Overview
Odors play an important role in our quality of life and well being, a role that is compromised by the effect of disease, drugs, aging, and environmental onslaught on the olfactory epithelium (OE). Olfactory receptor neurons (ORNs) in OE provide the primary input to the olfactory system, such that disruption of the cellular processes leading to the activation of the ORNs inevitably compromises olfactory function. The organization of the olfactory periphery in mammals is emerging to be more complex than originally thought. An often underappreciated aspect of that complexity is that odorants have 2 odorant-specific modes of signaling, that is, they inhibit as well as excite the ORNs. The ubiquity of this phenomenon across diverse animal species suggests that having opponent input into the olfactory system is fundamental to how olfaction works, yet in contrast to our canonical understanding of how odorants excite mammalian ORNs, our understanding of how odorants inhibit these cells is still unclear. Odorants potentially can inhibit ORNs through multiple not necessarily mutually exclusive mechanisms but compelling new data argue that phosphoinositide (PI) signaling at least in part mediates inhibitory odorant input to mammalian ORNs. The idea that inhibitory input is mediated through a distinct input pathway argues for the fundamental importance of inhibitory input to olfactory coding in mammals and identifies an additional possible cellular basis for olfactory dysfunction.
Emerging complexity in the olfactory periphery
There is a growing interest in the organizational complexity of the peripheral olfactory system. Emerging evidence for organizational complexity in the peripheral olfactory systems of diverse animals suggests that it is a general property of olfaction. Lobsters, for example, express at least 2 functional subpopulations of ORNs; canonical phaso-tonically discharging cells comingle with a subpopulation of rhythmically bursting ORNs that are entrained by odorants in a concentration-dependent manner (Bobkov and Ache 2007). Insects express at least 2 subpopulations of ORNs, each expressing a different type of olfactory receptor (OR). ORNs in one type of sensillum express homologs of ionotropic glutamate receptors (Benton et al. 2009), whereas those in other olfactory sensilla express members of a unique family of 7-transmembrane ORs that function as receptor channels (Benton et al. 2006; Sato et al. 2008; Smart et al. 2008; Wicher et al. 2008).
The mammalian sense of smell is organized into functional subsystems in addition to the main OE, including the vomeronasal organ, the septal organ of Masera, the Grueneberg ganglion, and the trigeminal system—noses within noses (Munger 2009). Functionally different subsets of cells occur in the OE itself in addition to the canonical ORNs. These so far include transient amino acid receptor (TAAR)-expressing neurons, GC-D neurons, transient receptor potential (TRP)-expressing cells, and V1R-expressing cells. Organizational complexity in the mammalian olfactory periphery has been the topic of a number of recent reviews (Breer et al. 2006; Ma 2007; Munger et al. 2009) so it won’t be detailed here beyond noting that more likely remains to be learned. For example, to what extent does organizational complexity extend to the canonical ORNs themselves? Proteomic and gene expression analyses of mammalian olfactory cilia reveal that elements of numerous known signaling and regulatory pathways occur in olfactory cilia, that is, the transduction compartment, which at least sets the molecular stage for more complex regulation of the output of these cells than is typically appreciated (Klimmeck et al. 2008; Mayer et al. 2008, 2009; McClintock et al. 2008).
Odorants inhibit as well as excite ORNs
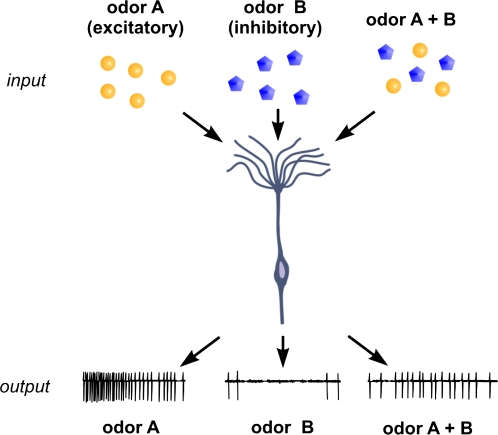
One aspect of organizational complexity is already known to be inherent in canonical mammalian ORNs. ORNs in many animals, including mammals, report to the central nervous system (CNS) with 2 odorant-specific modes of signaling (review: Ache and Young 2005). Simply said, ORNs can be excited by one odorant and inhibited by another (Figure 1). We refer to such odorant-specific opponent input to mammalian ORNs as “inhibition,” although other terminology such as suppression, hypoadditivity, and masking have been used to describe odorant-evoked decreases in the output in vertebrate ORNs (e.g., Kurahashi et al. 1994; Sanhueza et al. 2000; Duchamp-Viret et al. 2003; Takeuchi et al. 2009).
Figure 1.
Diagram of a hypothetical mammalian ORN that is excited by odor A, inhibited by odor B, and intermediately excited by a mixture of odor A and B. Note that the output of such an ORN would integrate the relative proportion of odors A and B. This figure appears in color in the online version of Chemical Senses.
There are several important functional implications of having ORNs report to the CNS with 2, odorant-specific modes of signaling. One is that the molecular receptive range (MRR) of ORNs has to include odorants that inhibit the cell, not just those that excite it. As many ORNs have relatively low levels of spontaneous activity, the MRR of an ORN or, more correctly, the MRR of the OR it expresses is biased toward odorants that excite the cell when tested individually, as it typically the case in these determinations and doesn’t reflect the true MRR of the cell. Another implication is that ORNs often respond less (or even not at all) to mixtures of odorants than they do to one or more of the components of the mixture, even when adjusted for the concentration-response function of the cell, a phenomenon known as mixture suppression. Mixture suppression has, among other things, practical consequences when searching for odor-responsive neurons using complex mixtures because it can actually reduce the number of odor-responsive units found. Most importantly, having ORNs report to the CNS with 2, odorant-specific modes of signaling implies that ORNs are actually capable of integrating the signal they send to the brain (e.g., Sanhueza et al. 2000; Duchamp-Viret et al. 2003; Ache and Young 2005). Figure 1 can be used to illustrate the integrative potential of ORNs by considering how the output of the hypothetical ORN would vary in response to mixtures of different ratios of the excitatory and inhibitory odorants. Although there is some evidence for ephaptic interaction among mammalian primary sensory ORNs at high stimulus concentrations (Scott and Sherrill 2008), mammalian ORNs, as held for ORNs in general (e.g., Dobritsa et al. 2003), are thought to project independently to the CNS without synaptic interaction in the periphery. Thus, the integrative properties of the cell presumably are inherent in the OR and the transduction machinery of the cell.
Unfortunately, this important aspect of olfaction often gets relegated to the back burner for reasons that are unclear, especially in regards to mammalian olfaction. Combinatorial coding is generally agreed to be the basis of odorant recognition and discrimination (e.g., Malnic et al. 1999), and in combinatorial coding, the absence or reduction of a signal is as meaningful as the presence of one. The low spontaneous activity of ORNs is often used to argue the difficulty of even conceiving inhibitory input because inhibition couldn’t be transmitted to the CNS. Yet, single odorants can inhibit the spontaneous activity of mammalian ORNs recorded extracellularly under physiological conditions in vivo in freely breathing rats (Duchamp-Viret et al. 1999) and can hyperpolarize mouse ORNs in loose-patch recordings (Delay and Restrepo 2004), both findings that counter this argument. Moreover, given the large convergence ratios in the olfactory bulb, even an extremely small bias in the inherent discharge of each ORN of a given type could potentially be multiplied many fold postconvergence. Because most natural odorants are complex mixtures, however, the probability is high that any one cell is coactivated by multiple odorants, allowing inhibition to be expressed not only or even primarily by modulating spontaneous activity by rather by tempering the net excitation of the cell, as illustrated in Figure 1. Although the incidence of inhibition in mammalian ORNs is often reported to be low (e.g., Duchamp-Viret et al. 1999), suggesting that it could be relatively unimportant even if it played a role in coding, the incidence has not been rigorously tested. The low levels of spontaneous activity of ORNs make it difficult to detect reductions in activity in the absence of concurrent excitation. Inhibition is relatively common in Drosophila ORNs, for example, where the incidence of spontaneous activity is higher (Hallem and Carlson 2006). In order to more accurately determine the true incidence of inhibition, it is important to screen odorant pairs or mixtures not single odorants. Most importantly, perhaps, odorant-specific opponent input presumably is fundamental to olfactory signaling because it occurs in most species of animals with defined olfactory systems and even in unicellular animals that lack systems-level organization (review: Ache and Young 2005).
Potential mechanisms of odorant-evoked inhibitory signaling
We have yet to fully understand how inhibitory signaling occurs in mammalian ORNs or indeed in any ORN. One cannot exclude that odorants interact in the perireceptor milieu, for example, by one odorant changing the affinity of another for a soluble odorant-binding proteins (OBPs), but there is no evidence to support that idea to date, and many relevant experiments are done in physiological saline lacking OBPs. Alternatively, odorants potentially could inhibit ORNs by directly blocking one or more elements involved in activating the cell because odorants can block both voltage-gated channels (Sanhueza and Bacigalupo 1999) and the olfactory cyclic nucleotide-gated (CNG) channel (Chen et al. 2006) in vertebrate ORNs. Although direct blockade of ion channels could potentially confound experiments targeted at understanding inhibitory input, it is unclear how direct blockade of such channels could account for odorant-specific reductions in output because canonical ORNs in mammals are all assumed to express the same or similar complement of voltage-gated channels, as well as the olfactory CNG channel.
Given that odorants typically are complex mixtures, inhibition could also result from weak agonists (antagonists) competing with strong agonists for the same OR, that is, from competitive inhibition. For many years, olfactory antagonists were unknown, but the revelation that mammalian ORs were members of the G protein–coupled receptor (GPCR) superfamily (Buck and Axel 1991) stimulated the search for compounds or odorants that would interact competitively with mammalian ORs. Several studies (Firestein and Shepherd 1992; Spehr et al. 2003; Araneda et al. 2004; Oka et al. 2004; Abaffy et al. 2007) have now identified odorants that can induce the parallel, saturating right shift in the dose-response function of the output of native ORNs or heterologous cells expressing ORs that is strongly suggestive of competitive antagonism (e.g., Tallarida and Jacob 1979). Although the exact mechanism underlying this interaction remains to be elucidated, it appears that odorants can inhibit the output of ORNs by competing for the same OR.
As mentioned earlier, single odorants can inhibit the spontaneous output of ORNs, which clearly cannot reflect competitive inhibition unless contaminating background odorants were driving the ongoing discharge (always a possibility given the difficulty of rigorously controlling odorant stimulation). Single odorants can also inhibit the activation of rodent ORNs by forskolin (Ukhanov K, personal communication). Because forskolin activates adenylyl cyclase independently of the OR, this finding excludes the possibility of competitive inhibition. A recent attempt to model the experimentally determined output of mammalian ORNs to binary odor mixtures finds that almost 50% of the cells don’t fit a model in which the odorants compete for a common binding site but rather are better fit by a model in which noncompetitive interaction modulates the OR (Rospars et al. 2008). These findings collectively argue for one or more noncompetitive mechanisms of inhibition.
In Drosophila at least, it is clear that inhibition is a property of a particular odorant–OR combination and not a property of the cell per se because expressing a nonnative OR in an “empty” ORN confers upon the ORN the odorant-evoked inhibition observed in the donor ORN that normally expresses that OR (Hallem and Carlson 2004). Noncompetitive inhibition could occur by having the inhibitory odorant stabilize the OR in an inactive state, as proposed by Carlson, for example, to explain odor-evoked inhibition in insect ORNs (Hallem and Carlson 2004; Yao et al. 2005). Alternately, the inhibitory odorant could stabilize the OR in a specific conformation that selectively interacts with an intracellular signaling complex that, when activated, opposes excitation. Such ligand-induced selective signaling (LiSS), among other names, was originally put forth 15 years ago (Kenakin 1995) but is rapidly becoming a generic theme for GPCRs. Rather than viewing ligand binding as consistently eliciting a specific intracellular signal, it has become increasingly clear that the nature of the ligand (and the dynamically changing intracellular environment) alters the flavor of the signaling for many different GPCRs (Rosenbaum et al. 2009; Millar and Newton 2010). The multiple G proteins found in mammalian olfactory cilia (Schandar et al. 1998; Mayer et al. 2008) is consistent with the possibility that LiSS may extend to olfactory GPCRs. If so, what might be the nature of such an inhibitory input signaling pathway?
PI signaling as a potential mediator of inhibitory input to mammalian ORNs
Considerable controversy dating back more than 10 years has surrounded the question of whether PI signaling plays a role in mammalian olfactory transduction (e.g., Schild and Restrepo 1998; Gold 1999). We now know that PI signaling is more complex than appreciated at the time of the controversy. Earlier studies of PI signaling in olfaction focused solely on the canonical PI turnover pathway, in which phospholipase C (PLC) metabolizes phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2] into the signaling molecules diacylglycerol and inositol 1,4,5-triphosphate (IP3). In parallel, however, PI(4,5)P2 can be acted upon by PI 3-kinase (PI3K) to produce phosphatidylinositol (3,4,5) trisphosphate (PIP3). In addition to requiring the same substrate, PI(4,5)P2, recent findings suggest that PLC signaling can regulate the activation of PI3K (e.g., Huang et al. 2007; Kortholt et al. 2007; Kolsch et al. 2008). Moreover, PIs once thought to be no more than metabolic substrates for the pathway can also assume signal function and modulate the activity of ion channels and transporters (review: Hilgemann et al. 2001).
The discovery of 3-PIs in particular, and their synthesis by a family of PI3Ks, has fostered important insight into cell signal transduction (reviews: Toker and Cantley 1997; Fruman et al. 1998; Zhang and Majerus 1998). In higher animals, there is a relatively large constitutive pool of phosphatidylinositol (3) phosphate in resting cells, in contrast to endogenously low levels of phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] and PIP3 that are transiently and rapidly elevated in response to external stimuli (review: Hawkins et al. 2006). PI3Ks can be activated by a wide array of ligands acting through both tyrosine kinases and GPCRs, depending on the particular isoform involved (review: Hawkins et al. 2006). From these findings, it can be concluded that a complete understanding of PI signaling in any system, including ORNs, requires PI3K-mediated signaling to be considered in concert with PLC-mediated signaling.
More recent findings suggest the need to revisit the question of whether PI signaling plays a role in mammalian olfactory transduction. Some mammalian ORNs express TRPM5 and PLCβ2 (Lin et al. 2007), and TRP channels are a common downstream target of PLC-mediated signaling in other systems (e.g., Liu and Liman 2003; Nilius et al. 2008). Exogeneous PIs, and especially PIP3, can negatively regulate the olfactory CNG channel (Zhainazarov et al. 2004) through complex interaction between PIP3 and Ca2+/calmodulin at the N-terminus of the channel (Brady et al. 2006). PI3K can modulate odor-activated increases in intracellular Ca2+ in acutely dissociated rodent ORNs in a cyclic nucleotide-dependent manner, the direction of which correlates with negative regulation of the CNG channel (Spehr et al. 2002). The latter findings were interpreted to suggest that when stimulated by odorant mixtures of sufficient complexity to contain both excitatory and inhibitory odorants for a given ORN, activation of the olfactory CNG channel in that cell would be reduced in a PI3K-dependent manner (Spehr et al. 2002; Zhainazarov et al. 2004). This interpretation is consistent with evidence that the olfactory CNG channel mediates inhibitory olfactory responses in mouse ORNs (Delay and Restrepo 2004) and that odorants shown to mask human perception can block the olfactory CNG current in native amphibian ORNs (Takeuchi et al. 2009).
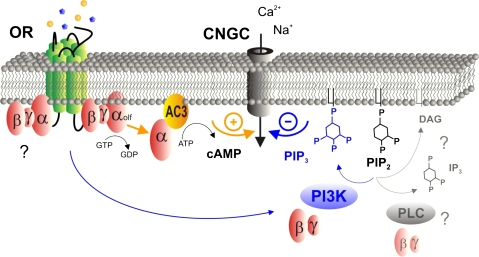
As PI3K-dependent signaling controls many constitutive functions in cells, including in mammalian ORNs (Moon et al. 2009), it is important to establish that any PI3K-dependent mechanisms implicated in olfactory transduction act sufficiently fast to regulate the electrophysiological output of the cells. Emerging evidence suggests this may be so. Odorants rapidly and transiently activate PI3K in rodent olfactory cilia in vitro and in the dissociated OE (Klasen et al. 2009). Blocking G protein–coupled isoforms of PI3K can modulate the earliest phase, that is, the latency, rise time, and peak magnitude, of the electrophysiological response of forskolin-sensitive rodent ORNs to complex odorants (Ukhanov et al. 2010). The potentially important implications of these findings for activation of the OR along with many more detailed mechanistic questions remain to be explored, but it is possible to suggest a working model that is consistent with these findings (Figure 2).
Figure 2.
Potential model for phosphoinositide-3-kinase (PI3K)-dependent opponent input to rodent olfactory receptor neurons (ORNs). One component of a binary odorant mixture (yellow circles) excites the cell through the canonical cyclic nucleotide signaling pathway: the olfactory receptor (OR) activates a G-protein (Gαolf), which in turn activates adenylyl cyclase (AC3) to produce cyclic adenosine monophosphate (cAMP) that activates the olfactory cyclic nucleotide gated channel (CNGC). The other component (blue stars), potentially binding to the same OR, inhibits the cell by activating phosphoinositide 3-kinase (PI3K) through the beta/gamma subunit of the same or still unknown G-protein. Activation of PI3K produces phosphatidylinositol (3,4,5) trisphosphate (PIP3), which negatively regulates sensitivity of the CNGC to cAMP and reduces the net output of the ORN. The involvement of phospholipase C (PLC, faded) to produce inositol-1,4,5-phosphate (IP3) and diacylglycerol (DAG) remains to be explored.
Future directions
Understanding more about the nature of inhibitory odorants will be important to better understanding the role of inhibitory input in odor coding. Are there inhibitory odorants per se, that is, is inhibition odorant-dependent or do odorants that inhibit one cell excite another, that is, is inhibition is cell-dependent and there aren’t inhibitory odorants per se? These 2 alternatives are not necessarily mutually exclusive because cell-dependent inhibition could be limited to a subset of odorants that were strictly inhibitory. However, limited data argue against strict odorant dependency in mammalian ORNs. Single odorants shown to inhibit one rodent ORN can activate another (Ukhanov et al. 2010). Limited in vitro data showing that odorants reported earlier to activate adenylyl cyclase in mammalian olfactory ciliary membranes at least partially overlap with those that activate PI3K (Klasen et al. 2009), as might be expected if inhibition was cell- and PI3K-dependent. These findings would be consistent with findings in Drosophila where it is reasonably clear that there is no such thing as an inhibitory odorant (Hallem and Carlson 2006). More work, ideally using high-throughput screening, is needed to rigorously address this fundamentally important question.
Better understanding the role of inhibitory input in odor coding will also come from knowing the extent to which inhibitory input is expressed across ORNs. Do all canonical ORNs mediate opponent input or only a functional subset of ORNs? Identifying even one inhibitory odorant in the MRR of any particular cell is time consuming, however, especially without high-throughput screening. Localization of inhibitory-specific signaling components as they are identified will offer a more efficient way to address this question. Toward this end, 2 different isoforms of PI3K implicated in inhibiting the output if rodent ORNs could be localized to many, if not most, ORNs in the murine OE (Brunert et al. 2010), but more work is needed to address this question. It is important to consider, too, that findings for one animal may not generalize across species because the relative contribution of inhibitory input to odor coding could potentially be an evolutionary variable that correlates with the reliance of a particular species on its odor world to survive.
We tend to think of excitatory and inhibitory input as distinct phenomena, but it is probably more appropriate to view excitation and inhibition as 2 points on what in reality is a continuum of odorant-specific input to ORNs. This idea is already inherent in considering the ORN as an integrating unit in which inhibition tempers the magnitude of excitation in a graded manner (Figure 1). However, the integrative function may include further dimensionality that determines not only the magnitude but also the pattern of the output because the extent to which inhibition is activated presumably would result in different rates of excitation and shape the dynamics of the output. Toward this end, different single odorants evoke different patterns of excitatory output in the same insect ORN (Wilson R, personal communication). Just how this occurs, and whether this intriguing finding translates to mammalian ORNs, remains to be determined, but one could envision that each ligand in the MRR of a mammalian OR activates the OR in a ligand-specific manner in which inhibition contributed to different patterns of excitation based on the extent to which a particular ligand coactivated inhibition. Different mechanisms of inhibition (e.g., competitive inhibition vs. a noncompetitive mechanism) could enhance this dimensionality. Rapid temporal changes in input are increasingly appreciated to contribute to the odorant-specific patterns of activity evoked in the olfactory bulb (e.g., Soucy et al. 2009). Changes in the time of onset or the rate of rise of the discharge of ORNs presumably would shape those patterns in a manner that contributed to coding at the first olfactory relay. Insight into this exciting idea will require more understanding of the molecular basis by which odorants bind to and activate ORs.
Because disruption of any of the cellular processes leading to the activation of the ORNs inevitably impairs olfactory function, we need to fully understand these processes. Knowing that ORNs have 2, odorant-specific modes of signaling and determining the cellular basis of how odorants inhibit as well as excite ORNs adds to this understanding. The emerging evidence that PI signaling at least in part mediates inhibitory odorant input to mammalian ORNs and contributes to the output of the cells evoked by natural, complex odorants argues for the fundamental importance of inhibitory input to olfactory coding in mammals and identifies an additional possible cellular basis for olfactory dysfunction.
Funding
Research from our laboratory that contributed to this article was supported by the National Institute on Deafness and Other Communication Disorders [DC005995 and DC001655].
Acknowledgments
I would like to thank the members of my laboratory for their many helpful, instructive discussions on olfactory signaling. I thank Drs Kirill Ukhanov, Daniela Brunert, and Rachel Wilson for critically reading a draft of the manuscript.
References
- Abaffy T, Malhotra A, Luetje CW. The molecular basis for ligand specificity in a mouse olfactory receptor: a network of functionally important residues. J Biol Chem. 2007;282:1216–1224. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48(3):417–430. doi: 10.1016/j.neuron.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Araneda RC, Peterlin Z, Zhang X, Chesler A, Firestein S. A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J Physiol. 2004;555:743–756. doi: 10.1113/jphysiol.2003.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobkov YV, Ache BW. Intrinsically bursting olfactory receptor neurons. J Neurophysiol. 2007;97:1052–1057. doi: 10.1152/jn.01111.2006. [DOI] [PubMed] [Google Scholar]
- Brady JD, Rich ED, Martens JR, Karpen JW, Varnum MD, Brown RL. Interplay between PIP3 and calmodulin regulation of olfactory cyclic nucleotide-gated channels. Proc Natl Acad Sci U S A. 2006;103(42):15635–15640. doi: 10.1073/pnas.0603344103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breer H, Fleischer J, Strotmann J. The sense of smell: multiple olfactory subsystems. Cell Mol Life Sci. 2006;63(13):1465–1475. doi: 10.1007/s00018-006-6108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunert D, Klasen K, Corey EA, Ache BW. PI3Kgamma-dependent signaling in mouse olfactory receptor neurons. Chem Senses. 2010;35:301–308. doi: 10.1093/chemse/bjq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Chen TY, Takeuchi H, Kurahashi T. Odorant inhibition of the olfactory cyclic nucleotide-gated channel with a native molecular assembly. J Gen Physiol. 2006;128:365–371. doi: 10.1085/jgp.200609577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay R, Restrepo D. Odorant responses of dual polarity are mediated by cAMP in mouse olfactory sensory neurons. J Neurophysiol. 2004;92:1312–1319. doi: 10.1152/jn.00140.2004. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Duchamp-Viret P, Chaput MA, Duchamp A. Odor response properties of rat olfactory receptor neurons. Science. 1999;284(5423):2171–2174. doi: 10.1126/science.284.5423.2171. [DOI] [PubMed] [Google Scholar]
- Duchamp-Viret P, Duchamp A, Chaput MA. Single olfactory sensory neurons simultaneously integrate the components of an odour mixture. Eur J Neurosci. 2003;18:2690–2696. doi: 10.1111/j.1460-9568.2003.03001.x. [DOI] [PubMed] [Google Scholar]
- Firestein S, Shepherd GM. Neurotransmitter antagonists block some odor responses in olfactory receptor neurons. Neuroreport. 1992;3:661–664. doi: 10.1097/00001756-199208000-00001. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Gold GH. Controversial issues in vertebrate olfactory transduction. Annu Rev Physiol. 1999;61:857–871. doi: 10.1146/annurev.physiol.61.1.857. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004;117:965–979. doi: 10.1016/j.cell.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34(Pt 5):647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- Hilgemann DW, Feng S, Nasuhoglu C. The complex and intriguing lives of PIP2 with ion channels and transporters. Sci STKE. 2001;2001(111):RE19. doi: 10.1126/stke.2001.111.re19. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhou H, Mahavadi S, Sriwai W, Murthy KS. Inhibition of Galphaq-dependent PLC-beta1 activity by PKG and PKA is mediated by phosphorylation of RGS4 and GRK2. Am J Physiol Cell Physiol. 2007;292(1):C200–C208. doi: 10.1152/ajpcell.00103.2006. [DOI] [PubMed] [Google Scholar]
- Kenakin T. Agonist-receptor efficacy. II. Agonist trafficking of receptor signals. Trends Pharmacol Sci. 1995;16:232–238. doi: 10.1016/s0165-6147(00)89032-x. [DOI] [PubMed] [Google Scholar]
- Klasen K, Corey EA, Kuck F, Wetzel CH, Hatt H, Ache BW. Odorant-stimulated phosphoinositide signaling in mammalian olfactory receptor neurons. Cell Signal. 2009;22:150–157. doi: 10.1016/j.cellsig.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimmeck D, Mayer U, Ungerer N, Warnken U, Schnolzer M, Frings S, Mohrlen F. Calcium-signaling networks in olfactory receptor neurons. Neuroscience. 2008;151(3):901–912. doi: 10.1016/j.neuroscience.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Kolsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121(Pt 5):551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A, King JS, Keizer-Gunnink I, Harwood AJ, Van Haastert PJ. Phospholipase C regulation of phosphatidylinositol 3,4,5-trisphosphate-mediated chemotaxis. Mol Biol Cell. 2007;18(12):4772–4779. doi: 10.1091/mbc.E07-05-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi T, Lowe G, Gold GH. Suppression of odorant responses by odorants in olfactory receptor cells. Science. 1994;26:118–120. doi: 10.1126/science.8016645. [DOI] [PubMed] [Google Scholar]
- Lin W, Margolskee R, Donnert G, Hell SW, Restrepo D. Olfactory neurons expressing transient receptor potential channel M5 (TRPM5) are involved in sensing semiochemicals. Proc Natl Acad Sci U S A. 2007;104(7):2471–2476. doi: 10.1073/pnas.0610201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Liman ER. Intracellular Ca2+ and the phospholipid PIP2 regulate the taste transduction ion channel TRPM5. Proc Natl Acad Sci U S A. 2003;100(25):15160–15165. doi: 10.1073/pnas.2334159100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M. Encoding olfactory signals via multiple chemosensory systems. Crit Rev Biochem Mol Biol. 2007;42(6):463–480. doi: 10.1080/10409230701693359. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Mayer U, Kuller A, Daiber PC, Neudorf I, Warnken U, Schnolzer M, Frings S, Mohrlen F. The proteome of rat olfactory sensory cilia. Proteomics. 2009;9(2):322–334. doi: 10.1002/pmic.200800149. [DOI] [PubMed] [Google Scholar]
- Mayer U, Ungerer N, Klimmeck D, Warnken U, Schnolzer M, Frings S, Mohrlen F. Proteomic analysis of a membrane preparation from rat olfactory sensory cilia. Chem Senses. 2008;33(2):145–162. doi: 10.1093/chemse/bjm073. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Glasser CE, Bose SC, Bergman DA. Tissue expression patterns identify mouse cilia genes. Physiol Genomics. 2008;32(2):198–206. doi: 10.1152/physiolgenomics.00128.2007. [DOI] [PubMed] [Google Scholar]
- Millar RP, Newton CL. The year in G protein-coupled receptor research. Mol Endocrinol. 2010;24:261–274. doi: 10.1210/me.2009-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon C, Liu BQ, Kim SY, Kim EJ, Park YJ, Yoo JY, Han HS, Bae YC, Ronnett GV. Leukemia inhibitory factor promotes olfactory sensory neuronal survival via phosphoinositide 3-kinase pathway activation and Bcl-2. J Neurosci Res. 2009;87:1098–1106. doi: 10.1002/jnr.21919. [DOI] [PubMed] [Google Scholar]
- Munger SD. Olfaction: noses within noses. Nature. 2009;459(7246):521–522. doi: 10.1038/459521a. [DOI] [PubMed] [Google Scholar]
- Munger SD, Leinders-Zufall T, Zufall F. Subsystem organization of the mammalian sense of smell. Annu Rev Physiol. 2009;71:115–140. doi: 10.1146/annurev.physiol.70.113006.100608. [DOI] [PubMed] [Google Scholar]
- Nilius B, Owsianik G, Voets T. Transient receptor potential channels meet phosphoinositides. EMBO J. 2008;27(21):2809–2816. doi: 10.1038/emboj.2008.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Omura M, Katauka H, Touhara K. Olfactory receptor antagonism between odorants. EMBO J. 2004;23(1):120–126. doi: 10.1038/sj.emboj.7600032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobika BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospars JP, Lansky P, Chaput M, Duchamp-Viret P. Competitive and noncompetitive odorant interactions in the early neural coding of odorant mixtures. J Neurosci. 2008;28(10):2659–2666. doi: 10.1523/JNEUROSCI.4670-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanhueza M, Bacigalupo J. Odor suppression of voltage-gated currents contributes to the odor-induced response in olfactory neurons. Am J Physiol. 1999;277:C1086–C99. doi: 10.1152/ajpcell.1999.277.6.C1086. [DOI] [PubMed] [Google Scholar]
- Sanhueza M, Schmachtenberg O, Bacigalupo J. Excitation, inhibition, and suppression by odors in isolated toad and rat olfactory receptor neurons. Am J Physiol Cell Physiol. 2000;279(1):C31–C39. doi: 10.1152/ajpcell.2000.279.1.C31. [DOI] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Schandar M, Laugwitz Kl, Boekhoff I, Kroner C, Gudermann T, Schultz G, Breer H. Odorants selectively activate distinct G protein subtypes in olfactory cilia. J Biol Chem. 1998;273(27):16669–16677. doi: 10.1074/jbc.273.27.16669. [DOI] [PubMed] [Google Scholar]
- Schild D, Restrepo D. Transduction mechanisms in vertebrate olfactory receptor cells. Physiol Rev. 1998;78(2):429–466. doi: 10.1152/physrev.1998.78.2.429. [DOI] [PubMed] [Google Scholar]
- Scott JW, Sherrill L. Effects of odor stimulation on antidromic spikes in olfactory sensory neurons. J Neurophysiol. 2008;100(6):3047–3085. doi: 10.1152/jn.90399.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38:770–780. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Soucy ER, Albeanu DF, Fantana AL, Murthy VN, Meister M. Precision and diversity in an odor map on the olfactory bulb. Nat Neurosci. 2009;12(2):210–220. doi: 10.1038/nn.2262. [DOI] [PubMed] [Google Scholar]
- Spehr M, Giesselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- Spehr M, Wetzel CH, Hatt H, Ache BW. 3-phosphoinositides modulate cyclic nucleotide signaling in olfactory receptor neurons. Neuron. 2002;33(5):731–739. doi: 10.1016/s0896-6273(02)00610-4. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Ishida H, Hikichi S, Kurahashi T. Mechanism of olfactory masking in the sensory cilia. J Gen Physiol. 2009;133(6):583–601. doi: 10.1085/jgp.200810085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallarida RJ, Jacob LS. The dose-response relation in pharmacology. New York: Springer-Verlag; 1979. p. 207. [Google Scholar]
- Toker A, Cantley LC. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387(6634):673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- Ukhanov K, Corey EA, Brunert D, Klasen K, Ache BW. Inhibitory odorant signaling in mammalian olfactory receptor neurons. J Neurophysiol. 2010;103:1114–1122. doi: 10.1152/jn.00980.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25(37):8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhainazarov AB, Spehr M, Wetzel CH, Hatt H, Ache BW. Modulation of the olfactory CNG channel by Ptdlns(3,4,5)P3. J Membr Biol. 2004;201(1):51–57. doi: 10.1007/s00232-004-0707-4. [DOI] [PubMed] [Google Scholar]
- Zhang X, Majerus PW. Phosphatidylinositol signalling reactions. Semin Cell Dev Biol. 1998;9(2):153–160. doi: 10.1006/scdb.1997.0220. [DOI] [PubMed] [Google Scholar]