Abstract
Variation in taste perception of different chemical substances is a well-known phenomenon in both humans and animals. Recent advances in the understanding of sweet taste signaling have identified a number of proteins involved in this signal transduction. We evaluated the hypothesis that sequence variations occurring in genes encoding taste signaling molecules can influence sweet taste perception in humans. Our population consisted of unrelated individuals (n = 160) of Caucasian, African–American, and Asian descent. Threshold and suprathreshold sensitivities of participants for sucrose were estimated using a sorting test and signal detection analysis that produced cumulative R-index area under the curve (AUC) scores. Genetic association analysis revealed significant correlation of sucrose AUC scores with genetic variation occurring in the GNAT3 gene (single point P = 10−3 to 10−4), which encodes the taste-specific Gα protein subunit gustducin. Subsequent sequencing identified additional GNAT3 variations having significant association with sucrose AUC scores. Collectively, GNAT3 polymorphisms explain 13% of the variation in sucrose perception. Our findings underscore the importance of common genetic variants influencing human taste perception.
Keywords: association, genetic variations, gustducin, sucrose
Introduction
Humans display substantial variation in the perception of natural and artificial chemical compounds for all known taste modalities, including sweet, bitter, umami, and sour tastes (Blakeslee and Salmon 1935; Breslin 2000; Kim et al. 2003; Chen et al. 2009). Whereas the environment plays a major role in determining individual differences in recognition thresholds for saltiness (Wise et al. 2007), inherited factors significantly contribute to the individual differences in the perceived intensity for bitter substances (Hansen et al. 2006) and recognition thresholds for bitter and sour tastes (Wise et al. 2007).
There are modest interindividual differences in the detection and recognition thresholds for sweeteners (Blakeslee and Salmon 1935; Kahn 1951; Okoro et al. 2000). The responses to sugars including sucrose tend to be unimodally distributed in the population (Blakeslee and Salmon 1935; Fushan et al. 2009). Although studies of sweet food preferences have revealed heritable differences (Bretz et al. 2006, Keskitalo et al. 2007), there have been no family studies published regarding threshold and perceived intensity measures of sweet perception, and Mendelian transmission has not been reported for variation in sweet taste sensitivity. A previous population-based study using psychophysical measures revealed that genetic variants at the TAS1R3 locus are strongly associated with sucrose perception (Fushan et al. 2009). However, these variants explain only 16% of the variation in these measures, and thus, the impact of genetic factors in human sweet taste variation is not fully understood.
There are a number of reported observations that the phenylthiocarbamide recognition thresholds are closely related with taste detection and recognition thresholds for sucrose (Hong et al. 2005; Chang et al. 2006). This could indicate the existence of partially common mechanisms influencing the threshold sensitivity variations for these substances because bitter and sweet taste signaling share common downstream pathways.
Recent advances in the understanding of mammalian taste transduction mechanisms have identified several signaling molecules, including gustducin (a G-protein α subunit involved in signal transduction of sweet, bitter, and umami tastes), G-protein polypeptides beta 3 and gamma 13, phospholipase C-β2, inositol triphosphate receptor, and, most recently, the transient receptor potential–like channel M5 (TRPM5, Gilbertson et al. 2000; Margolskee 2002). Expression patterns of these proteins were shown to be predominantly restricted to the taste sensory epithelium (Zhang et al. 2003). Thus, functional variations in these genes could result in phenotypic variation mainly within the chemosensory apparatus, whereas other organ systems would be unaffected.
We evaluated the hypothesis that common genetic variations occurring in the genes encoding taste signaling molecules can influence sweet taste perception in humans. Nucleotide sequence variations at the following gene loci were tested for the association with sucrose sensitivity: GNG13 (16p13), GNB3 (12p13), PLC-β2 (15q15), ITPR3 (6p21), TRPM5 (11p15), and GNAT3 (7q21), along with TAS1R1 (1p36), which encodes the umami-specific component of the TAS1R1/TAS1R3 taste receptor (Li et al. 2002).
Materials and methods
Subjects
Participants were enrolled with written informed consent under National Institutes of Health Combined Neuroscience Institutional Review Board protocol 01-DC-0230 and contained individuals (69 males and 91 females) who identified themselves as Caucasian (n = 103), Asian (n = 41), or African–American (n = 16). African–American individuals had origins in the sub-Saharan racial groups of Africa.
Psychophysical measurements
Measurements of sensitivity to sucrose were performed as described previously (Fushan et al. 2009). A series of preliminary trials empirically determined that solutions of 0, 0.5%, 1%, 2%, 2.4%, 2.8%, 3.2%, 3.6%, and 4% sucrose (Sigma, dissolved in deionized water) produced the best discrimination curves in a representative subpopulation of our subjects. Each concentration is used to calculate a detection threshold for a given sucrose interval (i.e., 0–0.5%, 0.5–1%, etc.) (see Supplementary Methods for further details). Each subject participated in 1 experiment that consisted of 6 replications performed over 3 sessions. Subjects were asked to complete 2 replications of the ranking test per session with a mandatory 5-min break between replications. Individual sessions were separated by at least a 24-h period. Subjects were presented with 20 ml of each of the solutions in randomized order and could ask for more at any time during the experiment. Panelists were asked to sample each of the solutions and rank them in order from least to most sweet. To minimize adaptation effects, subjects rinsed with water between each sample.
Phenotype modeling
Data from the 6 replications were pooled for each subject. For each pairwise sucrose concentration (0–0.5%, 0.5–1%, etc.), the R-index (hereafter referred to as Rp-index; see Supplementary Figure 1) was calculated as described in O'Mahony et al. (1992). The Rp-index is simply the R-index calculated between solutions p and p + 1; it ranges from 0.5 to 1 and reflects discrimination performance of a subject by accounting for individual differences in decision-making criteria. As such, it is an estimate of the magnitude of difference between 2 difficult to discriminate stimuli. Thus, the higher the Rp-index the greater the taste sensitivity as reflected by better performance on a sweetness discrimination task. Rp-indices were then used to derive the cumulative R-index (Rc) by summing the Rp-indices derived at each concentration step according to the equation below:
where 0.5 = chance detection for comparing 0% sucrose against itself and j {1, 2 … 8} and p = 1, 2, … j.
Rcj is a quantitative measure of ability to detect a signal in a background of noise and across a number of different intensities of the signal (after subtracting a chance level). Rc reflects the cumulative sensitivity of a subject across a given sucrose concentration range, and it can be used to estimate the theoretical suprathreshold sensitivity at each concentration across the range because it is derived from the summing of the Rp calculated at each concentration interval. For example, for the 2 given concentration intervals (i.e., 0–0.5% and 0.5–1%), we can calculate 2 pairwise R-indices, Rp1 and Rp2. Thus, the Rc-index for the second interval will be (Rp1 − 0.5) + (Rp2 − 0.5). Because Rc is obtained by summing Rp1 and Rp2, it also reflects a subject's ability to discriminate the 1% stimulus from 0%. For each subject, all Rc-indices across the range were then plotted. The format of the resulting graph is equivalent to a concentration response curve with increasing sucrose concentrations receiving larger Rc scores (see Supplementary Figure 1). The area under the Rc curve (AUC) was then calculated for each individual. The AUC is a unitary measure of overall discriminability across the full concentration range tested. The AUC was used as the dependent variable for assessing the association of genotype with phenotype.
Single nucleotide polymorphism marker selection
Our goal was to evaluate genes likely to be involved in the human sweet taste transduction pathway, including discovery of new functional polymorphisms and assessment of the phenotypic impact of known variants. For selected genes, we first surveyed recent reviews for known polymorphisms reported to be related to functional variations. In addition, we searched the dbSNP database (www.ncbi.nlm.nih.gov/sites/entrez?db=snp) for single nucleotide polymorphisms (SNPs) in transcribed regions of these genes with minor allele frequency (MAF) > 0.05, which allowed us to capture the main haplotypes in these genes and to exclude rare genotypes. Such SNPs from dbSNP were frequent and had been validated in multiple research projects, such as the HapMap project (www.hapmap.org). The following additional criteria were used for selection of tag SNPs: location in untranslated regions putatively important for gene regulation, exon/intron boundaries which could be associated with multiple messenger RNA (mRNA) isoforms producing exonic variations, or amino acid alterations in proteins.
Overall, 233 SNP markers were selected: 12 for TAS1R1, 30 for GNG13, 19 for GNB3, 10 for PLC-β2, 18 for ITPR3, 17 for TRPM5, and a total of 127 SNPs for the GNAT3 and CD36 loci. Detailed description of these SNPs can be found in Supplementary Tables 1–11.
Genotyping
SNPs were genotyped using the Applied Biosystem SNPlex Technology using an Applied Biosystems 3130xl DNA Analyzer and the GeneMapper 4.0 software (Applied Biosystems). Random individuals (n = 96) were genotyped in duplicate to assess genotyping accuracy.
Quality control for individual genotyping
For quality control, the following sequential criteria were applied: SNPs were omitted from analysis if poor genotype clusters prevented GeneMapper 4.0 (Applied Biosystems) software from making calls. For each SNP, low peak height genotypes (<25% of the average peak height) were removed because poor-quality samples often exhibit high background that SNPlex can mistake as heterozygotes. It is important to control for this as an excess of heterozygotes will artificially inflate the type-I error rate in Hardy–Weinberg equilibrium (HWE) tests. Any SNPs with less than 95% of the samples auto-called by the software were then either rescored manually or discarded if clustering confidence was low. Only genotypes with a GeneMapper Quality Score greater than 98% were subsequently used for analysis. In the DNA samples in this study, the call rate was 99%, the reproducibility rate was 100%, and the concordance rate was 99.8%. Reproducibility was determined by comparing blinded replicates on plates and by rerunning entire plates.
SNP imputation
Imputation of additional SNP markers was done for each subject population using information on SNP variations in the HapMap samples (HapMap release 23) to infer missing genotypes “in silico.” HapMap data were used from the following 4 human populations: Centre d'Etude du Polymorphisme Humain (Utah residents with ancestry from northern and western Europe) (CEU, 90 samples), Japanese in Tokyo, Japan (JPT, 45 samples), Han Chinese in Beijing, China (CHB, 45 samples), and Yoruba in Ibadan, Nigeria (YRI, 90 samples). Imputation of additional SNP markers was done for each subject population using genetic variation data from the corresponding HapMap population: CEU genetic variation data set for the Caucasian subject population, combined JPT and CHB genetic variation data set for the Asian subject population, and YRI genetic variation data set for the African–American subjects.
Model parameters were estimated using MACH v1.0.15 (Li et al. 2009), prioritized for high call rate (>99%) and absence of deviations from HWE (P > 0.001), MAF > 0.05, and non-outlier status in EIGENSTRAT principal component analysis (Price et al. 2006). With these model parameters, we used a hidden Markov model as implemented in MACH 1.0.15 to impute allele dosage, defined as the expected number of copies of the minor allele (a fractional value between 0 and 2) of autosomal SNPs. The average posterior probabilities for the most likely genotypes were reported as quality scores.
Statistical analysis
Before statistical analysis, all SNP markers were examined for the departure from HWE using a permutation version of the exact test implemented in PowerMarker 3.25 software (Liu and Muse 2005). Technical reasons, such as assay nonspecificity resulting in genotyping errors, and natural selection can inflate the type-I error rate, reducing the power of association analyses (Hosking et al. 2004). Therefore, HWE coefficients were calculated for the whole subject population (see above) and for each subject population separately in order to identify potential population substructure. No SNP markers with strong deviations from HWE (P < 0.001) were identified in these tests. SNP markers with MAF < 0.05 were excluded from subsequent statistical analysis.
The significance of association between each genotyped or imputed SNP and AUC scores was assessed using the Wald parametrical statistical test as implemented in PLINK v1.07 software (Purcell et al. 2007). SNP markers with strong deviations from HWE (P < 0.001) or with MAF < 0.05 were excluded from the statistical analysis. A permutation version of the exact test implemented in PowerMarker 3.25 software (Liu and Muse 2005) was used to examine deviations from HWE. Pointwise significance of association was then calculated using permutation procedure with 106 simulations. Deviations from the line of equality indicated either that the theoretical distribution was incorrect or that the sample contained values generated in some other manner (e.g., by a true association). The 233 observed SNP markers used for the association analysis represent 7 gene loci and cannot be considered independent from each other. The Bonferroni method was used to control the level of type-I error for multiple testing correction; however, the Bonferroni method assumes independence of individual tests. Thus, a type-I error value of 0.0002 (0.05/233) was too conservative in our analysis because it fails to include information about linkage disequilibrium (LD) relationships between the markers. Therefore, we evaluated LD relationships between SNP markers within the individual gene loci (data not shown) and identified 11 LD blocks across the whole data set. Because these LD blocks are essentially independent of each other, we reduced the level of type-I error threshold to a nominal value of 0.005 (0.05/11 ≈ 0.005). Also, we used a Q–Q plot to identify the level of P value at the point where the empirical distribution starts to deviate from the theoretical values (P ≈ 0.005). Deviations from the line of equality indicated either that the theoretical distribution was incorrect or that the empirical distribution contained values generated in some other manner (e.g., by a true association). Thus, SNPs with permutated P values < 0.005 were considered as significant in our association analysis. The fraction of genetic variance explained by the SNPs was assessed through the standard coefficient of determination (R2).
Haploview 4.1 (Barrett et al. 2005) was used to compute pairwise LD statistics for the SNP markers. The values of the standardized disequilibrium coefficient (D′ = D/D max, proportion of observed LD out of maximum possible LD) and correlation coefficient (r2) were calculated. The genetic structure of the populations and basic parameters of molecular diversity were calculated using the PowerMarker 3.25 software (Liu and Muse 2005). Median-joining network was constructed using software Network (www.fluxus-engineering.com).
The effect of haplotypes on sucrose perception was evaluated using linear regression analysis with AUC scores as the dependent variable and haplotypes as the independent variable. Inferring of haplotype phases was performed using a Bayesian statistical method and recombination model as implemented in the PHASE v2.1 software (Stephens et al. 2001). An omnibus test implemented in PLINK v1.07 software was performed comparing the alternate (each particular haplotype has a unique effect) versus the null (all other haplotypes combined without significant effect).
The proportions of variance in the outcome explained by the regression model were estimated based on the value of determination coefficient (R2) and F-test statistics. Two-way SNP–SNP interactions were investigated using multivariate logistic models as implemented in PLINK v1.07 software.
Sequencing
The sequence region of chromosome 7 (79934373–79992564 bp) containing 13386 bp sequence upstream of the GNAT3 ATG site and 44806 bp downstream was polymerase chain reaction (PCR) amplified from the genomic DNA using 6 overlapping primer pairs (Supplementary Table 13). PCR reactions were done using LA Taq DNA polymerase (TaKaRa Bio USA) under conditions as follows: 98 °C for 10 s and 68 °C for 20 min for a total of 30 cycles. PCR products were completely sequenced using BigDye Terminator v3.1 chemistry and a 3130xl Genetic Analyzer (Applied Biosystems).
Cell culture
NCI-H716 cells (CCL-251, American Type Culture Collection [ATCC]) were grown as a suspension in RPMI 1640 medium (ATCC) supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C with 5% CO2.
5′ rapid amplification of cDNA ends
Total RNA was extracted from the NCI-H716 cells using RNAeasy kit (Qiagen) according to the manufacturer's instructions. To determine the 5′ end of the GNAT3 mRNA, 5′ rapid amplification of cDNA ends (RACE) was performed using an Invitrogen 5′ RACE System (catalog no. 18374-058) with modifications. The first-strand complementary DNA (cDNA) was synthesized from 1 μg of total RNA from the NCI-H716 cells using a gene-specific primer GSP1 (Supplementary Table 13) followed by RNase treatment according the manufacturer's specifications. The resulting cDNA was purified using a Simple Nucleic Acid Purification column and tailed with deoxycytidine triphosphate and terminal deoxynucleotidyl transferase. PCR was used to amplify the dC-tailed cDNA using the abridged anchor primer and the GSP2 primer (Supplementary Table 13). The PCR product was reamplified by using abridged universal amplification primer and the nested gene-specific primer GSP3. The final PCR products were purified with a QIAquik PCR purification kit (Qiagen), double digested with NheI and HindIII, and individually inserted into NheI/HindIII-linearized pGL4.10 vector (Promega).
Bioinformatic analyses
TRANSFAC 7.0 database (Matys et al. 2003) was used to search for the potential cis-regulatory elements within the GNAT3 noncoding regions. The cutoff selection level for the matrix group was set at 0.85. Multiple sequence alignments were performed using CLUSTAL W program (Thompson et al. 1994). Manual correction of alignments was carried out using Geneious software (http://www.geneious.com/).
Results and discussion
Sample population and sucrose phenotypic data
The objective of this study was to evaluate the impact of genetic variations in genes encoding sweet taste signaling molecules on human sucrose perception. The subject population included 160 normal unrelated individuals of Caucasian (n = 103), Asian (n = 41), and African–American (n = 16) descent. Peri- and suprathreshold sensitivities of these individuals to sucrose were evaluated using sensory signal detection theory utilizing R-index measures (O'Mahony 1992).
Application of signal detection theory provided detailed information about subjects’ perception abilities in comparison to other testing methodologies. Signal detection theory views the detection of a stimulus (threshold measurement) as the outcome of a decision-making process dependent on both the degree of confidence the individual has in their judgment as well as the accuracy of the decision (Delwiche and O'Mahony 1996). The signal detection approach treats taste threshold measurements as both a perceptual and a cognitive process and thereby controls for individual differences in decision-making criteria (Delwiche and O'Mahony 1996).
Participants were asked to rank 9 sucrose solutions ranging from 0% to 4% sucrose in order from least to most sweet. The concentration step between 2 close solutions represented a detection threshold potentially recognizable by the majority of subjects (see Supplementary Methods for the further details). Eight pairwise R-indices (Rp) were calculated for each subject based on the data from 6 replications of this test. Rp-index reflects a subject's discrimination performance of 2 stimuli (Brown 1974; O'Mahony 1992). Although we generated data at 8 concentration differences across the range, we summarized each subject's cumulative performance across the entire concentration range by calculating a cumulative R-index (Rc). The Rc at a given concentration is derived by summing all Rps (after controlling for chance detection) derived across the range of solutions preceding that concentration. Thus, the value of Rc reflects the cumulative sensitivity across a given concentration range.
The AUC produced statistically reproducible measures for the taste sensitivities and reflects discrimination performance of these subjects across the whole concentration range. Physiologically, AUC is a more broad measure than a single R-index. Individual Rp-indices do not reflect the unequal weight of errors made by subjects at threshold versus suprathreshold concentrations (i.e., a failure to distinguish 0.5% from 1% sucrose interval is quite different than the failure to discriminate 0.5% from 4% sucrose). Cumulative Rc-indices, and hence AUC scores, will do so. For example, a subject having difficulties in discriminating 1% versus 0.5% sucrose solutions may have perfect performance over the other sucrose intervals. In this case, the AUC score of this subject will be relatively high. On the other hand, a discrimination error made between solutions of large difference (i.e., 0.5% vs. 4% sucrose) means that this subject will demonstrate poor performance at any of the other concentration intervals across this range. This will dramatically affect the AUC score. Thus, 2 subjects with equal individual Rp-indices at a particular sucrose interval can have quite different AUC values over the full concentration range tested. Therefore, the AUC is a quantitative measure that reflects psychophysical abilities to discriminate stimuli at threshold and also suprathreshold concentrations.
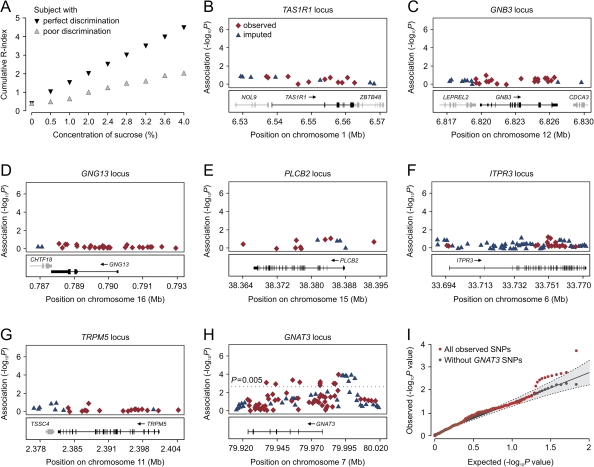
Figure 1A demonstrates examples of Rc-indices plotted as a function of concentration for the 2 representative subjects with different discrimination abilities. The AUCs produced statistically reproducible measures for the taste sensitivities and reflects discrimination performance of these subjects across the whole concentration range. AUC scores followed a Gaussian distribution in our subject population (data not shown) and were used as the dependent variable in parametrical statistical analysis for assessing the correlation between genotypes and phenotype.
Figure 1.
The results of single-SNP association analysis. (A) Cumulative Rc-indices of 2 subjects plotted as a function of sucrose concentrations. Gray inverted triangles, subject with perfect discrimination abilities; red triangles, subject with low discrimination abilities. The vertical axis is the cumulative R-index, the horizontal axis shows sucrose concentration intervals. (B–H) Association results for SNPs from 7 candidate gene loci (indicated on the top of each graph). Statistical significance of SNPs at each locus is shown on the −log (P) scale as a function of chromosomal position (NCBI build 36). Genotyped SNPs at each locus are shown in red rhombs. Imputed SNPs indicated by blue triangles. Bottom panel shows genes at each locus as annotated in the University of California–San Cruz Genome Browser Annotation Database as of 16 December 2008. (I) Quantile–quantile plot of tails of the P value distribution for the sucrose AUC scores. Scatter plot of the observed ordered −log P values versus the −log expected ordered P values under the complete null. Dashed lines and gray area indicate the 0.025 and 0.975 pointwise quantiles of the ordered P value under the complete null distribution. The percentiles depicted with dashed lines were calculated using a beta approximation for the distribution of ordered statistics of uniform variates and assuming independence across tests. The gray area was obtained with 1000 permutations. The plot in red is for the P values for all genotyped SNPs, whereas the plot in black is for P values excluding all SNPs within the GNAT3 region.
Results of single-SNP association analysis
A total of 233 SNPs residing at the 7 candidate gene loci were genotyped and analyzed for the association with sucrose AUC scores. The detailed characteristics of these SNPs and data on statistical tests are listed in Supplementary Tables 1–7. For some of these variants, human genetic association studies have already suggested or confirmed functional effects on various phenotypes. For example, it has been shown that GNB3 genetic variations are associated with essential hypertension (Benjafield et al. 1998; Wang et al. 2004) (Supplementary Table 1), whereas ITPR3 SNPs were found to correlate with type 1 diabetes (Roach et al. 2006; Qu et al. 2008) (Supplementary Table 3). Therefore, the purpose of our study was to discover new potentially functional polymorphisms and to assess the physiological impact of both known and newly discovered polymorphisms on sweet taste sensitivity.
Using information on observed SNP markers, we also inferred other common SNPs from the HapMap database in our genetic variation data set. Imputation resulted in an additional 190 SNP polymorphisms with MAF > 0.05 (data not shown), which potentially increased the discovery power of our genotyping panel.
A total of 423 SNPs that were directly genotyped or imputed were tested for the correlation with AUC score variance under the general genetic model. The graphical representation of the results of the statistical analysis is shown in Figure 1. We failed to observe statistically significant correlation of genotyped or imputed SNP alleles with sucrose AUC score variance at the TAS1R1, GNB3, GNG13, PLC-β2, ITPR3, or TRPM5 gene loci (Figure 1B–H, Supplementary Tables 1–6).
Eleven genotyped SNPs residing at the GNAT3 locus, which encodes the taste-specific Gα protein subunit gustducin, displayed significant association with AUC score variation (P < 0.005) (Figure 1H). The results of the statistical analyses for these SNPs are listed in Table 1 and Supplementary Table 7. In addition, we observed significant correlation of several imputed markers with sucrose AUC scores (Supplementary Table 8, see below).
Table 1.
GNAT3 SNP polymorphisms having significant association with sucrose AUC scores
| dbSNP | Coordinates of SNP |
Trait means (genotype: mean ± standard deviation) | Wald test P | R2 | ||
| Chromosome 7a | ATGb | Gene | ||||
| rs7792845 | 79989304 | −10127 | Upstream | CC/CT/TT: 5.9 ± 1.4 ¦ 6.8 ± 1.6 ¦ 7.2 ± 1.0 | 1.00 × 10−4 | 0.095 |
| rs940541 | 79988529 | −9352 | Upstream | CC/CT/TT: 6.2 ± 1.5 ¦ 6.6 ± 1.4 ¦ 7.5 ± 1.0 | 3.50 × 10−3 | 0.058 |
| rs1107660 | 79988067 | −8889 | Upstream | GG/GT/TT: 6.2 ± 1.6 ¦ 6.6 ± 1.4 ¦ 7.5 ± 1.0 | 1.51 × 10−3 | 0.068 |
| rs1107657 | 79987954 | −8776 | Upstream | CC/CT/TT: 6.1 ± 1.6 ¦ 6.7 ± 1.4 ¦ 7.5 ± 1.0 | 1.11 × 10−3 | 0.072 |
| rs1524600 | 79976239 | 2940 | Intron 1 | TT/TC/CC: 4.5 ± 1.4 ¦ 6.1 ± 1.5 ¦ 6.8 ± 1.5 | 1.40 × 10−3 | 0.072 |
| rs6467217 | 79976114 | 3065 | Intron 1 | CC/CT/TT: 5.1 ± 1.0 ¦ 6.0 ± 1.5 ¦ 6.8 ± 1.5 | 3.40 × 10−3 | 0.061 |
| rs6970109 | 79976010 | 3169 | Intron 1 | AA/AC/CC: 5.1 ± 1.0 ¦ 6.0 ± 1.5 ¦ 6.8 ± 1.5 | 2.70 × 10−3 | 0.062 |
| rs6975345 | 79961935 | 17244 | Intron 1 | CC/CT/TT: 5.0 ± 1.5 ¦ 6.2 ± 1.4 ¦ 6.7 ± 1.5 | 3.00 × 10−3 | 0.063 |
| rs10242727 | 79957666 | 21513 | Intron 2 | GG/AG/AA: 5.0 ± 1.6 ¦ 6.2 ± 1.4 ¦ 6.8 ± 1.5 | 1.50 × 10−3 | 0.071 |
| rs6467192 | 79945734 | 33445 | Intron 4 | AA/AG/GG: 5.1 ± 1.5 ¦ 6.2 ± 1.5 ¦ 6.7 ± 1.5 | 3.26 × 10−3 | 0.061 |
| rs6961082 | 79938905 | 40374 | Intron 5 | AA/AC/CC: 5.0 ± 1.3 ¦ 6.0 ± 1.4 ¦ 6.7 ± 1.5 | 2.40 × 10−3 | 0.064 |
March 2006 human reference sequence (NCBI build 36.1).
Gene coordinates of SNPs relative to known translation start site.
Figure 1I shows the quantile–quantile plot of the P values for association. Under the null hypothesis of no association, P values should follow a uniform distribution. However, the quantile–quantile plot of the logarithms of P values showed a strong deviation from the null distribution, likely owing to strong association observed within the GNAT3 locus. After removal of all SNPs from the GNAT3 gene region, the distribution of the logarithms of the P values fits the null distribution except at the tail of distribution (P ≈ 0.005) where the observed P values are smaller than the expected under the null hypothesis.
Gustducin is believed to be an essential component of the mammalian sweet taste transduction pathway (Huang et al. 1999; Ruiz-Avila et al. 2001; Clapp et al. 2008). Gustducin-deficient mouse knockout models have diminished or complete loss of behavioral and electrophysiological responses to sweet substances, indicating the importance of this gene for mammalian sweet taste transduction (He et al. 2002; Glendinning et al. 2005). The role of gustducin in human sweet taste signaling is less studied; however, histochemical and biochemical studies suggest that the function of this protein is very similar to that in animal orthologs (Takami et al. 1994).
Association of significant GNAT3 SNPs with sucrose AUC scores
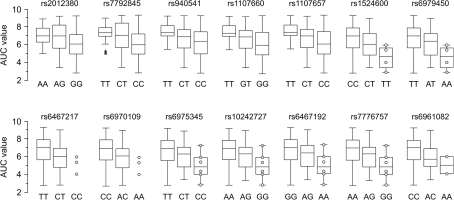
The results of association analysis for the 11 significant SNPs are shown in Table 1. The SNP with the highest association, rs7792845 (permuted P = 1 × 10−4), is located 10 kb upstream of the GNAT3 coding sequence (CDS) and explains 9.5% of the phenotypic variance. Of the 10 other SNPs, 3 (rs940541, rs1107660, and rs1107657) were located 8–9 kb upstream of the GNAT3 ATG translation start site, 4 (rs1524600, rs6467217, rs6970109, and rs6975345) were situated in intron 1, and the remaining 3 SNPs (rs10242727, rs6467192, and rs6961082) were located in introns 2, 4, and 5, respectively (Table 1, Supplementary Table 7). Statistical analysis failed to identify significant association of markers located in coding regions of GNAT3 with sucrose AUC scores (Supplementary Table 7).
In Figure 2, we have plotted the AUC scores as a function of significant SNP genotypes. Each additional copy of the variant alleles is associated with a difference in the mean phenotype. For example, the mean AUC value for the rs7792845 C/C genotype is 5.9, which is significantly different from that of the T/T genotype, which is 7.2. The heterozygote C/T genotype resulted in an intermediate average AUC value of 6.8 (Table 1). This trend suggests additivity of allele effects under the assumption of HWE in this subject population. We observed this effect in Caucasian, Asian, and African–American subject populations (Supplementary Tables 8–10) and in male and female subjects in these populations separately (Supplementary Tables 12 and 13). Thus, the associations between GNAT3 genetic variations and sucrose AUC scores were reproducible between subjects of different ancestry and gender.
Figure 2.
Box–Whisker plots showing the distributions of AUC scores among genotypic groups of the 14 significant SNPs (indicated above each plot). Genotypes are listed at the bottom of each plot. Empty bars correspond to 25% and 75% percentiles and vertical lines outside of the bars indicate value range. Black lines inside of bars are statistical medians. Asterisks indicate outliers. Circles show rare observations. Vertical axis is the AUC score.
LD relationships between significant SNPs
The significant SNPs listed in Table 1 span an ∼55-kb region at the GNAT3 locus. To better understand the relationship between the AUC score–associated SNPs at the GNAT3 locus, LD analysis was assessed in our subject population. LD analysis was performed with Haploview (Barrett et al. 2005) using the block definition proposed by Gabriel and coworkers (Gabriel et al. 2002). With the combined genotype data set, we generated 2 measures of LD: the square of correlation coefficient (r2) and D prime (D′). Our LD analysis revealed several LD blocks within the GNAT3 gene region (Supplementary Figure 3).
SNPs rs2012380 and rs7792845, which are located in the region 10–13 kb upstream of the ATG translation start site and were in LD with each other (r2 = 89, D′ = 97), represented a separate LD block (Supplementary Figure 3). Strong LD also existed between SNPs rs940541, rs1107660, and rs1107657 (r2 = 81–90, D′ = 94–100), which are situated in the 8–9 kb upstream gene region. The LD block represented by these 3 SNPs demonstrated a moderate correlation with the LD block harboring SNPs rs2012380 and rs7792845 (mean r2 = 0.43; Supplementary Figure 3). Thus, these 2 LD blocks were not completely independent from each other. In addition, the high- and low-sensitivity alleles of rs2012380, rs7792845, rs940541, rs1107660, and rs1107657 demonstrated a similar distribution in the sample population, indicating that these SNPs may underlie the same functional effect.
Another group of markers in nearly complete LD included SNPs rs1524600, rs6467217, rs6970109, rs6979450, rs7776757, rs7808426, rs6975345, rs10242727, rs6467192, and rs6961082 (r2 = 86–97, D′ = 100). Thus, all these markers represented a separate ∼38-kb LD block (from +2543 to +40274 bp) that includes exons 2–5 and corresponding intronic sequences of the GNAT3 gene (Supplementary Figure 3). This LD block was in nearly complete linkage equilibrium with LD blocks harboring SNPs rs2012380, rs7792845, rs940541, rs1107660, and rs1107657 (mean r2 = 0–0.07; Supplementary Figure 5).
Examination of the GNAT3 neighboring regions for association with sucrose AUC scores
A region on chromosome 7q was recently linked to components of metabolic syndrome (MetS) in several genome-wide linkage studies (Arya et al. 2002; An et al. 2005). Within this region is the gene CD36, which encodes a membrane receptor for long-chain fatty acids and lipoproteins which can impact a variety of conditions linked with MetS, including insulin resistance, inflammation, and atherosclerosis and may be involved in the etiology of type 2 diabetes mellitus (Ma et al. 2004; Corpeleijn et al. 2006; Love-Gregory et al. 2008). CD36 is located ∼90 kb upstream of the GNAT3 CDS and spans 76 kb. The influence of natural selection has resulted in high CD36 genetic variability in populations of different ancestry (Sabeti et al. 2006; Ayodo et al. 2007). CD36 has several documented alternative promoters that result in the production of at least 3 different mRNA isoforms (Andersen et al. 2006). SNP polymorphisms located in these regulatory regions were demonstrated to predict clinical outcomes (Supplementary Table 11).
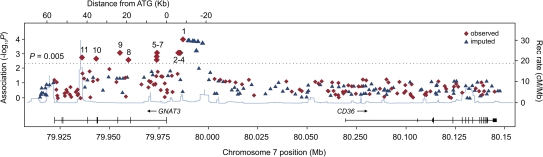
We genotyped a set of polymorphisms across a 140-kb region on chromosome 7q centered around CD36 to evaluate the effect of genetic variation occurring in this region on sucrose AUC scores. These additional markers included 15 SNPs located in the intergenic region between GNAT3 and CD36 and 80 SNPs located within the CD36 locus. Although only 53 of these 80 SNPs were polymorphic in our sample population (Supplementary Table 11), these SNPs represented all main haplotype blocks in this chromosomal region (data not shown). We also included functional CD36 SNPs having known associations with clinical outcomes as described in the literature (Supplementary Table 11).
The results of association analysis for this region are shown in Figure 3. These SNPs had no correlation with sucrose AUC scores, suggesting that the variance observed in this phenotype is unlikely to be due to effects of variation in neighboring genes.
Figure 3.
The results of single-SNP association analyses for chromosomal region 7q. Statistical significance values of SNPs are shown on the −log (P) scale as a function of chromosomal position (NCBI build 36). Genotyped SNPs are shown in red rhombs. Imputed SNPs indicated by colored triangles. Genes and the position of exons, as well as the direction of transcription, are noted below the plot (data from University of California–Santa Cruz genome browser). The most significant SNPs are green diamonds. Estimated recombination rate is plotted in cyan to reflect the local LD structure (data from HapMap).
Identification of additional GNAT3 sequence variants having significant correlation with sucrose AUC scores
Imputation analysis provided the ability to test the hypothesis that other genetic variation also may have significant correlation with phenotype. Association analysis revealed a number of imputed SNPs located upstream of the GNAT3 locus having significant correlation with AUC score variance (Supplementary Table 8). This indicated the possibility that other genetic variations residing at the GNAT3 locus also may have significant association with phenotype. At the same time, we did not identify observed or imputed SNPs located beyond the 18-kb upstream sequence region that had a significant correlation with phenotype (Figure 3).
To verify the results of imputation analysis (Supplementary Table 8) and to identify other genetic variants that may have significant correlation with phenotype, we sequenced the GNAT3 gene region extending from −18 to +40 kb in our entire subject population. This included nucleotide sequence extending from ∼18 kb upstream of the ATG translation start site, the region containing the putative promoter, all the exons 1–5 as well as introns 1–4, and part of intron 5. The results of this sequencing allowed us to validate the results of our association analysis for the previously genotyped SNPs. No genotyping errors were detected for the observed SNPs residing in this sequence region.
We identified 38 additional SNPs with MAF > 0.05 absent from our initial genotyping panel (data not shown). No frequent sequence variations were identified in the GNAT3 protein coding regions. The subsequent statistical analysis showed that only 1 of the 7 imputed SNPs, rs2012380, had significant correlation with phenotype (permuted P = 2.73 × 10−4), whereas the others failed (Supplementary Table 8). SNP rs2012380 is located 12772 bp upstream of the start of the GNAT3 CDS and was in strong LD with rs7792845 (r2 = 89, D′ = 97).
Among the other SNPs identified by sequencing, 3 demonstrated significant correlation with AUC score variance: rs6979450 (P = 4.31 × 10−3), rs7776757 (P = 3.21 × 10−3), and rs7808426 (P = 3.42 × 10−3) (Table 1, Supplementary Table 7). All 3 SNPs were located in intron 1 of GNAT3 and demonstrated strong LD with other SNPs from the 38-kb LD block (D′ = 1) (Supplementary Figure 3).
Associations between race and gender with AUC scores
In order to elucidate the influence of ethnicity and gender on sucrose sensitivities in our sample population, we compared the distribution of AUC scores in subjects based on their ancestral origin and/or sex. The results are shown in Supplementary Figure 2. No significant difference in distribution of AUC scores was observed for subjects of Caucasian and Asian ancestries (Supplementary Figure 2A,B,C). The distribution of AUC scores in the African–American subject population was slightly different from those observed in Caucasian and Asian subject populations (P = 0.05). This could indicate that African–American subjects perceive sucrose at higher concentration thresholds than subjects of Caucasian and Asian ancestries. However, the number of African–American subjects was relatively small (n = 16), and this suggestive observation will require further investigation. No statistically significant differences in AUC score variations were observed between male and female subjects in the combined sample population (Supplementary Figure 2D) and in the Caucasian and Asian sample populations individually (Supplementary Figure 2E,F, respectively). Thus, gender and racial origin had no statistically significant associations with the distribution of AUC scores in our sample population.
Haplotype association analysis
The use of haploytpes defined by tagging SNPs provides potentially greater power to detect association compared with individual SNPs themselves (Liu et al. 2008). However, the statistical power of haplotype-based association analysis is challenged by a trade-off between the benefits of modeling abundant variation and the cost of the extra degrees of freedom for modeling the multimarker variations (Liu et al. 2008).
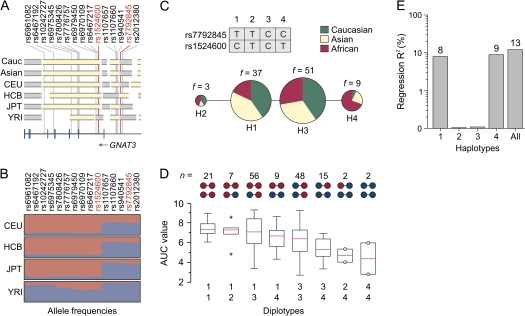
SNPs that were significantly correlated with sucrose AUC scores (Table 1) represented 3 distinct LD blocks spanning much of the GNAT3 sequence region (Figure 4A and see above). We further evaluated the hypothesis that haplotypes defined by particular combinations of high- and low-sensitivity alleles display an additional association with phenotype.
Figure 4.
The results of haplotype association analysis. (A) A graphical presentation of the LD block structure of a 79-kb region across the 7q21 (79.925–80.004 Mb) in Caucasian and Asian sample populations and in 4 HapMap populations (indicated on the left). Individual LD blocks are in gray and yellow. The bottom plot shows the locations of the GNAT3 exons and introns. The positions of 15 significant SNPs identified in this study are indicated by red lines below their rs numbers. (B) Allele frequencies of significant SNPs in 4 HapMap populations (indicated on the left). Individual alleles are represented by colored rectangles. The heights of 2 rectangles indicate relative allele frequencies in population. Red, allele associated with high AUC scores; blue, allele associated with low AUC scores. (C) Median-joining network of 4 haplotypes, color coded according to regions of origin. Haplotypes are indicated by numbers 1–4. The sizes of colored circles correspond to the frequency of haplotype in the entire population (also indicated on the top of each circle). Tag SNPs used for haplotype inference are also indicated. (D) Box–Whisker plots demonstrate distributions of AUC scores as a function of haplotype combinations. Haplotype combinations are indicated on the bottom of the plot. Upper scheme indicates presence of high- (red) and low (blue)-sensitivity alleles in the particular haplotypes. Number of subjects with particular haplotype combinations (n) is also indicated on the top. Empty bars correspond to 25% and 75% percentiles and vertical lines outside of the bars indicate value range. Red lines inside of bars are statistical medians. Asterisks indicate outliers. Circles show rare observations. Vertical axis is the AUC score. (E) The fraction of genetic variance explained by 4 haplotypes. Vertical log axis is a coefficient of determination (regression R2). Haplotypes are indicted on the bottom.
Haplotype inference utilizing the variation patterns of the 15 significant SNPs resulted in the identification of a total of 24 unique GNAT3 haplotypes in our subject population (data not shown). However, many of these occurred at very low frequency. To avoid the analytical problems associated with dimensionality in our data set and to capture the common diversity of the GNAT3 haplotypes, we selected 2 tag SNPs (rs7792845 and rs1524600, sequenced in all individuals) from 2 independent LD blocks (see above) using an association-based criterion and the highest regression R2 values. Thus, rs7792845 represented SNPs rs2012380, rs940541, rs1107660, and rs1107657, whereas SNP rs1524600 represented all the other significant SNP markers from the other LD block (Figure 4A). In addition, we checked variation patterns of significant SNPs in the 4 HapMap populations to avoid the possibility of stratification of the selected SNPs (Figure 4B).
The basis of this design is that even when individual haplotypes defined by specific SNPs do not correlate perfectly with tagged SNPs, haplotype combinations might do so, and these combinations can be identified by selection of the appropriate coefficients in a linear regression. Such a strategy reduces the number of tests and thus false-positive results. SNPs rs7792845 and rs1524600 had the strongest association with sucrose AUC scores, had no significant stratification in the worldwide human populations, and better explained phenotypic variance in comparison to other markers from the corresponding LD blocks. Thus, we concluded that these tag SNPs were sufficient to represent all the other significant SNPs.
We used these 2 SNPs to infer haplotypes and their respective frequencies by a Bayesian statistical method under the general recombination model (Stephens et al. 2001). Bayesian predictions depicted a total of 4 different haplotypes in the sample population (Figure 4C). Two of these haplotypes were common (H1 and H3) in non-African sample populations, with frequencies greater than 30% (Table 2). The remaining haplotypes (H2 and H4) had frequencies of less than 10% in the entire sample population (Figure 4C). H1 represented a combination of the alleles of rs7792845 T and rs1524600 C that were associated with high sensitivity to sucrose (Table 2, Figure 4C). Conversely, in the H4 haplotype, both rs7792845 C and rs1524600 T alleles were associated with low AUC scores. H2 and H3 defined alternative combinations of high- and low-sensitivity alleles (Figure 4C). It is interesting to note that low-sensitivity H4 was much more frequent in the African–American subject population than in the Caucasian and Asian sample populations (Figure 4C, Table 2).
Table 2.
Association analysis of common GNAT3 haplotypes
| Characteristics | Haplotype |
|||
| 1 | 2 | 3 | 4 | |
| rs7792845 allele | T | T | C | C |
| rs1524600 allele | C | T | C | T |
| Frequency (%) | ||||
| Caucasian | 38 | 4 | 54 | 4 |
| Asian | 41 | 1 | 45 | 13 |
| African | 16 | 4 | 38 | 42 |
| F statistic | ||||
| F | 15 | 0.05 | 2.5 | 17 |
| P | 1 × 10−5 | 0.8 | 0.1 | 6 × 10−6 |
| Regression R2 | 0.08 | — | — | 0.09 |
To test whether these haplotypes display a significant association with sucrose AUC scores, we incorporated each GNAT3 haplotype as an explanatory factor in a linear regression model and carried out an omnibus test (Liu et al. 2008). The synthetic haplotypes served as a baseline haplotype in our regression model because the heterogeneous group is not biologically meaningful. By assuming additive effects for all the haplotypes, we fitted a full haplotype model for the 4 haplotypes using the combined haplogroup as baseline. Under these conditions, H1 and H4 demonstrated significant association with sucrose AUC scores (P = 1 × 10−5 and 6 × 10−6, respectively) (Table 2). However, there was no evidence for correlation of H2 and H3 with phenotype (P = 0.8 and 0.1, respectively). A global regression test statistic had 3 degrees of freedom and was highly significant (P = 4.75 × 10−6). We estimated that haplotypes H1 and H4 explain roughly 13% of AUC score variations in the sample population (Figure 4E).
Figure 4D and Supplementary Figure 4 demonstrate the effect of different diplotypes on sucrose AUC scores in the subject populations. Subjects carrying 1 or 2 copies of the high-sensitivity haplotype H1 (combinations H1/H1 and H1/H2) had highest sensitivities to sucrose. In contrast, subjects carrying a copy of the low-sensitivity haplotype (combinations H4/H4, H2/H4, and H3/H4) had reduced AUC scores. Interestingly, subgroups of individuals carrying combinations H1/H4 and H3/H3 demonstrated similar distributions of AUC scores. H1/H4 represents a combination of high- and low-sensitivity haplotypes, whereas haplotype H3 is presumably derived from recombination between H1 and H4. Together these results suggest that although the GNAT3 locus harbors variants that contribute to taste perception, the relationships are complex.
As a whole, our analysis identified 2 “risk” haplotypes of the GNAT3. The presence of a copy of one of them (which consists of a combination of high-sensitivity alleles of rs7792845 and rs1524600) is associated with higher sensitivity to sucrose. The alternative haplotype represented by a combination of low-sensitivity alleles of both SNPs is associated with reduced sensitivity to sucrose. In addition, haplotypes represented by other combinations of alleles of tagging SNPs (“low–high” and “high–low”) had no significant effect on phenotype. However, the number of homozygote subjects carrying identical copies of these variant haplotypes was insufficient for adequate statistical analysis. Thus, it remains unclear whether the effect of a particular haplotype is due to additive effect of alleles residing at different LD blocks or is due to specific combinations of alleles at these blocks.
Putative mechanisms underlying GNAT3 SNP associations
Gustducin is believed to be an essential component involved in the mammalian sweet taste transduction pathway (Huang et al. 1999; Ruiz-Avila et al. 2001; Clapp et al. 2008). Gustducin-deficient mouse knockout models have diminished or complete loss of behavioral and electrophysiological responses to sweet substances, indicating the importance of this protein for mammalian sweet taste transduction (He et al. 2002; Glendinning et al. 2005). Even single amino acid mutations in gustducin can result in significant loss of responsiveness to sweetness in the laboratory animal (Ruiz-Avila et al. 2001). The role of gustducin in human sweet taste signaling is less studied; however, histochemical and biochemical studies suggest that the function of this protein is very similar to that in animal orthologs (Takami et al. 1994).
Our sequencing analysis failed to identify sequence variations in the known protein coding regions of the human gustducin gene. The most significantly associated SNPs, including rs7792845 and rs2012380, were located more than 8 kb upstream of the GNAT3 CDS (Table 1). Eukaryotic core and proximal promoters typically lie upstream of the gene and can have regulatory elements several kilobases away from the transcriptional start site (TSS) (Heintzman and Ren 2007; Venters and Pugh 2009). Thus, the results of association analysis may indicate a regulatory role for the nucleotide sequences harboring SNPs rs7792845 and rs2012380 as well as other SNPs residing in this region (Table 1). The available literature does not provide information about the location of transcription initiation site for the human gustducin mRNA. Thus, a 5′ UTR of the mRNA (and, therefore, the core promoter) or alternative protein coding exons may be located in this sequence region.
To better understand this, we examined GNAT3 mRNA isolated from NCI-H716 cells by 5′ RACE analysis. The NCI-H716 cell line was derived from enteroendocrine L cells of the human colon. It was previously shown that enteroendocrine L cells express sweet taste receptors and other components of the taste transduction pathway (including gustducin), indicating a role for chemical sensation in these tissues (Rozengurt et al. 2006; Jang et al. 2007).
The results of the 5′ RACE analysis are summarized in Supplementary Table 12. The RACE product included first 462 nt (exons 1–4) from a total of 1065 nt of the cDNA listed in the National Center for Biotechnology Information (NCBI) database (accession numbers BC147016 and BC147017). Within this region, no sequence variations were detected in the NCI-H716 cDNA in comparison to the reported cDNA sequence. RACE identified 8 alternative TSS within the 150-bp sequence located upstream of the ATG translation start site. A TSS located at nucleotide −125 was identified in 31% of cDNA clones and thus can be considered as a strong transcription initiator in NCI-H716 cells (Supplementary Table 12). No additional UTRs beyond this 150-bp sequence were detected in 24 RACE products. This suggests that GNAT3 mRNA contains neither an unknown 5′ UTR in the cDNA nor alternative protein CDSs in NCI-H716 cells. Also, these data suggest that the location of core GNAT3 promoter resides close to this 150-bp upstream sequence.
Therefore, we hypothesize that the distal sequence region harboring SNPs rs7792845, rs2012380, or any other significant SNPs (Table 1) located in this region may contain regulatory elements interacting with the proximal promoter elements, at least in NCI-H716 cells.
Supplementary Figures 6 and 7 show patterns of evolutionary conservation between the human 70-kb region of chromosome 7 containing GNAT3 and the corresponding nucleotide sequences from the genomes of other vertebrates. No large evolutionarily conserved elements (>350 bp long, identity >77%) occur at the location of the human SNPs that are significantly associated with phenotype. However, multiple alignment of nucleotide sequences surrounding SNPs rs7792845, rs1524600, rs6467217, rs6970109, rs6975345, rs10242727, rs6467192, rs6979450, and rs6961082 with those from the other vertebrates demonstrated a high degree of interspecies sequence similarity (identity >80% across the 100 bp) (data not shown). This is consistent with a conservation of genomic function in these regions. We hypothesize that SNPs residing at these sequence regions can result in changes in mRNA expression.
Concluding remarks
Our experiments tested for association between genetic variations at the genes encoding taste signaling molecules and human sucrose sensitivity in vivo. Our data show that genetic variations occurring at the GNAT3 locus are significantly correlated with sucrose sensitivity as measured by AUC scores. Fifteen significant SNPs were identified in the noncoding regions of the GNAT3 gene, including 5 SNPs located within the region 9–13 kb upstream of the CDS. In addition, we evaluated multiple polymorphisms across this 140-kb region of chromosome 7q but did not identify any additional associations. Genetic variations in GNAT3 gene region may be associated with promoter activity of the GNAT3 gene and therefore with variations in GNAT3 mRNA levels in humans carrying different alleles.
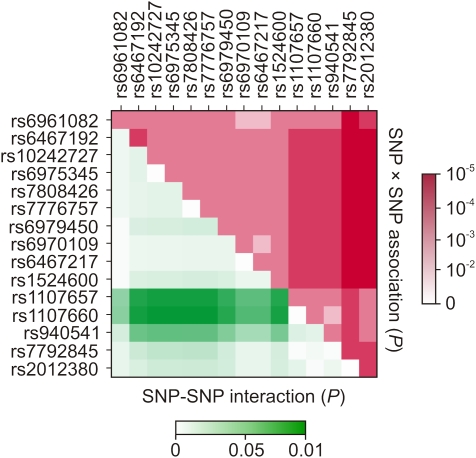
Accurate and properly regulated transcription of eukaryotic genes requires a complex system that involves promoter and enhancer sequences which interact with a large number of different proteins, both near the transcription initiation site and at more distant sites (Venters and Pugh 2009). Sequence regions harboring significantly associated GNAT3 SNPs could bind accessory proteins that may facilitate interactions between activators and coactivators during GNAT3 gene transcription initiation. In support of this hypothesis, we identified evidence of an interaction (P < 0.05) between SNPs located upstream of the CDS (rs1107657, rs1107660, and to a less extent rs940541) and intronic SNPs (Figure 5). This could indicate functional interaction and synergism between the corresponding sequence regions in the transcription of the GNAT3 gene. We hypothesize that alleles of SNPs associated with high sensitivity to sucrose lead to normal formation of accessory complex and transcription initiation. On the other hand, low-sensitivity alleles may prevent binding of accessory proteins, which result in a reduced level of GNAT3 gene transcription.
Figure 5.
Matrix plot demonstrating the results of SNP-SNP association SNP-SNP interaction analyses. Upper triangle shows P values of association for pairwise combinations of significant GNAT3 SNPs. Each square represents the magnitude of P value for a single pair of markers, with a red color indicating highest level of significance and white color indicating lowest level of significance (scale on the right). The squares on the medial diagonal correspond to P values of a single-SNP association analysis. Lower triangle shows probability of interactions between significant SNPs. Each square represents the magnitude of P value for a pair of markers, with a green color indicating highest level of significance and white color indicating lowest level of significance (scale on the bottom).
Genetic variations occurring in the GNAT3 explain 13% of the variation in sucrose perception in our subject population, which has representation from worldwide human populations. Therefore, this could explain an additional substantial fraction of the genetic contribution to differences in sweet taste perception in different human populations (Fushan et al. 2009). In addition, gustducin is an essential component involved in the signal transduction of both bitter and umami tastes (Spielman 1998; He et al. 2004). Thus, we hypothesize that these genetic variations in this gene may also influence these tastes among different individuals.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
This work was supported by National Institute on Deafness and Other Communication Disorders/National Institutes of Health (Z01-000046-09) and Givaudan Flavors Corporation (CRADA DC-CR-06-01).
Supplementary Material
Acknowledgments
We thank the research volunteers for their participation and Joseph Kleinman and Lindsay Bellegia for assistance with administration of taste sensory tests. C.T.S. and J.P.S. note that they are employees of Givaudan Flavors Corporation.
References
- An P, Freedman BI, Hanis CL, Chen YD, Weder AB, Schork NJ, Boerwinkle E, Province MA, Hsiung CA, Wu X, et al. Genome-wide linkage scans for fasting glucose, insulin, and insulin resistance in the National Heart, Lung, and Blood Institute Family Blood Pressure Program: evidence of linkages to chromosome 7q36 and 19q13 from meta-analysis. Diabetes. 2005;54:909–914. doi: 10.2337/diabetes.54.3.909. [DOI] [PubMed] [Google Scholar]
- Andersen M, Lenhard B, Whatling C, Eriksson P, Odeberg J. Alternative promoter usage of the membrane glycoprotein CD36. BMC Mol Biol. 2006;7:8. doi: 10.1186/1471-2199-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya R, Blangero J, Williams K, Almasy L, Dyer TD, Leach RJ, O'Connell P, Stern MP, Duggirala R. Factors of insulin resistance syndrome–related phenotypes are linked to genetic locations on chromosomes 6 and 7 in nondiabetic Mexican-Americans. Diabetes. 2002;51:841–847. doi: 10.2337/diabetes.51.3.841. [DOI] [PubMed] [Google Scholar]
- Ayodo G, Price AL, Keinan A, Ajwang A, Otieno MF, Orago AS, Patterson N, Reich D. Combining evidence of natural selection with association analysis increases power to detect malaria-resistance variants. Am J Hum Genet. 2007;81:234–242. doi: 10.1086/519221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benjafield AV, Jeyasingam CL, Nyholt DR, Griffiths LR, Morris BJ. G-protein beta3 subunit gene (GNB3) variant in causation of essential hypertension. Hypertension. 1998;32:1094–1097. doi: 10.1161/01.hyp.32.6.1094. [DOI] [PubMed] [Google Scholar]
- Blakeslee A, Salmon T. Genetics of sensory thresholds: Individual taste reactions for different substances. Proc. Natl. Acad. Sci. USA. 1935;21:84–90. doi: 10.1073/pnas.21.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin PA. Human gustation. In: Finger TE, Silver WL, Restrepo D, editors. The neurobiology of taste and smell. New York: Wiley-Liss; 2000. pp. 423–461. [Google Scholar]
- Bretz WA, Corby PM, Melo MR, Coelho MQ, Costa SM, Robinson M, Schork NJ, Drewnowski A, Hart TC. Heritability estimates for dental caries and sucrose sweetness preference. Arch Oral Biol. 2006;51:1156–1160. doi: 10.1016/j.archoralbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Brown J. Recognition assessed by rating and ranking. Br J Psychol. 1974;65:13–22. [Google Scholar]
- Chang WI, Chung JW, Kim YK, Chung SC, Kho HS. The relationship between phenylthiocarbamide (PTC) and 6-n-propylthiouracil (PROP) taster status and taste thresholds for sucrose and quinine. Arch Oral Biol. 2006;51:427–432. doi: 10.1016/j.archoralbio.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Chen QY, Alarcon S, Tharp A, Ahmed OM, Estrella NL, Greene TA, Rucker J, Breslin PA. Perceptual variation in umami taste and polymorphisms in TAS1R taste receptor genes. Am J Clin Nutr. 2009;90:770S–779S. doi: 10.3945/ajcn.2009.27462N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapp TR, Trubey KR, Vandenbeuch A, Stone LM, Margolskee RF, Chaudhari N, Kinnamon SC. Tonic activity of Galpha-gustducin regulates taste cell responsivity. FEBS Lett. 2008;582:3783–3787. doi: 10.1016/j.febslet.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpeleijn E, van der Kallen CJ, Kruijshoop M, Magagnin MG, de Bruin TW, Feskens EJ, Saris WH, Blaak EE. Direct association of a promoter polymorphism in the CD36/FAT fatty acid transporter gene with Type 2 diabetes mellitus and insulin resistance. Diabet Med. 2006;23:907–911. doi: 10.1111/j.1464-5491.2006.01888.x. [DOI] [PubMed] [Google Scholar]
- Delwiche J, O'Mahony M. Changes in secreted salivary sodium are sufficient to alter salt taste sensitivity: use of signal detection measures with continuous monitoring of the oral environment. Physiol Behav. 1996;59:605–611. doi: 10.1016/0031-9384(95)02120-5. [DOI] [PubMed] [Google Scholar]
- Fushan AA, Simons CT, Slack JP, Manichaikul A, Drayna D. Allelic polymorphism within the TAS1R3 promoter is associated with human taste sensitivity to sucrose. Curr Biol. 2009;19:1288–1293. doi: 10.1016/j.cub.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gilbertson TA, Damak S, Margolskee RF. The molecular physiology of taste transduction. Curr Opin Neurobiol. 2000;10:519–527. doi: 10.1016/s0959-4388(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Glendinning JI, Bloom LD, Onishi M, Zheng KH, Damak S, Margolskee RF, Spector AC. Contribution of alpha-gustducin to taste-guided licking responses of mice. Chem Senses. 2005;30:299–316. doi: 10.1093/chemse/bji025. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Reed DR, Wright MJ, Martin NG, Breslin PA. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCL, and caffeine. Chem Senses. 2006;31:403–413. doi: 10.1093/chemse/bjj044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Danilova V, Zou S, Hellekant G, Max M, Margolskee RF, Damak S. Partial rescue of taste responses of alpha-gustducin null mice by transgenic expression of alpha-transducin. Chem Senses. 2002;27:719–727. doi: 10.1093/chemse/27.8.719. [DOI] [PubMed] [Google Scholar]
- He W, Yasumatsu K, Varadarajan V, Yamada A, Lem J, Ninomiya Y, Margolskee RF, Damak S. Umami taste responses are mediated by alpha-transducin and alpha-gustducin. J Neurosci. 2004;24:7674–7680. doi: 10.1523/JNEUROSCI.2441-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N and, Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol Life Sci. 2007;64:386–400. doi: 10.1007/s00018-006-6295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Chung JW, Kim YK, Chung SC, Lee SW, Kho HS. The relationship between PTC taster status and taste thresholds in young adults. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;99:711–715. doi: 10.1016/j.tripleo.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Hosking L, Lumsden S, Lewis K, Yeo A, McCarthy L, Bansal A, Riley J, Purvis I, Xu CF. Detection of genotyping errors by Hardy-Weinberg equilibrium testing. Eur J Hum Genet. 2004;12:395–399. doi: 10.1038/sj.ejhg.5201164. [DOI] [PubMed] [Google Scholar]
- Huang L, Shanker YG, Dubauskaite J, Zheng JZ, Yan W, Rosenzweig S, Spielman AI, Max M, Margolskee RF. Ggamma13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatonium. Nat Neurosci. 1999;2:1055–1062. doi: 10.1038/15981. [DOI] [PubMed] [Google Scholar]
- Jang HJ, Kokrashvili Z, Theodorakis MJ, Carlson OD, Kim BJ, Zhou J, Kim HH, Xu X, Chan SL, Juhaszova M, et al. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci USA. 2007;104:15069–15074. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn SG. Taste perception-individual reactions to different substances. IL Acad Sci Trans. 1951;44:263–269. [Google Scholar]
- Keskitalo K, Knaapila A, Kallela M, Palotie A, Wessman M, Sammalisto S, Peltonen L, Tuorila H, Perola M. Sweet taste preferences are partly genetically determined: identification of a trait locus on chromosome 16. Am J Clin Nutr. 2007;86:55–63. doi: 10.1093/ajcn/86.1.55. [DOI] [PubMed] [Google Scholar]
- Kim U-K, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Liu N, Zhang K, Zhao H. Haplotype-association analysis. Adv Genet. 2008;60:335–405. doi: 10.1016/S0065-2660(07)00414-2. [DOI] [PubMed] [Google Scholar]
- Love-Gregory L, Sherva R, Sun L, Wasson J, Schappe T, Doria A, Rao DC, Hunt SC, Klein S, Neuman RJ, et al. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum Mol Genet. 2008;17:1695–1704. doi: 10.1093/hmg/ddn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Bacci S, Mlynarski W, Gottardo L, Soccio T, Menzaghi C, Iori E, Lager RA, Shroff AR, Gervino EV, et al. A common haplotype at the CD36 locus is associated with high free fatty acid levels and increased cardiovascular risk in Caucasians. Hum Mol Genet. 2004;13:2197–2205. doi: 10.1093/hmg/ddh233. [DOI] [PubMed] [Google Scholar]
- Margolskee RF. Molecular mechanisms of bitter and sweet taste transduction. J Biol Chem. 2002;277:1–4. doi: 10.1074/jbc.R100054200. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro EO, Brisibe F, Jolayemi ET, Hadizath Taimagari G. Taste sensitivity to sodium chloride and sucrose in a group of adolescent children in Northern Nigeria. Ethn Dis. 2000;10:53–59. [PubMed] [Google Scholar]
- O'Mahony M. Understanding discrimination tests: a user-friendly treatment of response bias, rating, and ranking R-index tests and their relationship to signal detection. J Sens Stud. 1992;7:1–47. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu HQ, Marchand L, Szymborski A, Grabs R, Polychronakos C. The association between type 1 diabetes and the ITPR3 gene polymorphism due to linkage disequilibrium with HLA class II. Genes Immun. 2008;9:264–266. doi: 10.1038/gene.2008.12. [DOI] [PubMed] [Google Scholar]
- Roach JC, Deutsch K, Li S, Siegel AF, Bekris LM, Einhaus DC, Sheridan CM, Glusman G, Hood L, Lernmark A, et al. Genetic mapping at 3-kilobase resolution reveals inositol 1,4,5-triphosphate receptor 3 as a risk factor for type 1 diabetes in Sweden. Am J Hum Genet. 2006;79:614–627. doi: 10.1086/507876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozengurt N, Wu SV, Chen MC, Huang C, Sternini C, Rozengurt E. Colocalization of the alpha-subunit of gustducin with PYY and GLP-1 in L cells of human colon. Am J Physiol Gastrointest Liver Physiol. 2006;291:G792–G802. doi: 10.1152/ajpgi.00074.2006. [DOI] [PubMed] [Google Scholar]
- Ruiz-Avila L, Wong GT, Damak S, Margolskee RF. Dominant loss of responsiveness to sweet and bitter compounds caused by a single mutation in alpha-gustducin. Proc Natl Acad Sci USA. 2001;98:8868–8873. doi: 10.1073/pnas.151235798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Schaffner SF, Fry B, Lohmueller J, Varilly P, Shamovsky O, Palma A, Mikkelsen TS, Altshuler D, Lander ES. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- Spielman AI. Gustducin and its role in taste. J Dent Res. 1998;77:539–544. doi: 10.1177/00220345980770040601. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami S, Getchell TV, McLaughlin SK, Margolskee RF, Getchell ML. Human taste cells express the G protein alpha-gustducin and neuron-specific enolase. Brain Res. 1994;22:193–203. doi: 10.1016/0169-328x(94)90047-7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang S, Lin R, Jiang X, Cheng Z, Turdi J, Ding J, Wu G, Lu X, Wen H. GNB3 gene C825T and ACE gene I/D polymorphisms in essential hypertension in a Kazakh genetic isolate. J Hum Hypertens. 2004;18:663–668. doi: 10.1038/sj.jhh.1001700. [DOI] [PubMed] [Google Scholar]
- Wise PM, Hansen JL, Reed DR, Breslin PA. Twin study of the heritability of recognition thresholds for sour and salty taste. Chem Senses. 2007;32:749–754. doi: 10.1093/chemse/bjm042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.