Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) remains an important cause of cancer death. Changes in apoptosis signaling in pancreatic cancer result in chemotherapy resistance and aggressive growth and metastasizing. The aim of this study was to characterize the apoptosis pathway in pancreatic cancer computationally by evaluation of experimental data from high-throughput technologies and public data bases. Therefore, gene expression analysis of microdissected pancreatic tumor tissue was implemented in a model of the apoptosis pathway obtained by computational protein interaction prediction.
Methodology/Principal Findings
Apoptosis pathway related genes were assembled from electronic databases. To assess expression of these genes we constructed a virtual subarray from a whole genome analysis from microdissected native tumor tissue. To obtain a model of the apoptosis pathway, interactions of members of the apoptosis pathway were analysed using public databases and computational prediction of protein interactions. Gene expression data were implemented in the apoptosis pathway model. 19 genes were found differentially expressed and 12 genes had an already known pathophysiological role in PDAC, such as Survivin/BIRC5, BNIP3 and TNF-R1. Furthermore we validated differential expression of IL1R2 and Livin/BIRC7 by RT-PCR and immunohistochemistry. Implementation of the gene expression data in the apoptosis pathway map suggested two higher level defects of the pathway at the level of cell death receptors and within the intrinsic signaling cascade consistent with references on apoptosis in PDAC. Protein interaction prediction further showed possible new interactions between the single pathway members, which demonstrate the complexity of the apoptosis pathway.
Conclusions/Significance
Our data shows that by computational evaluation of public accessible data an acceptable virtual image of the apoptosis pathway might be given. By this approach we could identify two higher level defects of the apoptosis pathway in PDAC. We could further for the first time identify IL1R2 as possible candidate gene in PDAC.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the 8th most common cancer in the western world [1]. Its mortality almost equals its incidence rate of 6.3/100,000 [2]. Despite combined modality therapy pancreatic carcinoma shows a unsatisfactory response to treatment [3]. Recently, a comprehensive genomic analysis of Jones et al. could identify apoptosis as a core signaling pathway in pancreatic cancer. The pathway was genetically altered in most of 24 primary pancreatic cancer cell lines [4]. Clinicopathologically, this defective apoptosis signaling contributes to the tumor's poor response to chemotherapy, ionizing radiation and immunotherapy [5] and affects the metastasizing capacity and growth rate of the tumor [6], [7]. Therefore, understanding of apoptosis resistance is a prerequisite for improving cancer therapy.
Apoptosis, or cell death program, can be activated by various mechanisms within the extrinsic and the intrinsic pathway. While activation of cell death receptors leads to the engagement of the extrinsic pathway, the intrinsic pathway is activated by mitochondria during cellular stress, both resulting in an activation of caspases [8].
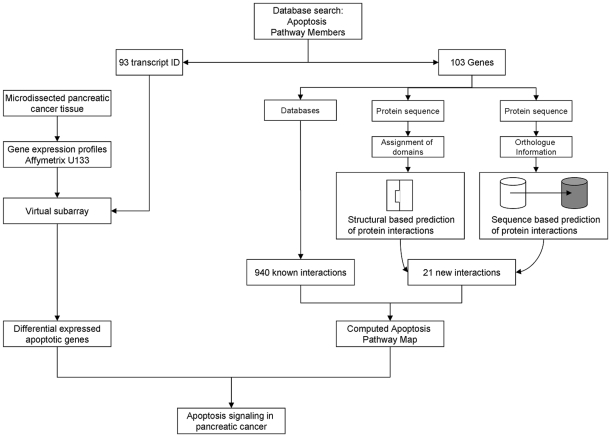
Today, the apoptosis pathway is one of the best investigated intracellular pathways. However, interpretation of experimental data is hindered by the multitude of signaling molecules and complex interactions of the pathway. In this study we tried to approach the cell death pathway in pancreatic cancer by a computational analysis of experimental data from highthroughput technologies and public databases. We tried to use the great amount of information to model the intracellular information flow of the apoptosis pathway in pancreatic cancer. For a graphic display of the study design see Figure 1 .
Figure 1. Graphic display of the study design.
The implementation of gene expression data into a model of the apoptosis pathway obtained by protein interaction databases and protein interaction prediction showed a consistent pattern of higher-level defects in the intrinsic pathway and on the level of cell death receptors that can potentially result in the phenotype of apoptosis resistance in pancreatic cancer.
Results
Computational construction of the apoptosis pathway map
Interactions of the 103 apoptosis associated genes from our database search were initially evaluated by screening of protein-protein interaction databases. The search resulted in 940 known interactions. Those interactions represented experimentally proven interactions between defined proteins. This data was used to construct a pathway map, as mentioned above ( Figure 2 ).
Figure 2. Pathway map of the apoptosis pathway.
The nodes in these graphs represent receptors, ligands, effectors, kinases and transcription factors, while each edge describes a relation between these species. In the upper part of the figure the direct apoptosis induction is shown (A), whereas in the lower part the modulation through gene expression is depicted (B). Black interactions signify known protein interactions from databases. For better view we did not display all of the 940 known interactions, please see File S3 for a list of all interactions. Blue edges signify computationally predicted interactions for all 103 apoptosis-associated genes with a high evidence level.
In a second step we tried to find previously unknown interactions between the 103 apoptosis-associated genes. We therefore had to assign structural families to the gene products because most of the structures were previously unknown. The structural assignment and family classification for the apoptotic associated genes resulted in the assignment of 53 genes. Applying the interface conservation evaluation to possible interactions between the products of those 53 genes resulted in 21 novel interactions (for examples see File S2 , for whole data see File S3 ).
Those novel interactions represent putative interactions which are not yet experimentally proven. All new interactions were implemented in the first map of the cell death pathway ( Figure 2 ).
GeneChip results
We constructed a virtual subarray to identify gene expression changes of the 93 apoptotic genes for which an identifier could be obtained. To evaluate the performance of this approach we compared the results for the apoptotic gene set with whole genome and virtual subarray analysis. Of 23 probe sets identified with the virtual subarray analysis only 18 were detected in the whole genome analysis. The mean expression intensities of the probesets detected only by subarray analysis was 125 compared to 346 for probe sets detected with both methods, indicating that the virtual subarray analysis was more sensitive. The 23 probesets represented 19 differentially expressed genes ( Figure 3 ). Of these, 11 genes were overexpressed and 8 were underexpressed in PDAC compared to microdissected normal ductal cells. Among the nineteen genes, 12 were already reported by other groups in PDAC ( Table 1 ).
Figure 3. Analysis of apoptosis-associated gene expression in PDAC.
Heat map of 19 microdissected PDACs (marked red), 13 samples of microdissected normal ductal cells (marked green), and 13 established pancreatic tumor cell lines (marked magenta) using the 93 differential gene set and a Euclidian distance matrix. Normal stromal cells served as internal quality control (marked blue).
Table 1. Results of the GeneChip analysis.
| No. | Affy ID | Name | Gene Symbol | Function | FC | Ref. in PDAC |
| Upregulated genes | ||||||
| 13 | 205403_at | IL1-R2 | IL1R2 | Decoy-receptor | 7.1 | |
| 36 | 202094_at 202095_s_at | Survivin/BIRC5 | BIRC5 | Apoptosis inhibitor | 5.0 | [19], [32] |
| 14 | 219423_x_at 210847_x_at | DR3 | TNFRSF25 | Cell death receptor | 4.1 | [10] |
| 65 | 204285_s_at 204286_s_at | NOXA | PMAIP1 | MP | 3.1 | |
| 38 | 220451_s_at | Livin/BIRC7 | BIRC7 | Apoptosis inhibitor | 2.9 | |
| 56 | 241722_x_at | Mcl-1 | MCL1 | MP | 2.8 | [33], [34] |
| 18 | 1729_at | TRADD | TRADD | Signal molecule | 2.4 | [35] |
| 21 | 201587_s_at | Irak, pelle | IRAK1 | Signal molecule | 2.2 | [35] |
| 10 | 206467_x_at | DcR3 | TNFRSF6B | Decoy receptor | 2.1 | [36], [37] |
| 72 | 226530_at | BMF | BMF | MP | 2.1 | |
| 29 | 220034_at | Irak, pelle | IRAK3 | Signal molecule | 2.0 | [35] |
| Downregulated genes | ||||||
| 81 | 210657_s_at | ARTS | SEPT4 | MP | 0.49 | |
| 2 | 201466_s_at | AP-1 | JUN | Signal molecule | 0.48 | [38] |
| 35 | 225858_s_at | XIAP | BIRC4 | Apoptosis inhibitor | 0.48 | [19], [32] |
| 4 | 207643_s_at | TNF-R1 | TNFRSF1A | Cell death receptor | 0.46 | [37] |
| 68 | 201848_s_at 201849_at | BNIP3 | BNIP3 | MP | 0.37 | [39], [40] |
| 45 | 240437_at | Caspase 9 | CASP9 | Protease | 0.27 | [41] |
| 23 | 205558_at | TRAF 6 | TRAF6 | Signal molecule | 0.19 | |
| 76 | 211152_s_at | HtrA2/Omi | HTRA2 | MP | 0.13 | |
Upregulated genes (fold-change >2, q<5%) are listed in the upper part, the downregulated genes in the lower part of the table. The numbers represent the order of the genes in our supplementary data 2 (MP = mitochondrial protein; FC = Fold change).
Verification of differential expression
We selected two genes, Livin/BIRC7 and IL1R2, for validation by quantitative RT-PCR and/or immunohistochemistry. We confirmed an significant upregulation in PDAC cells or IL1R2 (p = 0.035) and LIVIN/BIRC7 (p = 0.01) ( Figure 4 A,B). Parallel to RT-PCR 16 samples from patients with PDAC and 16 normal pancreatic tissues were stained for Livin/BIRC7. 89% of the PDAC cells were tested positive for Livin/BIRC7, in contrast to only 62% of the of the normal ductal cells (p = 0.001). PDAC tissue showed also more intensive staining than normal tissue, and the results were statistically significant (p<0.001) ( Figure 4 C,D ).
Figure 4. Validation of differential expressed genes by quantitative RT-PCR.
The graphs display the results of the quantitative RT-PCR in normal tissue of the pancreas and pancreatic adenocarcinoma of Livin/BIRC7 (t-test with p = 0.01) (A) and IL1R2 (t-test with p = 0.035) (B). Immunohistochemical staining for Livin/BIRC7 in benign pancreas and invasive adenocarcinoma. Pancreatic carcinoma (arrow) showing intensive cytoplasmic staining (original magnification x100)(C). Benign ductal epithelium shows a noticeable fainter staining (arrow) (original magnification ×40)(D). * indicates p-value <0.05.
Discussion
Pancreatic cancer is a malignancy with very poor prognosis and no significant improvement in therapy over the last 30 years [1]. Recently, a comprehensive genomic analysis identified apoptosis as a core signaling pathway in pancreatic cancer [4].
The apoptosis pathway is one of the best investigated intracellular pathways. However, the pathway comprises a multitude of signaling molecules and displays complex interactions. This leaves results of experimental studies hard to interpret. In this study we tried to approach the cell death pathway in pancreatic cancer by a computational analysis of experimental data from high-throughput technologies and by evaluation of public databases. With the help of these technologies we tried to better understand the great amount of information and to make statements about disturbances in the information flow in the apoptosis pathway in pancreatic cancer. Our results were compared to previous publications on apoptosis in pancreatic cancer.
103 apoptosis associated genes were identified by database search. To assess interactions between the identified apoptosis associated genes we evaluated databases and computationally predicted protein interactions. The fact that apoptosis pathway is one of the best known pathways was mirrored by the great amount of experimental proven protein interactions in public databases. Our approach yielded 940 interactions and we could further identify 21 previously unknown interactions computationally by the sequence-based and structure-based prediction of protein interactions. Especially MCL-1, CASP 3, TRADD, SEPT 4, AIF, CALM and TRAF 6 showed branchings in the signaling cascade previously unknown. Although we could not experimentally proof those interactions, this is an evidence for the complexity of the information flow within this pathway. By setting cell death receptors as starting point for the signaling pathway we constructed a model of the pathway to visualize the interactions.
Gene expression analysis of the apoptosis-associated genes was performed using a virtual subarray in a set of microdissected tissue from normal pancreatic ducts and PDAC. Comparing the data from the subarray with whole genome analysis revealed considerably more probe sets to be differentially expressed using the virtual subarray approach. Interestingly the probe sets not identified by whole genome analysis displayed lower expression values, demonstrating that the construction of a virtual subarray might result in an enhanced sensitivity for detection at the lower end of gene expression intensities. This is mainly due to the smaller number of probe sets tested, reducing the possible noise of fluctuation during the analysis.
Gene expression analysis showed 19 differentially expressed genes. Of these, 11 genes were overexpressed and 8 were underexpressed in PDAC compared to microdissected normal ductal cells. Among the nineteen genes, 12 were already reported by other groups in PDAC.
Survivin, Livin, MCL-1, and DcR3 were upregulated and showed very good accordance to previous reports (see File S1 ). TNF-R1, BNIP3, and Caspase 9 were downregulated, those genes also showed good accordance to previous studies (see File S1 ).
However, XIAP, a member of the IAP family of proteins was downregulated in our analysis contrary to previous studies. This discrepancy might be due to tumor heterogeneity, a fundamental facet of all solid tumors [9]. It might also be due to the differences in study designs, because most of the previous studies on XIAP used pancreatic carcinoma cell lines, while we used microdissected native tumor tissue.
However, our data yielded interesting result, as we found two major foci of dysregulations within the cell death pathway.
One of the major foci was at the level of cell receptors. Generally, there seems to be a downregulation of cell death receptors, and an upregulation of decoy-receptors. The downregulation of the cell death receptor TNRF-1 and the upregulation of the Fas-decoy receptor DcR3 in our data was already reported earlier [10]. This dysregulation is meant to help the tumor evade the immune system, because of the diminished sensibility towards apoptotic ligands [11], [12].
Another decoy receptor, IL1R2, was chosen for further validation, because upregulation of this receptor was not reported previously. By means of quantitative RT-PCR we could for the first time validate an upregulation of IL1R2 in pancreatic cancer. IL1, the ligand of IL1R2 is known to be secreted by pancreatic cancer cells [13]. It has important physiological functions in inflammation and proliferation but can also trigger apoptosis through activation of IRAK and MyD88 [14], [15], [16]. While the microenvironment could benefit from the angiogenetic and the proliferative properties of IL1, the decoy-receptor might protect pancreatic cancers from apoptosis induced by the immune response [17].
The second major focus of dysregulations was found on the level of post-mitochondrial regulatory proteins, the inhibitors of apoptosis proteins (IAPs). This group of proteins inhibits the function of caspases and the apoptosome and thereby interferes both with the extrinsic and the intrinsic pathway. The IAPs are already known for their important role in carcinogenesis of other tumor entities and also PDAC [18], [19]. In our study, we found an upregulation of Survivin/BIRC5 and Livin/BIRC7 in microdissected tumor-tissue and we could validate the dysregulation of Livin/BIRC7 by quantitative RT-PCR and IHC.
The dysregulations in the group of IAPs might have a high clinical relevance, because the intrinsic pathway normally mediates the cytotoxic effect of irradiation and many chemotherapeutics [20], [21].
The computational analysis of the apoptosis pathway in PDAC thereby rendered a good accordance of our results with previous experimental references on apoptosis in pancreatic cancer. Using existing raw data from high-throughput technologies, we could partly reproduce experimental data. This data was put in the context of the complex intracellular apoptosis signaling by computational interaction analysis. Although a great amount of information can be assessed fast and descriptive by our approach, it is economically challenging to experimentally prove the findings. This must be considered a major disadvantage of our approach.
In Conclusion, the present study shows that by computational evaluation of data from gene expression analysis and public databases an acceptable virtual image of the apoptosis pathway might be given. Comparison of our data to previous publications rendered good accordance. By this approach we could identify defects at the level of cell death receptors and the inhibitor of apoptosis proteins, which might underlie the phenotype of distinct apoptosis resistance in PDAC. We could further for the first time identify IL1R2 as possible candidate gene.
Materials and Methods
Interaction prediction of the apoptosis pathway members
The apoptosis pathway related genes were assembled from electronic databases, such as the Kyoto Encyclopedia of Genes and Genomes (www.genome.ad.jp/kegg), Gene Data Base of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) and GeneMAPP (www.genmapp.org). Keywords for the search were “apoptosis”, “cell death”, “cell death pathway”, “cell death receptor” (see File S1 ).
To evaluate interactions of the apoptosis associated proteins we initially queried databases with known protein-protein interactions such as NetPro (www.molecularconnections.com), SCOPPI (www.scoppi.org) and HPRD (www.hprd.org).
To find novel interactions we used two different methods. First, we used the structure-based prediction of protein interactions (see File S2 ). Most of the 103 apoptosis-associated genes were of unknown structure. We initially used the Genomic Threading Database (GTD) as a fold recognition method to assign structural families to the gene products [22]. Domains of proteins were then defined by the Structural Classification of Proteins, SCOP. Two domains are considered interacting if there are at least 5 residue pairs within 5 Å, in accord to the interface definitions [23]. Only domain-assignments with certain and high confidence by GTD were considered. To predict potential interactions of two given domains we then used SCOPPI [24]. This database provided evident domain-domain interactions, which served as structural templates for our original assigned domains. Two proteins are considered interacting if each contains a domain where there is a structural evidence for such a domain-domain interaction according to SCOPPI. The potential interactions were evaluated by analysis of the interface conservation. Information of the residues in the interface was again obtained from the SCOPPI database. The original protein sequence was aligned against the SCOPPI template sequence, a conservation of more than 30% of the interface residues was assumed to be sufficient to share the same interaction partner.
Second, we used a sequence-based prediction of protein interactions (see File S2 ). Therefore, we used NetPro, an expert curated and annotated database containing around 100,000 protein-protein interactions, for the prediction of an interaction of our proteins in question. Using this orthologous information and BLAST we searched for homologous interactions (>80% sequence identity) for a given protein pair. We only provide new interactions which were not confirmed before with NetPro or HPRD [25], [26]. To construct our pathway map, we set the cell death receptors as starting points of the signaling cascade. Interacting proteins were defined as downstream signaling proteins. Proteins which are known cell death receptor ligands were displayed as extra-cellular proteins.
Gene expression analysis
For the construction of the virtual subarray data sets E-MEXP-950 and E-MEXP-1121 was used [27], [28].
Affymetrix probe set identifiers were obtained from Ensembl resulting in 189 probeset identifiers for 93 genes. For 10 genes no identifier could be obtained (see File S1 ). The Cel Files obtained from the Affymetrix MAS 5.0 software were used for further analysis. The Cel Files were loaded into dChip2006 (http://www.dchip.org), then normalized, and expression values were calculated using the PM/MM model. The expression values of the 189 probesets were exported and further explored using SAM (http://www-stat.stanford.edu/~tibs/sam/) and Excel (Microsoft, Redmond, WA). We scored genes as differentially expressed if they met the following criteria: a fold change >2 and a q value <5%. Heat maps were generated using dChip.
Reverse Transcription Polymerase Reaction (RT-PCR)
1 ng of cDNA was used for a TaqMan assay (Applied Biosystems, Weiterstadt, Germany). The genes were amplified with a TaqMan Universal PCR Master Mix according to the manufacturer's instructions, with an ABI PRISM 5700 Sequence Detection System using gene specific primer and probes. Gene expression was quantified by the comparative cT-Method, normalizing cT-values to a housekeeping gene (β-actin) and calculating the relative expression values using the following primers: RT-PCR: BIRC7/Livin: ACT GAC CAG CCC TGA TTC C and CTC CAG GGA AAA CCC ACT TT; Actin: AAG CCA CCC CAC TTC TCT CTA A and AAT GCT ATC ACC TCC CCT GTG T; IL1R2: ATC AGC TTC TCT GGG GTC AA and GGT AGG CGC TCT CTA TGT GG [29].
Immunohistochemistry
For immunohistochemistry, a tissue microarray (TMA) containing 16 PDAC samples was constructed. Of this TMA 5 µm sections were prepared using silanized slides (Menzel Gläser, Braunschweig, Germany). Immunohistochemistry for Livin/BIRC7 was performed using the streptavidin-biotin-peroxidase method as described previously and antigen retrieval was carried out in a microwave oven (250 W for 30 min in a citrate solution pH 6.0) [30], [31]. The primary antibody used was a mouse monoclonal antibody against the Livin/BIRC7 protein (#40958, Active Motif, Rixensart, Belgium). Normal colon mucosa and colorectal carcinoma were used as a positive control. As a negative control specimens were incubated without the primary antibody. Afterwards the slides were briefly counterstained with hematoxylin. Stained and unstained PDAC cells or ductal cells were counted and the ratio was generated. The staining intensity was evaluated semi-quantitatively by one pathologist (A.H.) without knowledge of the histopathologic and molecular data in 3 grades (negative, moderate and strong).
Statistical analysis
For statistical analysis the t test and the chi square-test of “SPSS 13.0” for Windows were used.
Literature search
To better assess changes in gene expression seen in our data, we conducted a comprehensive literature search on molecular defects of apoptosis in pancreatic cancer. Keywords were the name of the gene or protein together with the term “pancreatic carcinoma”, “pancreatic cancer”, “pancreas cancer” or “pancreatic ductal adenocarcinoma”. Included were studies on the level of the genome, gene expression and protein/functional studies on tumor tissue and/or cell lines. The literature search comprised publications until November 2009. See also File S1 .
Supporting Information
Data from our comprehensive literature search for the role of apoptosis-associated genes in pancreatic cancer. The table displays all genes, which were considered in our study. Please note that for most of the studies on the level of DNA, which means mutational studies, no quantitative statement concerning expression was made. Included were studies on the level of the genome, gene expression and protein/functional studies on tumor tissue and/or cell lines. The literature search comprised publications until December 2009. (--/- = less/slightly less expression than in normal tissue; +/- = expression depending on sample/cell-line, no general statement possible; 0 = no difference of expression to normal tissue/normal function of protein in experimental studies; ++/+ = higher/slightly higher expression than in normal tissue; ? = no quantitative statement in this study.)
(1.49 MB DOC)
Characteristics of our protein interaction prediction (A). Examples of structural models of three possible new interactions in the cell death pathway (more than 30% sequence and interface identity). The structural alignment between template and interacting protein structures is <2 Angstrom. 1 = Arts-Apollon; 2 = p16-ERK; 3 = p16-JNK (B).
(2.16 MB PPT)
Protein interactions from our database search and our interaction prediction analysis. Sheet one shows already known interactions from our database search. Sheet two shows putative interactions, proposed by our interaction prediction model.
(0.09 MB XLS)
Acknowledgments
This study was supported by Deutsche Krebshilfe and the MedDrive38 program of the medical faculty of Technische Universität Dresden. We like to thank Beatrix Jahnke for technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by Deutsche Krebshilfe and the MedDrive38 program of the medical faculty of Technische Universität Dresden. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Lowenfels AB, Maisonneuve P. Epidemiology and risk factors for pancreatic cancer. Best Pract Res Clin Gastroenterol. 2006;20:197–209. doi: 10.1016/j.bpg.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Wolff RA, Chiao P, Lenzi R, Pisters PW, Lee JE, et al. Current approaches and future strategies for pancreatic carcinoma. Invest New Drugs. 2000;18:43–56. doi: 10.1023/a:1006383831045. [DOI] [PubMed] [Google Scholar]
- 4.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JM, Attardi LD. The role of apoptosis in cancer development and treatment response. Nat Rev Cancer. 2005;5:231–237. doi: 10.1038/nrc1560. [DOI] [PubMed] [Google Scholar]
- 6.Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupnow BA, Knox SJ. The role of radiation-induced apoptosis as a determinant of tumor responses to radiation therapy. Apoptosis. 1999;4:115–143. doi: 10.1023/a:1009675028784. [DOI] [PubMed] [Google Scholar]
- 8.Rückert F, Pilarsky C, Saeger H, Grützmann R. Apoptotic Signaling in Pancreatic Cancer – Therapeutic Application. Current Cancer Therapy Reviews. 2009;5:122–133. [Google Scholar]
- 9.Heppner GH. Tumor heterogeneity. Cancer Res. 1984;44:2259–2265. [PubMed] [Google Scholar]
- 10.Ringel B, Ibrahim SM, Kohler H, Ringel J, Koczan D, et al. Apoptotic molecules in pancreatic carcinoma cell lines. Ann N Y Acad Sci. 1999;880:175–178. doi: 10.1111/j.1749-6632.1999.tb09521.x. [DOI] [PubMed] [Google Scholar]
- 11.Bernstorff WV, Glickman JN, Odze RD, Farraye FA, Joo HG, et al. Fas (CD95/APO-1) and Fas ligand expression in normal pancreas and pancreatic tumors. Implications for immune privilege and immune escape. Cancer. 2002;94:2552–2560. doi: 10.1002/cncr.10549. [DOI] [PubMed] [Google Scholar]
- 12.Ungefroren H, Voss M, Bernstorff WV, Schmid A, Kremer B, et al. Immunological escape mechanisms in pancreatic carcinoma. Ann N Y Acad Sci. 1999;880:243–251. doi: 10.1111/j.1749-6632.1999.tb09529.x. [DOI] [PubMed] [Google Scholar]
- 13.Arlt A, Vorndamm J, Muerkoster S, Yu H, Schmidt WE, et al. Autocrine production of interleukin 1beta confers constitutive nuclear factor kappaB activity and chemoresistance in pancreatic carcinoma cell lines. Cancer Res. 2002;62:910–916. [PubMed] [Google Scholar]
- 14.Dupraz P, Cottet S, Hamburger F, Dolci W, Felley-Bosco E, et al. Dominant negative MyD88 proteins inhibit interleukin-1beta/interferon-gamma -mediated induction of nuclear factor kappa B-dependent nitrite production and apoptosis in beta cells. J Biol Chem. 2000;275:37672–37678. doi: 10.1074/jbc.M005150200. [DOI] [PubMed] [Google Scholar]
- 15.Ruckdeschel K, Mannel O, Schrottner P. Divergence of apoptosis-inducing and preventing signals in bacteria-faced macrophages through myeloid differentiation factor 88 and IL-1 receptor-associated kinase members. J Immunol. 2002;168:4601–4611. doi: 10.4049/jimmunol.168.9.4601. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida Y, Kumar A, Koyama Y, Peng H, Arman A, et al. Interleukin 1 activates STAT3/nuclear factor-kappaB cross-talk via a unique TRAF6- and p65-dependent mechanism. J Biol Chem. 2004;279:1768–1776. doi: 10.1074/jbc.M311498200. [DOI] [PubMed] [Google Scholar]
- 17.Matsuo Y, Sawai H, Funahashi H, Takahashi H, Sakamoto M, et al. Enhanced angiogenesis due to inflammatory cytokines from pancreatic cancer cell lines and relation to metastatic potential. Pancreas. 2004;28:344–352. doi: 10.1097/00006676-200404000-00025. [DOI] [PubMed] [Google Scholar]
- 18.Wright CW, Duckett CS. Reawakening the cellular death program in neoplasia through the therapeutic blockade of IAP function. J Clin Invest. 2005;115:2673–2678. doi: 10.1172/JCI26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopes RB, Gangeswaran R, McNeish IA, Wang Y, Lemoine NR. Expression of the IAP protein family is dysregulated in pancreatic cancer cells and is important for resistance to chemotherapy. Int J Cancer. 2007. [DOI] [PubMed]
- 20.Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25:4798–4811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- 21.Schniewind B, Christgen M, Kurdow R, Haye S, Kremer B, et al. Resistance of pancreatic cancer to gemcitabine treatment is dependent on mitochondria-mediated apoptosis. Int J Cancer. 2004;109:182–188. doi: 10.1002/ijc.11679. [DOI] [PubMed] [Google Scholar]
- 22.Jones DT. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J Mol Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- 23.Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. A dataset of protein-protein interfaces generated with a sequence-order-independent comparison technique. J Mol Biol. 1996;260:604–620. doi: 10.1006/jmbi.1996.0424. [DOI] [PubMed] [Google Scholar]
- 24.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein-protein interfaces. Nucleic Acids Res. 2006;34:D310–314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, et al. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grutzmann R, Pilarsky C, Ammerpohl O, Luttges J, Bohme A, et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays. Neoplasia. 2004;6:611–622. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pilarsky C, Ammerpohl O, Sipos B, Dahl E, Hartmann A, et al. Activation of Wnt signalling in stroma from pancreatic cancer identified by gene expression profiling. J Cell Mol Med. 2008;12:2823–2835. doi: 10.1111/j.1582-4934.2008.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fink L, Seeger W, Ermert L, Hanze J, Stahl U, et al. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 30.Stoehr R, Wissmann C, Suzuki H, Knuechel R, Krieg RC, et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84:465–478. doi: 10.1038/labinvest.3700068. [DOI] [PubMed] [Google Scholar]
- 31.Karikari CA, Roy I, Tryggestad E, Feldmann G, Pinilla C, et al. Targeting the apoptotic machinery in pancreatic cancers using small-molecule antagonists of the X-linked inhibitor of apoptosis protein. Mol Cancer Ther. 2007 doi: 10.1158/1535-7163.MCT-06-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogler M, Durr K, Jovanovic M, Debatin KM, Fulda S. Regulation of TRAIL-induced apoptosis by XIAP in pancreatic carcinoma cells. Oncogene. 2007;26:248–257. doi: 10.1038/sj.onc.1209776. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto Y, Hosotani R, Wada M, Lee JU, Koshiba T, et al. Immunohistochemical analysis of Bcl-2, Bax, Bcl-X, and Mcl-1 expression in pancreatic cancers. Oncology. 1999;56:73–82. doi: 10.1159/000011933. [DOI] [PubMed] [Google Scholar]
- 34.Shi X, Liu S, Kleeff J, Friess H, Buchler MW. Acquired resistance of pancreatic cancer cells towards 5-Fluorouracil and gemcitabine is associated with altered expression of apoptosis-regulating genes. Oncology. 2002;62:354–362. doi: 10.1159/000065068. [DOI] [PubMed] [Google Scholar]
- 35.Grutzmann R, Saeger HD, Luttges J, Schackert HK, Kalthoff H, et al. Microarray-based gene expression profiling in pancreatic ductal carcinoma: status quo and perspectives. Int J Colorectal Dis. 2004;19:401–413. doi: 10.1007/s00384-003-0563-3. [DOI] [PubMed] [Google Scholar]
- 36.Tsuji S, Hosotani R, Yonehara S, Masui T, Tulachan SS, et al. Endogenous decoy receptor 3 blocks the growth inhibition signals mediated by Fas ligand in human pancreatic adenocarcinoma. Int J Cancer. 2003;106:17–25. doi: 10.1002/ijc.11170. [DOI] [PubMed] [Google Scholar]
- 37.Bai J, Sui J, Demirjian A, Vollmer CM, Jr, Marasco W, et al. Predominant Bcl-XL knockdown disables antiapoptotic mechanisms: tumor necrosis factor-related apoptosis-inducing ligand-based triple chemotherapy overcomes chemoresistance in pancreatic cancer cells in vitro. Cancer Res. 2005;65:2344–2352. doi: 10.1158/0008-5472.CAN-04-3502. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto S, Tomita Y, Hoshida Y, Morooka T, Nagano H, et al. Prognostic significance of activated Akt expression in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2004;10:2846–2850. doi: 10.1158/1078-0432.ccr-02-1441. [DOI] [PubMed] [Google Scholar]
- 39.Mahon PC, Baril P, Bhakta V, Chelala C, Caulee K, et al. S100A4 contributes to the suppression of BNIP3 expression, chemoresistance, and inhibition of apoptosis in pancreatic cancer. Cancer Res. 2007;67:6786–6795. doi: 10.1158/0008-5472.CAN-07-0440. [DOI] [PubMed] [Google Scholar]
- 40.Akada M, Crnogorac-Jurcevic T, Lattimore S, Mahon P, Lopes R, et al. Intrinsic chemoresistance to gemcitabine is associated with decreased expression of BNIP3 in pancreatic cancer. Clin Cancer Res. 2005;11:3094–3101. doi: 10.1158/1078-0432.CCR-04-1785. [DOI] [PubMed] [Google Scholar]
- 41.Mori T, Doi R, Toyoda E, Koizumi M, Ito D, et al. Regulation of the resistance to TRAIL-induced apoptosis as a new strategy for pancreatic cancer. Surgery. 2005;138:71–77. doi: 10.1016/j.surg.2005.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data from our comprehensive literature search for the role of apoptosis-associated genes in pancreatic cancer. The table displays all genes, which were considered in our study. Please note that for most of the studies on the level of DNA, which means mutational studies, no quantitative statement concerning expression was made. Included were studies on the level of the genome, gene expression and protein/functional studies on tumor tissue and/or cell lines. The literature search comprised publications until December 2009. (--/- = less/slightly less expression than in normal tissue; +/- = expression depending on sample/cell-line, no general statement possible; 0 = no difference of expression to normal tissue/normal function of protein in experimental studies; ++/+ = higher/slightly higher expression than in normal tissue; ? = no quantitative statement in this study.)
(1.49 MB DOC)
Characteristics of our protein interaction prediction (A). Examples of structural models of three possible new interactions in the cell death pathway (more than 30% sequence and interface identity). The structural alignment between template and interacting protein structures is <2 Angstrom. 1 = Arts-Apollon; 2 = p16-ERK; 3 = p16-JNK (B).
(2.16 MB PPT)
Protein interactions from our database search and our interaction prediction analysis. Sheet one shows already known interactions from our database search. Sheet two shows putative interactions, proposed by our interaction prediction model.
(0.09 MB XLS)