Abstract
Insects can detect a large range of odors with a numerically simple olfactory system that delivers high sensitivity and accurate discrimination. Therefore, insect olfactory receptors hold great promise as biosensors for detection of volatile organic chemicals in a range of applications. The array of olfactory receptor neurons of Drosophila melanogaster is rapidly becoming the best-characterized natural nose. We have investigated the suitability of Drosophila receptors as detectors for volatiles with applications in law enforcement, emergency response, and security. We first characterized responses of the majority of olfactory neuron types to a set of diagnostic odorants. Being thus able to correctly identify neurons, we then screened for responses from 38 different types of neurons to 35 agents. We identified 13 neuron types with responses to 13 agents. As individual Drosophila receptor genes have been mapped to neuron types, we can infer which genes confer responsiveness to the neurons. The responses were confirmed for one receptor by expressing it in a nonresponsive neuron. The fly olfactory system is mainly adapted to detect volatiles from fermenting fruits. However, our findings establish that volatiles associated with illicit substances, many of which are of nonnatural origin, are also detected by Drosophila receptors.
Keywords: biosensor, Drosophila melanogaster, drugs, explosives, olfactory receptors, toxic gasses
Introduction
Law enforcement, emergency response, and security agencies often need to detect concealed toxic, explosive, or otherwise illicit materials. Olfactory sensing has proven to be the quickest and most accurate method to do this in nonlaboratory situations. Detection systems that provide exquisite sensitivity and odor discrimination are animal olfactory systems, and thus animals such as dogs have been widely utilized to detect illicit materials. Many materials can be detected by trained dogs, although often it is not known which volatiles they use as cues (Harper et al. 2005). Their use is also limited by a number of other factors; they are expensive to train, need expert personnel to guide them on site, and detect only what they are trained to detect. In addition, their behavior can be highly variable, and there are many situations where it is impractical or unethical to use dogs. Although electronic noses provide adequate sensitivity and are able to differentiate odors for some applications, they cannot identify odors as reliably as natural noses can (Röck et al. 2008). An ideal detection system for illicit substances would be a technology that can harness the olfactory coding power of the animal olfactory system but without using actual animals.
To detect odors, animals employ a range of olfactory receptor neurons (ORNs) as the detector units, each usually expressing a single receptor that can bind odorants and activate the neuron (Touhara and Vosshall 2009). Information from each ORN class is collected in one of many olfactory glomeruli in the brain. The pattern recognition systems of the brain then extract information on odor identity, odor intensity, and its presence in space and time (Wilson and Mainen 2006). In mammalian ORNs, olfactory receptor (OR) genes encode proteins with 7 transmembrane domains (Buck and Axel 1991) that have been shown to be G-protein coupled receptors mediating odor sensitivity (Firestein 2001). The recent isolation of a rat OR sensitive to 2.4-dinitrotoluene, a component of explosives also detected by dogs (Radhika et al. 2007), suggests mammalian OR proteins can possibly be used as biosensors. However, functional studies are difficult, and ligand information is lacking for most of their ORs. In one recent large-scale functional study Saito et al. (2009) screened a large number of mouse and human ORs with 93 odorants. They found ligands for 52 mouse and 10 human ORs, but these represented only 20% of the receptors they screened.
In insects such as Drosophila melanogaster odors are similarly detected by different functional classes of ORNs in a combinatorial fashion (de Bruyne et al. 2001; Hallem and Carlson 2006). However, compared with mammals most insects have at least 20-fold fewer ORN classes and many fewer receptor genes (Touhara and Vosshall 2009). Moreover, in Drosophila, ligands have been identified for more than 60% of receptors (de Bruyne et al. 1999, 2001; Hallem and Carlson 2006; Kreher et al. 2008; Laissue and Vosshall 2008). With around 100 odorants tested on large numbers of both Drosophila and mouse receptors, the much lower rate of deorphaning in the latter (Saito et al. 2009) suggests that insect receptors may on average be more broadly tuned than mammalian ones.
Insect ORs hold great promise as biosensors in commercial devices for detection of volatile organic chemicals as, unlike mammalian receptors, they might not require linking to downstream secondary messenger systems. The largest and best-characterized family of insect receptors, encoded by the Or genes, have an inverted membrane topology compared with mammalian receptors (Benton et al. 2006; Smart et al. 2008) and do not primarily signal through G-proteins (Sato et al. 2008; Smart et al. 2008). The insect Or proteins form a novel class of heteromeric cation channels, directly gated by odorants (Sato et al. 2008; Wicher et al. 2008). In addition, a second, smaller family of ORs was recently discovered in D. melanogaster, these genes also encode ion channels, in this case related to ionotropic glutamate receptors and they have been called the IR family (ionotropic receptors, Benton et al. 2009). Insect ORs may thus be advantageous for use as detectors in a biosensor as they may not require any coexpression of downstream signaling molecules. However, we do not know to what extent insects are sensitive to industrial volatile organic chemicals. Can they rival the ability of trained dogs to detect and identify security risks? Like dogs, insects can also be trained to respond to volatiles in explosives, and bees have been successfully used to detect land mines in the field (King et al. 2003; Shaw et al. 2005; Rains et al. 2006).
Among insects, the adult olfactory system of D. melanogaster is presently the best characterized on all 3 levels of organization; functional characterization of ORN classes, receptor gene expression, and neuronal wiring. The ORNs express 48 Ors, 9 IRs, and 3 gustatory receptors (Grs). ORNs are distributed over 2 appendages, antennae, and maxillary palps, housed in basiconic, coeloconic, and trichoid sensilla. The response properties of single ORNs can be determined using electrophysiological recording (de Bruyne et al. 1999, 2001), and an in vivo expression system (Dobritsa et al. 2003) can be used to determine and confirm ligand information for individual Or proteins. These factors, combined with gene expression studies (Fishilevich and Vosshall 2005; Couto et al. 2005), have enabled the mapping of most receptors to neuron classes. Thus if a novel compound is found to stimulate a particular neuron, the receptor responsible can be determined.
Here, we investigate the usefulness of Drosophila ORs as sensors for materials that pose risks to security with the ultimate goal of incorporating them in an automated standoff detection device. We first use a diagnostic set of odorants to map the identity of ORN classes by in vivo electrophysiological recordings on antennae and maxillary palps, and in doing so provide the first in vivo data on responses from 2 new types of sensilla. Next, we screen a set of volatiles associated with various threats to security, many of which are synthetic chemicals of nonnatural origin, and show that insect ORs are able to detect a number of these compounds. We further demonstrate that we can confirm the receptors responsible for detecting volatiles of interest.
Materials and methods
Fly stocks and rearing conditions
We used a standard CS-5 strain used in many other olfactory studies (Helfand and Carlson 1989). All flies were reared on yeasted semolina/syrup medium in 40 mL vials at 22 °C and normal daylight. To express transgenes of targeted ORs in the empty neuron Δab3A (Dobritsa et al. 2003), we used the Δhalo mutation which removes the Or22a and Or22b genes normally expressed there and drove expression from a UAS-Or construct by an Or22a-GAL4 promotor construct. We crossed w; Δhalo/CyO; P{UAS-OrX}/ TM3 to w; Δhalo/CyO; P{Or22a-Gal4} to generate w; Δhalo; P{UAS-OrX}/P{Or22a-Gal4}. These flies were kindly given to us by John Carlson.
Electrophysiological recordings from single olfactory sensilla
The basic recording technique was described elsewhere (de Bruyne et al. 1999, 2001). A 4- to 10-day-old male fly was immobilized in a plastic pipette tip. Recordings were made from AgCl-coated silver wire inserted in saline filled glass capillaries (0.015 M KCl). One microelectrode was inserted through the wall of a single olfactory sensillum to contact the lymph surrounding the dendrites of the ORNs. The reference electrode was inserted in the eye. Signals were amplified 1000× via a 10× active probe fed into an AD converter with digital amplification (USB-IDAC, Syntech). Responses were analyzed off-line using Autospike software (Syntech). Odor responses were calculated as the change in action potentials firing rate (in spikes per second), that is, the difference between the number in the 500 ms during and prior to stimulation. For recordings of ac4 sensilla, in which action potential amplitude differences did not allow reliably attribution to the activity of a single neuron, we counted all spikes together. For all other sensilla, we include only those recordings for which we were able to separate the activity of the 2 or 4 neurons in a single sensillum. Sensillum identification was based on responses to a set of diagnostic odorants (see text). Olfactory responses to all 35 illicit agents were established from at least 3 sensilla from at least 2 different flies.
Odor stimulation
Stimulation with odorants was comparable with de Bruyne et al. (1999, 2001). Specifically, a glass tube held 5 mm from the preparation supplied continuous humidified air at 66 cm/s (zero grade, BOC). Volatiles were injected into the air for a 500-ms period from 5-mL disposable syringes holding 10 μL of odorant solution on filter paper, giving a headspace dilution factor of 10%. All odorants are listed in Table 1 with their provenance. The majority of odorants were obtained commercially (Fluka or Aldrich) and dissolved in a suitable solvent, either paraffin oil (Fluka), acetonitrile (BDH), or distilled water at 10−2 v/v. Several chemical samples were obtained courtesy of the Australian Federal Police.
Table 1.
Agents tested on Drosophila ORs
| No. | Name | CAS no. | Sourcea | Solb | Substancec |
| Toxic gasses and precursors | |||||
| 1 | Ammonia (hydroxide solution) | 1336-21-6 | Sig. 338818 | w | T + p/c Chloramine |
| 2 | Chlorine (as bleach) | — | White King | w | T + p/c Chloramine |
| 3 | Chlorine (calcium hypochlorite) | 7782-50-5 | Sig. 92401 | w | T + p/c Chloramine |
| 4 | Diazinon | 333-41-5 | Sig. 45428 | a | T (organophosphate) |
| 5 | Endosulfan | 115-29-7 | CSIRO | a | T (organochlorine) |
| 6 | Methyldiethanolamine | 105-59-9 | Sig. 471828 | po | p/c Nitrogen mustard BA |
| 7 | Ethyldiethanolamine | 139-87-7 | Sig. 112062 | po | p/c Nitrogen mustard BA |
| 8 | Diethyl phosphite | 762-04-9 | Sig. 32449 | po | p/c Organophosphate NA |
| 9 | Triethyl phosphite | 122-52-1 | Sig. 90540 | po | p/c Organophosphate NA |
| Explosive precursors and contaminants | |||||
| 10 | Acetone | 67-64-1 | Sig. 90872 | po | p/c Acetone peroxide E |
| 11 | Cyclohexanone | 108-94-1 | Sig. 398241 | po | p/c Plastic E |
| 12 | 2,3-Dimethyl-2,3-dinitrobutane | 3964-18-9 | Sig. 156345 | a | p/c Plastic or sheet E |
| 13 | 2-Ethyl-1-hexanol | 104-76-7 | Sig. 538051 | po | p/c Polymer-based E |
| 14 | Hexamine | 100-97-0 | Sig. H11300 | w | p/c Nitroamine E |
| 15 | Hydrogen peroxide | 7722-84-1 | M. 10366.0500 | w | p/c Acetone peroxide E |
| 16 | Methylethyl ketone | 78-93-3 | Sig. 02469 | po | p/c Methyl ethyl ketone peroxide E |
| 17 | Nitric acid | 7697-37-2 | M. 101687F | w | p/c Nitroamine E |
| 18 | Nitromethane | 75-52-5 | Sig. 02484 | a | p/c ANFO E |
| 19 | Potassium perchlorate | 7778-74-7 | Sig. 241830 | w | p/c Ammonium perchlorate E |
| 20 | Sodium perchlorate | 7601-89-0 | Sig. 410241 | w | p/c Ammonium perchlorate E |
| 21 | Sulfur | 7704-34-9 | Sig. 414980 | a | p/c Black powder E |
| Explosives | |||||
| 22 | Cyclotrimethylenetrinitramine (RDX) | 121-82-4 | AFP sample | a | E |
| 23 | Hexamethylene triperoxide-diamine (HMTD) | 283-66-9 | AFP sample | a | E |
| 24 | Nitroglycerin | 55-63-0 | AFP sample | a | E |
| 25 | Pentaerythritol tetranitrate (PETN) | 78-11-5 | AFP sample | a | E |
| 26 | Triacetone triperoxide (TATP) | 17088-37-8 | AFP sample | a | E |
| 27 | 2,4,6-Trinitrotoluene (TNT) | 118-96-7 | AFP sample | a | E |
| Drugs, drug precursors, and contaminants | |||||
| 28 | Acetic anhydride | 108-24-7 | Sig. 45830 | w | p/c Heroine |
| 29 | Benzaldehyde | 100-52-7 | Sig. B1334 | po | p/c Methamphetamine |
| 30 | Formamide | 75-12-7 | M. 1.09684.1000 | w | p/c Amphetamines |
| 31 | 1-phenyl-2-nitropropene | 705-60-2 | AFP sample | a | p/c Methamphetamine |
| 32 | Phenyl-2-propanone | 103-79-7 | AFP sample | po | p/c Methamphetamine |
| 33 | Safrole | 94-59-7 | AFP sample | po | p/c MDMA |
| 34 | Sassafras oil | — | AFP sample | po | Source of saffrole |
| 35 | 3,4-Methylenedioxy-N-methyl amphetamine (MDMA) | 69610-10-2 | AFP sample | w | D |
Product numbers are given for compounds obtained from Sigma-Aldrich (Sig.) or Merck (M.). AFP, Australian Federal Police, CSIRO, Commonwealth Sientific and Industrial Research Organization.
Solvents: w, water, po, paraffin oil, a, acetonitrile.
The illicit substance association: T, toxic gas, p/c, precursor/contaminant of, BA, blistering agent, NA, nerve agent, E, explosive, D, drug.
Results
Identification of Drosophila sensillum types and ORN classes
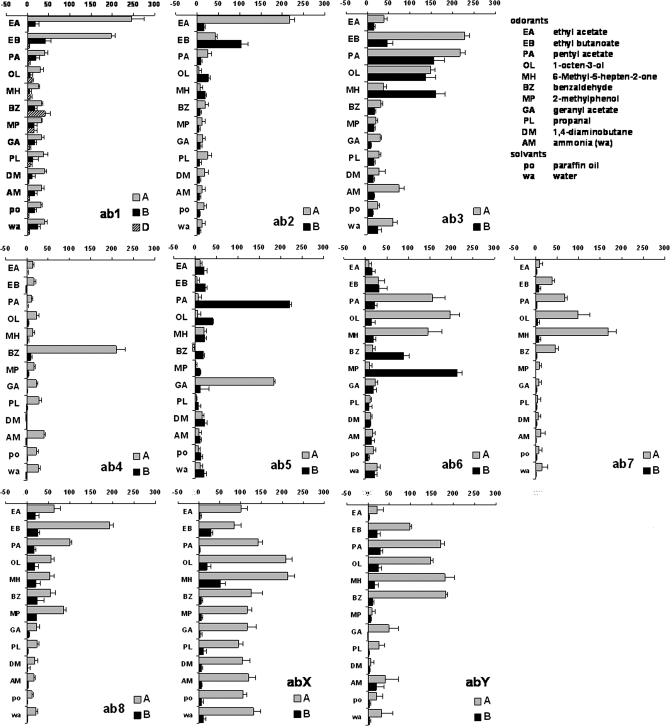
We recorded responses to a diagnostic set of 11 odorants from a total of 110 different sensilla of the basiconic and coeloconic category on the antennae and maxillary palps of Drosophila. These sensilla could be classified into 17 different types based on the responses from the neurons housed in them. The response profiles for the 10 antennal basiconic sensillum types are shown in Figure 1. Each of the sensillum types shows a clearly distinct response spectrum. The ab1 sensillum contains 4 ORNs, the A neuron responds strongly to ethyl acetate (EA) and ethyl butanoate (EB), and the D neuron typically has very small spikes and responds lightly to benzaldehyde (BZ). We have not included responses from the ab1C neuron in the Figure as this neuron responds exclusively to CO2 (de Bruyne et al. 2001), and while we did check for its presence, we did not test CO2 on all other sensilla. The ab2 sensilla are also characterized by responses to ethyl acetate (EA) and ethyl butanoate (EB), but in this case, the A neuron responds strongly to ethyl acetate (EA), whereas the B neuron responds strongly to ethyl butanoate (EB). The ab3 and ab8 sensilla each have a strong response to ethyl butanoate from the A neuron, but in ab3 sensilla, the B neuron responds to 6-methyl-5-hepten-2-one (MH). The ab4 and ab5 sensilla are easily identified by their narrowly tuned responses to benzaldehyde (BZ) and geranyl acetate (GA), respectively. Response spectra for the A neurons of ab6 and ab7 are somewhat similar, but in ab6 sensilla, the B neuron responds strongly to 2-methylphenol (MP).
Figure 1.
Identifying Drosophila ORNs by specific response profiles of identified ORNs in antennal basiconic (ab) sensilla. All odorants are at 10−2 dissolved in paraffin oil (po), except ammonia which was dissolved in water (wa). The ab1C neuron, excluded for clarity, does not respond to any of these odorants. n = 6–10, error bars are standard error of the mean.
In our recordings, we also identified 2 previously uncharacterized types of basiconic sensilla which we have named abX and abY. Each shows evidence of 2 active ORNs. One sensillum (abX) is characterized by high responses to the 2 solvents in the A neuron. This masks any putative responses to most odorants except 1-octen-3ol (OL) and 6-methyl-5-hepten-2-one (MH). The second sensillum type (abY) shows high responses to several odorants from the A neuron and only minor responses from the B neuron. The A neuron responds mainly to pentyl acetate (PA) 1-octen-3ol (OL) and 6-methyl-5-hepten-2-one (MH), odorants which evoke responses from several ORNs but is distinguished by its strong response to benzaldehyde (BZ).
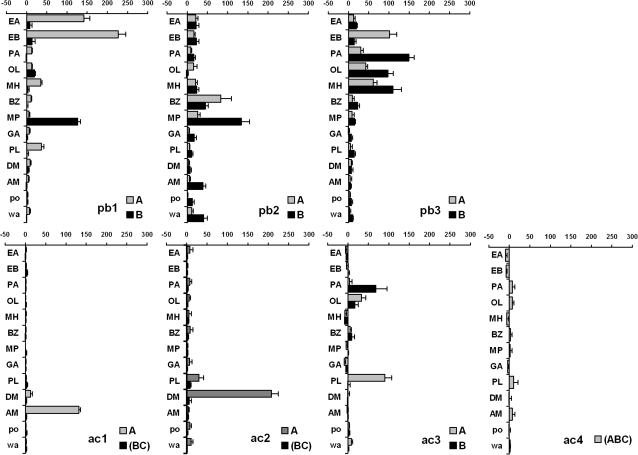
We also recorded from basiconic sensilla on the maxillary palps (Figure 2), testing the same set of odorants on the 2 appendages for the first time. The 3 palpal sensillum types are easily differentiated by distinctive responses from A and B neurons, respectively, to ethyl butanoate (EB) and 2-methylphenol (MP) in pb1, benzaldehyde (BZ) and 2-methylphenol (MP) in pb2, and ethyl butanoate (EB) and pentyl acetate (PA) in pb3.
Figure 2.
Identifying Drosophila ORNs by specific response profiles of identified ORNs in palpal basiconic (pb) and antennal coeloconic (ac) sensilla. For some ORNs, combined spike counts are presented because spikes could not be separated reliably. Otherwise as in Figure 1.
Finally, we included all 4 coeloconic sensillum types on the antennal surface in our analysis (Figure 2). The response spectra for the ac1 and ac2 sensillum types can be easily differentiated by unique responses to ammonia (AM) for ac1A and 1,4-diaminobutane (DM) for ac2A. There is a discrepancy between a physiological study by Yao et al. (2005) which identifies only 2 neurons in ac1 and ac2, and a receptor expression study by Benton et al. (2009) which finds 3 neurons in both these types. We have not detected the presence of a third neuron in either of these sensilla, possibly because it does not fire regular spontaneous spikes nor respond to any of the stimuli we presented. A response from the ac3A neuron to propanal (PL) characterizes the ac3 sensillum type. Finally, we were not always able to reliably separate spikes fired by the 3 neurons in ac4 and therefore present the data as combined spike counts.
Our recordings with this diagnostic set of odorants establish a simple protocol for recognizing sensilla housing 38 of the known 47 different ORN classes of Drosophila. From a number of studies performed over the past 5 years, it is possible to infer which Or, Gr, or IR gene underlies the physiological response for the majority of these ORN classes (Couto et al. 2005; Fishilevich and Vosshall 2005; Hallem and Carlson 2006; Kreher et al. 2008; Benton et al. 2009). These receptors are listed in Table 2.
Table 2.
Responses to illicit agents across Drosophila ORNs
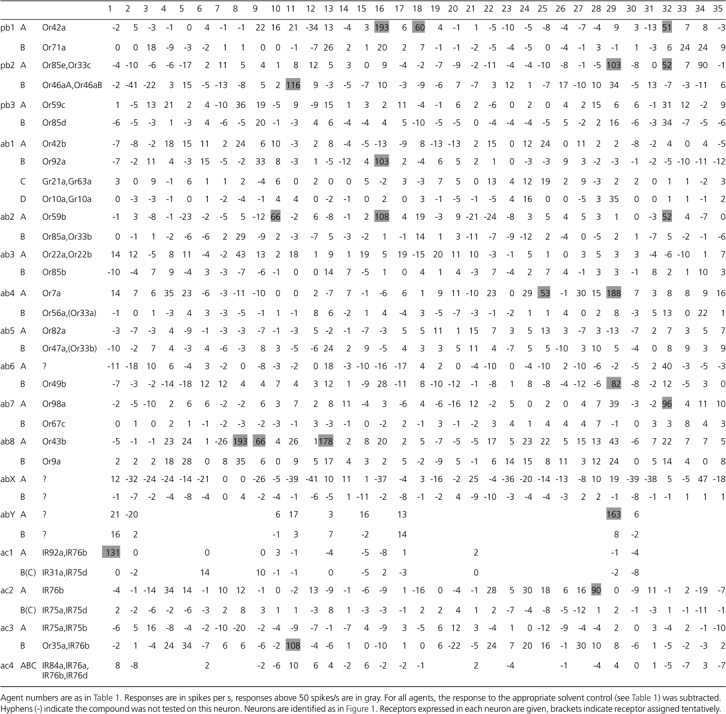 |
Drosophila receptors respond to illicit substances
In order to determine if Drosophila ORNs and thus receptors can respond to indicators of illicit substances, we then tested a set of 35 agents for responses from the 38 different neuron classes. These agents are known to be associated with toxic gasses, explosives, or illicit drugs (Table 1). The list includes some actual toxins and explosives, but also precursors that are a part of the production process, or associated contaminants that are more readily detected by detector dogs than are the actual agents. For instance, 2,3-dimethyldinitrobutane is a contaminant specifically added to plastic and sheet explosives (Harper et al. 2005). The aromatic volatiles benzaldehyde, phenyl-2-propanone, and 1-phenyl-2-nitropropanone are part of one particular method of methamphetamine synthesis (Dayrit and Dumlao 2004). The alkyl phosphites are precursors for organophosphate nerve agents (Francis et al. 2009), whereas the diethanolamines are precursors as well as hydrolysis products of nitrogen mustard blistering agents (Ohsawa and Seto 2006).
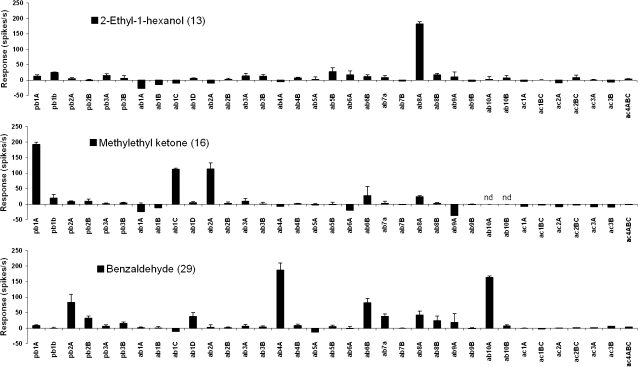
We tested all compounds at a 1% dilution. In addition, we set the threshold for a positive response at a robust 50 spikes/s to maximize the likelihood that responses we designate as positive will translate to detection of lower doses in the field. Out of a total of 1272 agent-neuron combinations, we found 22 positive responses, that is, 1.7% (Table 2). Of the 35 agents, we found positive responses to 13, 5 of these responses being high (>150 spikes/s). Some of the agents excited only one of the neuron classes we tested, whereas others elicited responses from several (Figure 3). For example, the ab8A neuron, expressing the Or43b receptor, is the only one to respond to 2-ethyl-1-hexanol (agent 13). In contrast, methyl ethyl ketone (agent 16) stimulates 3 different neuron classes and benzaldehyde (agent 29) at least 4 different neuron classes to varying degrees. However, responses to individual agents are still highly specific because each compound excites a different set of receptors.
Figure 3.
Responses across all ORNs to 3 selected agents. All responses are after subtraction of the solvent control (paraffin oil). nd, not tested on this neuron. n = 3–8, error bars are standard error of the mean.
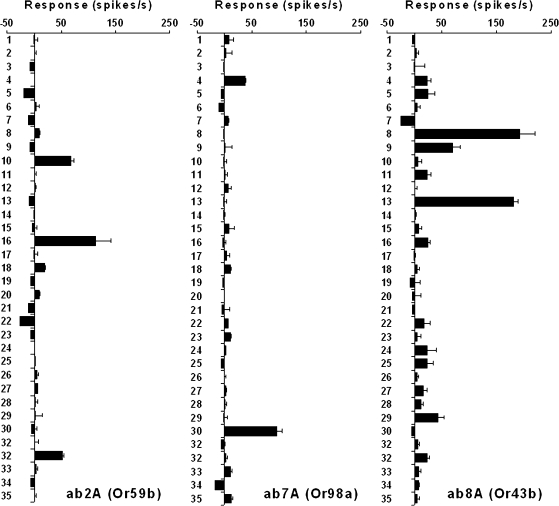
Representative examples showing the response profiles to the complete set of agents for 3 different neuron classes/receptors are given in Figure 4. The ab2A neuron, expressing Or59b, responds to the 2 small ketones methylethyl ketone (agent 16) and acetone (agent 10). The ab7A neuron, expressing Or98a, shows a robust response to phenyl-2-propanone (agent 32). The only other (minor) response is to benzaldehyde (agent 29) which is structurally similar. Finally, the ab8A neuron, expressing Or43b, is excited by several compounds but most prominently by 2 very different chemicals, the aliphatic alcohol 2-ethyl-1-hexanol (agent 13) and organophosphate diethyl phosphite (agent 8), an ester of phosphorous acid. The related triethyl phosphite (agent 9) also excites this neuron. To our knowledge, these 3 compounds have not been previously reported to excite olfactory neurons.
Figure 4.
Response spectra of 3 representative ORNs to all agents. All responses are after subtraction of the appropriate solvent control (paraffin oil). n = 3–8, error bars are standard error of the mean.
Responses from selected transgenes in in vivo expression
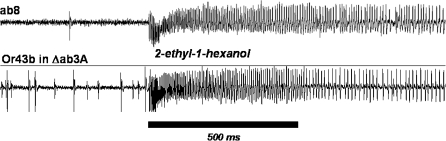
For future studies, it will be important to ensure that the correct receptor is identified that is responsible for the responses to particular agents of interest. We therefore performed experiments to confirm the allocation of responses to agents of interest to individual receptor genes for 2 of the neurons that gave responses of interest. We used the “empty neuron system” developed by Dobritsa et al. (2003) to determine the response from a single Or gene expressed in a nonnative neuron. This system utilizes a mutant strain of Drosophila in which the 2 native receptors of the ab3A neuron have been deleted, this creates a nonresponsive Δab3A in which other receptors can be expressed and functionally tested. The compounds 2-ethyl-1-hexanol (agent 13) and diethyl phosphite (agent 8) were detected by ab8A neurons, which have been shown to express Or43b. We confirmed that this receptor is in fact responsible for detecting these agents by expressing Or43b in the empty neuron. Recording from ab3 sensilla with transformed Δab3A neurons, we saw a robust response to 2-ethyl-1-hexanol (Figure 5A) and diethyl phosphite (not shown) when Or43b was expressed.
Figure 5.
Confirmation of the receptors responsible for the detection of agent 13 Traces of recordings (1.5 s) showing responses to 2-ethyl-1-hexanol mediated by the Or43b gene in its native sensillum ab8 (top trace) and when expressed in Δab3A neurons (bottom trace). Bar represents stimulus time.
Discussion
The Drosophila olfactory neuron array provides a rapid screening system for receptors for compounds of interest
The development of biosensors based on animal ORs requires the identification of receptors for compounds of interest. In many organisms, this can be very difficult as the identification of ligands for receptors requires expression in heterologous systems and such assays have proven technically difficult. In addition, for mammalian ORs the large number of receptors would make comprehensive screening very time consuming. The Drosophila ORs offer particular advantages for identifying receptors for compounds of interest. The response characteristics of many classes of ORNs have been characterized, receptor gene expression has been mapped to these ORN classes, and some ligand information is available for many receptors. This means that identification of receptors for compounds of interest can be accomplished by using in vivo recording to find an ORN class that responds to the compound.
Here, we have taken advantage of this property to show that a large number of Drosophila ORs can be screened in a rapid and inexpensive manner. Building on previously published data on ligand responses of a large number of ORN classes, we first established a protocol for distinguishing 17 olfactory sensilla housing 38 different neuron classes, which utilizes in vivo recording of ORN responses to a relatively small and easily tested set of 11 odorants. We then validated our system by establishing that we could use it to identify receptors for new compounds, including wholly synthetic chemicals that are not known to be natural products. We also demonstrate that once we identify a neuron that responds to a compound of interest, we can show that the receptor known to map to that neuron does in fact respond to the compound using an in vivo functional assay. This screening method and validation approach shows that the Drosophila receptors offer particular advantages for developing biosensors, especially when coupled with the fact that they may not require coexpression of downstream signaling components.
Identification of Drosophila receptors for illicit substances
We used our screening method on a set of 35 chemicals that are indicators of illicit substances and identified neurons, and hence candidate receptors, for 13 of them. Our results thus clearly show that insect ORs can be identified that respond to compounds of interest to law enforcement, security, and emergency response. We found responses for ammonia, diethyl phosphite, triethyl phosphite, acetone, cyclohexanone, 2-ethyl-1-hexanone, methyl ethyl ketone, nitromethane, acetic anhydride, pentaerythritol tetranitrate (PETN), benzaldehyde, phenyl-2-propanone, 1-phenyl-2-nitropropene, and sassafras oil. Some of the responses were high enough to suggest sensitivity at lower doses. However, none of the agents we tested evoked the very high responses (>200 spikes/s) that can be observed with some of the natural odorants that are known to excite particular ORNs at very low doses.
Most of the compounds for which we found responses are industrial products (except sassafras oil), but several also occur as natural products. For instance, acetone (agent 10) and methyl ethyl ketone (agent 16) are organic solvents used extensively in industry. Yet, they are also relatively abundant naturally occurring chemicals. The same can be said for benzaldehyde (agent 29), which is a drug precursor but also a well known natural flavor compound. We also tested sassafras oil (agent 34) which is a natural product that excited the pb2A neuron. Interestingly, Saffrole (agent 33), which usually makes up 80–90% of sassafras oil, does not excite this neuron. It is thus likely that the response is to one of the minor terpenoid or phenyl propanoid components of the oil.
Of particular interest were our findings that several Drosophila ORNs/receptors responded to wholly synthetic chemicals that are not known to be natural products. For instance, pb1A, which expresses the Or42a receptor, responds to nitromethane (agent 18) used in the manufacture of explosives. The ab4A neuron, which expresses Or7a, responds to the explosive PETN (agent 25). PETN is among a number of compounds in Table 2 that have very low volatility (Eiceman et al. 1997), making them unlikely candidates as odorants. However, contaminants or degradation products might be volatile enough to be detected.
The ab8A neuron responded to several compounds of interest, and we used the empty neuron assay to confirm that the Or43b receptor is responsible for the detection of 2 of them. Or43b detects the fatty alcohol 2-ethyl-1-hexanol (agent 13), which is one of the dominant odorants in polymer-based explosives that dogs can detect (Harper et al. 2005). 2-Ethyl-1-hexanol is also widely used as a solvent, whereas its esters are used as emollients and as UV absorbers in sunscreens. Its detection can therefore have applications in various industries. The Or43b receptor also responds to diethyl phosphite (agent 8).
Insect receptors in a biosensor
To what extent do the compounds that are detected by Drosophila ORNs represent generally “smellable” cues? Dogs use 2-ethyl-1-hexanol as a cue to find polymer-based explosives (Harper et al. 2005), and interestingly, we found a Drosophila receptor for this compound. The odorant present in cast-based explosives that dogs can detect is 2,4-dinitrotoluene (Harper et al. 2005), to which honeybees can also be trained to respond at concentrations comparable with dogs (Shaw et al. 2005). Thus, it may well be that insect receptors can be found for many of the cues that detector dogs use to find illicit substances.
Although trained bees and wasps can be used in the field (Shaw et al. 2005; Rains et al. 2006), there is no precedent for Drosophila. It is certainly possible to train Drosophila in the laboratory, but its long-term olfactory memories are less stable than in bees (Meller and Davis 1996). A more promising way to make use of the olfactory sensitivities demonstrated here is coupling receptor proteins to an electronic or optical readout in an automated standoff detection device. At present there is no established system to transduce activity of any OR to the electronics of a typical sensing device but several approaches have been explored. For instance, Hou et al. (2007) immobilized the rat I7 receptor within a lipid environment on an electrode, whereas Lee et al. (2009) used a planar electrode to measure extracellular potentials generated by HEK-293 cells expressing the same receptor. Fluorescence measurements of yeast cells that were engineered to couple the mammalian signaling components to GFP were used by Radhika et al. (2007) to characterize another rat receptor.
Insect receptors might prove more robust and, forming ion channels themselves, may need less biological components to function. Odor responses have been measured after heterologous expression of Drosophila Ors in various cell types such as insect Sf9 cells (Smart et al. 2008), HEK-293 cells (Wicher et al. 2008), and Xenopus oocytes (Sato et al. 2008) requiring only the addition of the coreceptor Or83b. The challenge is to express them in cellular or cell-free systems coupled to electronics similar to what has been achieved with vertebrate Ors. Their tuning width and sensitivities have evolved to suit a system of a limited number of detectors (ca. 50), perhaps making it more likely a machine replicate of an insect nose can be designed.
Odor coding across arrays of ORNs
As well as establishing and validating a screening protocol, our data also provides a nearly complete map of the coding of 11 biologically relevant odorants across the majority of ORNs that comprise the Drosophila adult olfactory system. To date no single olfactory system, vertebrate or invertebrate, has been characterized in sufficient detail to allow a complete understanding of odor coding. Our analysis covers 38 different ORN classes, more than any previous single study, but is still not complete. There is evidence from receptor mapping for at least 10 more ORN classes in sensilla of the trichoid and intermediate category (Couto et al. 2005), which we did not include in our study. Physiological studies on 7 of the 13 Or genes expressed in these sensilla show that some of our 11 diagnostic odors induce excitatory or inhibitory responses from them (Hallem and Carlson 2006). In future studies, the diagnostic set may need to be expanded to include these sensilla.
How many neuron classes make up the complete olfactory input to the fly's brain? The antennal lobe of the Drosophila brain has 54 glomeruli; the primary processing units for all sensory neuron classes on antennae and palps (Laissue and Vosshall 2008) but not all of them are olfactory. So far 47 different ORN classes are known to project there, each to one individual glomerulus. Thus a further 7 can be expected. At least 3 of the remaining glomeruli are known to receive axons from neurons in the arista that are not olfactory (Laissue and Vosshall 2008). The other 3 are most likely associated with sensilla of the sacculus, a 3-chambered invagination of the antennal surface (Shanbhag et al. 1995). Due to the inaccessibility of these sensilla, nothing is known about their physiology. It is not known how many of these are olfactory but some express IR genes (Benton et al. 2009). Thus our analysis comprises 38 of 47–51 ORN classes that make up the entire olfactory input to the brain of Drosophila.
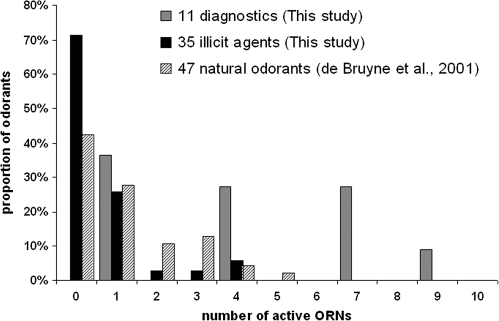
Assessments of odor coding across neurons are inevitably influenced by the choice of odors tested. In Figure 6, we compare the neuronal activity across Drosophila ORNs for 3 different sets of chemicals, including the 2 odor sets used in this study. First, we show our 11 natural diagnostic odorants that were chosen specifically to differentiate between Drosophila olfactory sensilla and were thus expected to excite at least one, preferably several ORNs. Second, we include our 35 agents that were chosen for their use as indicators of illicit substances, many of them not being natural stimuli. Third, a set of recordings from 16 ORNs in antennal basiconics (ab1–ab7) using a set of 47 natural odorants chosen for their generation of responses from ORNs in other insect species, prior to detailed knowledge of Drosophila ORNs (de Bruyne et al. 2001). The frequency distributions indicate the percentage of odor stimuli that excite a particular number of neurons. The results show that although the proportion of stimuli that do not elicit responses from any neurons is considerably higher in the set of 35 illicit agents than in the 47 natural odorants, the distribution is not dramatically different. This may indicate the shape of the probability distribution for future screens. The comparison also clearly shows the shifted distribution of the responses to the diagnostic set, which is highly biased toward responses from multiple ORNs and contains no odorants that fail to excite an ORN.
Figure 6.
Distributions of ORN responses to 3 different sets of odorants. The percentage of odor stimuli that evoke a response from n ORNs is plotted against n. ORN response is defined as >50 spikes/s above the solvent control.
Finally, our data may provide the first published response spectra from neurons of the ab9 and ab10 sensilla, which had previously been proposed to exist based on Or gene expression data (Couto et al. 2005). Responses we recorded from the A neuron in the abY sensillum closely resemble those recorded by Hallem and Carlson (2006) for Or67a which Couto et al. (2005) find in a sensillum type they call ab10. The (low) responses from the B neuron in abY also match the combined spectra of Or49a (Kreher et al. 2008) and Or85f (Hallem and Carlson 2006) that were shown to be coexpressed in neurons paired with Or67a-expressing neurons, that is, in the same sensillum. The data therefore suggest that our abY is identical to ab10 as defined by Couto et al. (2005) and that Or67a is expressed in ab10A neurons and Or49a and Or85f in ab10B neurons.
Is abX the ab9 sensillum? According to Couto et al. (2005) ab9 contains one neuron that expresses Or69a and Or69b and a second that expresses Or67b. For these 3 genes, there is only a published response spectrum for Or67b (Kreher et al. 2008), and this response does not correspond to either of the 2 neurons in abX. In particular, Or67b showed a high response to benzaldehyde (BZ), which we did not find in either neuron. The high response to the paraffin oil control we found for the A neuron also complicates a correct identification and we thus cannot assign the label ab9 to abX with any confidence.
Conclusion
Our study establishes a rapid protocol for finding receptors for novel compounds by screening Drosophila ORNs for responses. We demonstrate that several Drosophila ORNs detect compounds of interest to law enforcement, security, and emergency response. We confirm the receptors responsible for responses to several compounds using the “empty neuron” approach. These results should facilitate the use of insect receptors as biosensors for volatile organic compounds with applications in various branches of industry or government.
Funding
This work was supported by the National Security Science and Technology Unit of the Australian government under the Research Support for Counter Terrorism Program.
Acknowledgments
We thank Stephen Trowell for conceptualizing this study, and Chunyan Liao, Alisha Anderson, Michelle Michie, and Amalia Berna for assistance (Commonwealth Sientific and Industrial Research Organization Entomology, Canberra). We also thank John Chiefari and Sophie Dixon (Prime Minister and Cabinet), Susan Shahin (Emergency Management in Australia), and Paul Kirkbride (Australian Federal Police, Canberra). We are grateful to John Carlson (Yale University) for supplying the transgenic flies.
References
- Benton R, Sachse S, Michnik SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLOS Biol. 2006;4 doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson BJ. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005;15:1535–1547. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Dayrit FM, Dumlao MC. Impurity profiling of methamphetamine hydrochloride drugs seized in the Philippines. Forensic Sci Int. 2004;144:29–36. doi: 10.1016/j.forsciint.2004.03.002. [DOI] [PubMed] [Google Scholar]
- De Bruyne M, Clyne PJ, Carlson JR. Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci. 1999;19:4520–4532. doi: 10.1523/JNEUROSCI.19-11-04520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyne M, Foster K, Carlson JR. Odor coding in the Drosophila antenna. Neuron. 2001;30:537–552. doi: 10.1016/s0896-6273(01)00289-6. [DOI] [PubMed] [Google Scholar]
- Dobritsa A, van der Goes van Naters WM, Warr CG, Steinbrecht RA, Carlson JR. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- Eiceman GA, Preston D, Tiano G, Rodriguez J, Parmeter JE. Quantitative calibration of vapor levels of TNT, RDX, and PENT using a diffusion generator with gravimetry and ion mobility spectrometry. Talanta. 1997;45:57–74. doi: 10.1016/s0039-9140(97)00107-0. [DOI] [PubMed] [Google Scholar]
- Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15:1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Francis GJ, Milligan DB, McEwan MJ. Detection and quantification of chemical warfare agent precursors and surrogates by selected ion flow tube mass spectrometry. Anal Chem. 2009;81:8892–8899. doi: 10.1021/ac901486c. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Harper RJ, Almirall JR, Furton KG. Identification of dominant odor chemicals emanating from explosives for use in developing optimal training aid combinations and mimics for canine detection. Talanta. 2005;67:313–327. doi: 10.1016/j.talanta.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Helfand SL, Carlson JR. Isolation and characterization of an olfactory mutant in Drosophila with a chemically specific defect. Proc Natl Acad Sci U S A. 1989;86:2908–2912. doi: 10.1073/pnas.86.8.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Jaffrezic-Renault N, Martelet C, Zhang A, Minic-Vidic J, Gorojankina T, Persuy MA, Pajot-Augy E, Salesse R, Akimov V, et al. A novel detection strategy for odorant molecules based on controlled bioengineering of rat olfactory receptor I7. Biosens Bioelectron. 2007;22:1550–1555. doi: 10.1016/j.bios.2006.06.018. [DOI] [PubMed] [Google Scholar]
- King TL, Horine FM, Daly KC, Smith BH. Explosives detection with hard-wired moths. In: Braudaway DW, Zoughi R, editors. Proceedings of the 20th Instrumentation and Measurement Technology Conference; 2003 May 20–22. Piscataway. Piscataway (NJ): IEEE Publications; 2003. [Google Scholar]
- Kreher SA, Mathew D, Kim J, Carlson JR. Translation of sensory input into behavioral output via an olfactory system. Neuron. 2008;59:110–124. doi: 10.1016/j.neuron.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue PP, Vosshall LB. The olfactory sensory map in Drosophila. Adv Exp Med Biol. 2008;628:102–114. doi: 10.1007/978-0-387-78261-4_7. [DOI] [PubMed] [Google Scholar]
- Lee SH, Jun SB, Ko HJ, Kim SJ, Ko HJ, Park TH. Cell-based biosensor using microfabricated planar electrode. Biosens Bioelectron. 2009;24:2659–2664. doi: 10.1016/j.bios.2009.01.035. [DOI] [PubMed] [Google Scholar]
- Meller VH, Davis RL. Biochemistry of insect learning: lessons from bees and flies. Insect Biochem Molec Biol. 1996;26:327–335. doi: 10.1016/0965-1748(95)00100-x. [DOI] [PubMed] [Google Scholar]
- Ohsawa I, Seto Y. Determination of nitrogen mustard hydrolysis products, ethanolamines by gas chromatography-mass spectrometry after tert-butyldimethylsilyl derivatization. J Chromatogr A. 2006;11222:42–248. doi: 10.1016/j.chroma.2006.04.076. [DOI] [PubMed] [Google Scholar]
- Radhika V, Proikas-Cezanne T, Jayaraman M, Onesime D, Ha JH, Dhanasekaran DN. Chemical sensing of DNT by engineered olfactory yeast strain. Nat Chem Biol. 2007;3:325–330. doi: 10.1038/nchembio882. [DOI] [PubMed] [Google Scholar]
- Rains GC, Utley SL, Lewis JW. Behavioral monitoring of trained insects for chemical detection. Biotechnol Prog. 2006;22:2–8. doi: 10.1021/bp050164p. [DOI] [PubMed] [Google Scholar]
- Röck F, Barsan N, Weimar U. Electronic nose: current status and future trends. Chem Rev. 2008;108:705–725. doi: 10.1021/cr068121q. [DOI] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Shanbhag SR, Singh K, Singh RN. Fine structure and primary sensory projections of sensilla located in the sacculus of the antenna of Drosophila melanogaster. Cell Tissue Res. 1995;282:237–249. doi: 10.1007/BF00319115. [DOI] [PubMed] [Google Scholar]
- Shaw J, Seldomridge N, Dunkle D, Nugent P, Spangler L, Bromenshenk J, Henderson C, Churnside J, Wilson J. Polarization lidar measurements of honey bees in flight for locating land mines. Opt Express. 2005;13:5853–5863. doi: 10.1364/opex.13.005853. [DOI] [PubMed] [Google Scholar]
- Smart R, Kiely A, Beale M, Vargas E, Carraher C, Kralicek AV, Christie DL, Chen C, Newcomb RD, Warr CG. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38:770–780. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 2009;71:307–332. doi: 10.1146/annurev.physiol.010908.163209. [DOI] [PubMed] [Google Scholar]
- Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- Wilson RI, Mainen ZF. Early events in olfactory processing. Annu Rev Neurosci. 2006;29:163–201. doi: 10.1146/annurev.neuro.29.051605.112950. [DOI] [PubMed] [Google Scholar]
- Yao CA, Ignell R, Carlson JR. Chemosensory coding by neurons in the coeloconic sensilla of the Drosophila antenna. J Neurosci. 2005;25:8359–8367. doi: 10.1523/JNEUROSCI.2432-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]