from each pixel. Rather, molecular identity of an ion can be confirmed by acquiring MS/MS spectra from various locations in the sample. Figure 3 shows MS/MS spectra from the three ions that were acquired from liver. All three spectra contain fragments consistent with the structures of OLZ and metabolites. Just as importantly, fragment ions that cannot be attributed to OLZ or its metabolites are not observed.
Figure 3.
CID spectra of the proposed olanzapine (A), desmethyl metabolite (B), and hydroxymethyl metabolite (C) ions. The fragment ions observed correspond to known transitions of the respective structures.
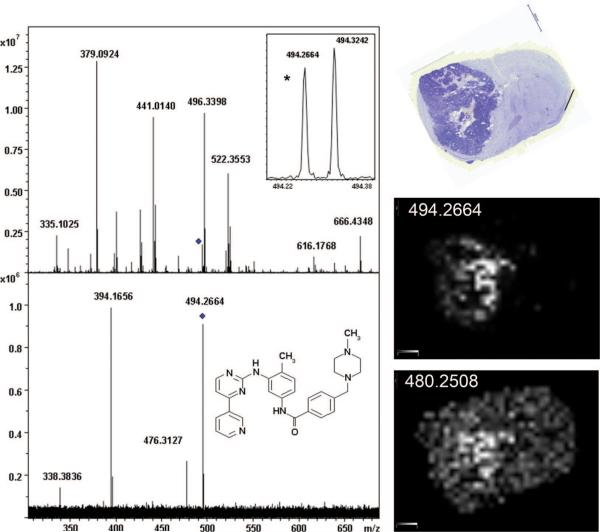
The antitumor drug, imatinib, was imaged from a mouse brain glioma. The glioma is clearly evident in the stained section shown in Figure 4, encompassing nearly half of the brain. The average spectrum is shown at the top of Figure 4 with an inset showing the m/z region of the protonated molecule of the drug. In this spectrum, ions were observed at m/z 494.2664 and 480.2506, potentially the [M + H]+ for imatinib and the desmethyl metabolite. Considering the computed m/z for protonated imatinib is 494.2663, this corresponds to a measurement error of <0.5 ppm. Images for the two ions are seen in Figure 4. With the use of the strategy described above, the elemental composition for each ion was determined to correspond to those of the respective compound. The measured ion images support this by showing both ions to be most abundant within the tumor. Consistent results have been observed in other IMS studies of this drug. Further, the fact that the image shows almost none of the presumed imatinib ion distributed outside of the glioma indicates that this ion is not endogenous to nontumor brain. The two isobaric ions of nominal m/z 494 were analyzed by MS/MS to confirm the presence of imatinib. The spectrum, shown in Figure 4, contains a strong ion at m/z 394.1656. Imatinib readily loses methylpiperazine to form an ion at m/z 394.1662. The fragment ion observed at m/z 476 can be attributed to a loss of water from the protonated molecule. The fragment ion at m/z 338 has not been observed previously from imatinib30 and is most likely originating from the isobaric ion detected at m/z 494.3242. The presumed metabolite peak was not measured with sufficient intensity to carry out MS/MS confirmation.
Figure 4.
FTICR images of imatinib and des-methyl metabolite from glioma/mouse brain (cresyl violet stained). Top spectrum: average of all spectra with expanded view of protonated molecule region (inset). Bottom spectrum: CID fragments from the nominal proposed imatinib ion.
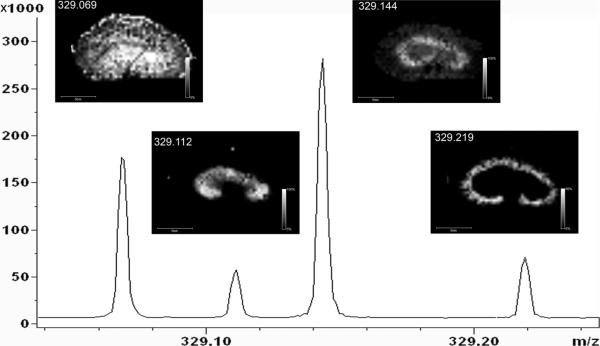
The ability to image many species from a single acquisition is a significant asset. Traditional MS/MS imaging suffers from many of the same shortcomings as imaging techniques like radiography and PET, namely, the technique monitors only a select few species that must be known a priori. The high resolving power of the FTICR allows thousands of ion images to be generated. Although the specificity of the examples presented here is high, it must nevertheless be acknowledged that the FTICR imaging strategy does not offer the same molecular specificity as achieved by acquiring an MS/MS spectrum at every pixel. In many cases, however, the specific elemental composition of an ion can sufficiently identify known compounds, particularly when the same ions are not observed from control samples. The ability to analyze thousands of ion images from a single data set promises to be a significant benefit. An example of high-mass-resolution imaging is shown in Figure 5. Here, the four major ions observed at the nominal m/z of the 2-hydroxymethyl metabolite of OLZ, detected within a range of 200 mDa, are sufficiently resolved that the respective ion images show clearly the unique distribution of each species. Collecting such images has not been possible before and makes FTICR an enabling platform from new studies for more extensive probing of metabolic processes.
Figure 5.
FTICR images of hydroxymethyl OLZ and three isobaric ions.
CONCLUSION
Image analysis using MALDI-FTICR is still early in development. A key factor which has hindered its application to imaging studies is an inherently low throughput. For example, the studies presented above contained 1000–1700 pixels and the typical imaging speed was ~4 pixels/min. This is dramatically slower than imaging systems based on time-of-flight (TOF) mass analyzers which are capable of 30–50 pixels/min. Fundamental factors such as longer detection time are necessary for high resolving power, but currently the majority of the time is consumed within the control electronics which will undoubtedly improve as the application matures. Improvements are also needed to increase the efficiency of handling, transfer, and processing of very large data sets which can be as much as an order of magnitude larger than similar TOF images.
High performance instruments like FTICR can offer a new perspective for MALDI imaging of drugs and metabolites. Imaging small molecules with FTICR provides many of the original experimental advantages of IMS analyses that are lost with the MS/MS strategy, that is, the simultaneous imaging of many ions without any prior knowledge of the target species. In this study, the analyses were directed toward specific drug and metabolite compounds. However, with the ability to generate ion images from the hundreds to thousands of peaks measured in a single-scan experiment, it is now possible to simultaneously investigate other small molecules that may be involved in critical metabolic processes
ACKNOWLEDGMENT
The authors gratefully acknowledge financial support from NIH, Grant 5R01-GM058008-08, and DoD, Grant W81XWH-05-1-0179
Footnotes
NOTE ADDED AFTER ASAP PUBLICATION There was an error in Figure 3 in the version published ASAP June 20, 2008; the corrected version published ASAP July 14, 2008.
References
- (1).Drexler DM, Garrett TJ, Cantone JL, Diters RW, Mitroka JG, Conaway MCP, Adams SP, Yost RA, Sanders M. J. Pharmacol. Toxicol. Methods. 2007;55:279–288. doi: 10.1016/j.vascn.2006.11.004. [DOI] [PubMed] [Google Scholar]
- (2).DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li LJ. J. Proteome Res. 2007;6:1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Taban IM, Altelaar AFM, Van der Burgt YEM, McDonnell LA, Heeren RMA, Fuchser J, Baykut G. J. Am. Soc. Mass Spectrom. 2007;18:145–151. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- (4).Verhaert PD, Conaway MCP, Pekar TM, Miller K. Int. J. Mass Spectrom. 2007;260:177–184. [Google Scholar]
- (5).Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. J. Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- (6).Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. J. Proteome Res. 2007;6:1295–1305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- (7).Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. Nat. Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- (8).Meistermann H, Norris JL, Aerni HR, Cornett DS, Friedlein A, Erskine AR, Augustin A, Mudry MCD, Ruepp S, Suter L, Langen H, Caprioli RM, Ducret A. Mol. Cell. Proteomics. 2006;5:1876–1886. doi: 10.1074/mcp.M500399-MCP200. [DOI] [PubMed] [Google Scholar]
- (9).Shimma S, Sugiura Y, Hayasaka T, Hoshikawa Y, Noda T, Setou M. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2007;855:98–103. doi: 10.1016/j.jchromb.2007.02.037. [DOI] [PubMed] [Google Scholar]
- (10).Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, Woods AS. J. Mass Spectrom. 2007;42:1093–1098. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Bird NC, Atkinson SJ, Clench MR, Mangnall D, Majeed AW. Clin. Exp. Metastasis. 2007;24:275–275. [Google Scholar]
- (12).Stoeckli M, Staab D, Schweitzer A. Int. J. Mass Spectrom. 2007;260:195–202. [Google Scholar]
- (13).Hsieh Y, Casale R, Fukuda E, Chen JW, Knemeyer I, Wingate J, Morrison R, Korfmacher W. Rapid Commun. Mass Spectrom. 2006;20:965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- (14).Hsieh Y, Chen J, Korfmacher WA. J. Pharmacol. Toxicol. Methods. 2007;55:193–200. doi: 10.1016/j.vascn.2006.06.004. [DOI] [PubMed] [Google Scholar]
- (15).Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Anal. Chem. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- (16).Reyzer ML, Caprioli RM. Curr. Opin. Chem. Biol. 2007;11:29–35. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- (17).Signor L, Varesio E, Staack RF, Starke V, Richter WF, Hopfgartner G. J. Mass Spectrom. 2007;42:900–909. doi: 10.1002/jms.1225. [DOI] [PubMed] [Google Scholar]
- (18).Kutz KK, Schmidt JJ, Li LJ. Anal. Chem. 2004;76:5630–5640. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- (19).Stemmler EA, Cashman CR, Messinger DI, Gardner NP, Dickinson PS, Christie AE. Peptides. 2007;28:2104–2115. doi: 10.1016/j.peptides.2007.08.019. [DOI] [PubMed] [Google Scholar]
- (20).Stemmler EA, Hsu YWA, Cashman CR, Messinger DI, de la Iglesia HO, Dickinson PS, Christie AE. Gen. Comp. Endrocrinol. 2007;154:184–192. doi: 10.1016/j.ygcen.2007.06.025. [DOI] [PubMed] [Google Scholar]
- (21).Grange AH, Brumley WC. J. Am. Soc. Mass Spectrom. 1997;8:170–182. [Google Scholar]
- (22).Grange AH, Osemwengie LI, Brilis GM, Sovocool GW. Environ. Forensics. 2001;2:61–74. [Google Scholar]
- (23).Sleighter RL, Hatcher PG. J. Mass Spectrom. 2007;42:559–574. doi: 10.1002/jms.1221. [DOI] [PubMed] [Google Scholar]
- (24).Tolmachev AV, Monroe ME, Jaitly N, Petyuk VA, Adkins JN, Smith RD. Anal. Chem. 2006;78:8374–8385. doi: 10.1021/ac0606251. [DOI] [PubMed] [Google Scholar]
- (25).Tremblay LB, Dittmar T, Marshall AG, Cooper WJ, Cooper WT. Mar. Chem. 2007;105:15–29. [Google Scholar]
- (26).Khatib-Shahidi S. Ph.D. Dissertation. Vanderbilt University; Nashville, TN: 2007. [Google Scholar]
- (27).Chiu JA, Franklin RB. J. Pharm. Biomed. Anal. 1996;14:609–615. doi: 10.1016/0731-7085(95)01651-1. [DOI] [PubMed] [Google Scholar]
- (28).Aravagiri M, Teper Y, Marder SR. Biopharm. Drug Dispos. 1999;20:369–377. doi: 10.1002/1099-081x(199911)20:8<369::aid-bdd200>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- (29).Chay SH, Herman JL. Arzneim.-Forsch./Drug Res. 1998;48:446–454. [PubMed] [Google Scholar]
- (30).Marc Marull BR. J. Mass Spectrom. 2006;41:390–404. doi: 10.1002/jms.1002. [DOI] [PubMed] [Google Scholar]