Abstract
Our lab has isolated hexameric peptides that are structure-selective ligands of Holliday junctions (HJ), central intermediates of several DNA recombination reactions. One of the most potent of these inhibitors, WRWYCR, has shown antibacterial activity in part due to its inhibition of DNA repair proteins. To increase the therapeutic potential of these inhibitors, we searched for small molecule inhibitors with similar activities. We screened 11 small molecule libraries comprising over nine million individual compounds and identified a potent N-methyl aminocyclic thiourea inhibitor that also traps HJs formed during site-specific recombination reactions in vitro. This inhibitor binds specifically to protein free HJs and can inhibit HJ resolution by RecG helicase, but only showed modest growth inhibition of bacterial with a hyperpermeable outer membrane; nonetheless, this is an important step in developing a functional analog of the peptide inhibitors.
Site-specific recombination (SSR) catalyzed by bacteriophage λ Integrase (Int) requires the formation and subsequent resolution of a 4-armed DNA intermediate known as a Holliday Junction (HJ) (Supplemental information Figure 1A, and reviewed in ref1). We have previously identified hexapeptide inhibitors from combinatorial peptide libraries that stabilized the HJ intermediate, or blocked recombination completely2, 3. The activity of many of these peptides, such as WRWYCR (Supplemental information Figure 1B), is not dependent upon interactions with any of the proteins required for recombination. In fact, these peptides inhibit several mechanistically distinct tyrosine recombinases3, 4, DNA cleavage and HJ resolution by Vaccinia virus topoisomerase5, 6, and structurally unrelated HJ processing enzymes like RecG helicase and RuvABC resolvase7. All of these activities are based on the ability of the peptides to bind to protein-free HJs, and with somewhat lesser affinity to other branched DNAs and replication forks7. Because HJs are central intermediates in several DNA repair pathways8, we reasoned that these peptides may interfere with DNA repair and thus have antibacterial activity. Indeed, Minimal Inhibitory Concentration (MIC) values range from 32–64 μg/ml in Gram negative bacteria and 4–32 μg/ml in Gram positive bacteria, including methicillin resistant Staphylococcus aureus (MRSA)4. Evidence from genetic and biochemical studies supports a model where the need for DNA repair causes the formation of Holliday junctions that are trapped by the peptides; in this way repair is blocked and the cells die4, 7, 9, 10.
DNA repair is a novel antibacterial target. Moreover, target-site resistance is unlikely to develop because HJs are generated in multiple independent ways8. Consistent with this hypothesis, we have been unable to isolate stable peptide resistant mutants. Due to the increasing problem of multiple drug resistance in bacteria, we wanted to identify small molecules with similar activities as the peptides. We performed screens of 11 small molecule libraries comprising over nine million individual compounds to identify functional analogs of the HJ trapping peptides. Here we present the identification and characterization of an N-methyl aminocyclic thiourea that is an active inhibitor of our in vitro recombination assays, binds specifically to protein free HJs, and inhibits a HJ-processing helicase.
Compound identification
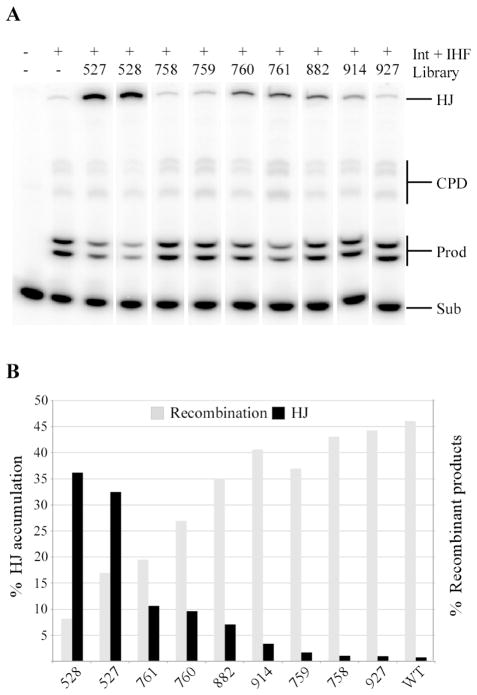
The mixture-based libraries we screened were synthesized at Torrey Pines Institute for Molecular Studies (reviewed in ref11). In general the compounds in the libraries we tested had low molecular weights and 3 diversity positions, or R groups, built on unique scaffolds (Figure 2 in Supplemental information). These scaffolds were first evaluated for inhibition of site-specific recombination without regard to individual R groups in the libraries by making a “mixture of mixtures.” For example, the 40 R1, 37 R2, and 80 R3 mixtures of the N-methyl aminocyclic thiourea library (DCR 528, Supplemental information Table 1) were pooled together to make one mixture comprising the total diversity of that library (40 R1 * 37 R2 * 80 R3 = 118,400 compounds). This “mixture of mixtures” was tested in Int-mediated recombination between two attL sites known as the bent-L2, 12 pathway at 10 μg/ml and quantitated with respect to accumulation of HJs (Figure 1). Individual compounds from the mixture are present in the recombination reactions at approximately 84 pg/ml (~225 pM for DCR 528 given an average molecular weight of 374 g/mol for compounds in that library)i. This concentration is well below the IC50 of even our most potent peptide inhibitor. However, as discussed previously2, 11, the inhibitory activity seen in the libraries is likely the result of a number of chemically similar compounds with characteristics and potency close to the most active inhibitors and this group increases the “effective concentration” of the inhibitors in the mixtures. As seen in Figure 1, the accumulation of HJs in treated reactions coincides with a reduction in recombinant products as catalysis in the context of the HJ is blocked (refer to Supplemental information, Figure 1A). Of the libraries screened, the N-benzyl aminocyclic thiourea scaffold (DCR 527) and the N-methyl aminocyclic thiourea scaffold (DCR 528) accumulated the most HJs and inhibited recombination to the greatest extent (Figure 1B). Therefore, we pursued both libraries.
Figure 1. Scaffold ranking.
(A) Lanes from Tris-Tricine-SDS gels showing recombination reactions in the presence of the small molecule mixtures at 10 μg/ml in bent-L recombination. “Sub” refers to the double radioactive end-labeled DNA substrate which recombines with an unlabeled partner to generate products, “Prod”. “CPD” refers to covalent protein-DNA complexes and “HJ” is the Holliday junction. (B) Quantitation of the reactions shown in Figure 1A for HJ accumulation (left Y-axis) and concurrent reduction in the formation of recombinant products (right Y-axis). Int and IHF are the proteins performing the recombination reaction.
Following scaffold selection, defined R group mixtures were screened to identify individual compounds with activity (reviewed in ref 11, 13). Each mixture has a “defined position” and individual compounds within that mixture all have the same functionality at that R group position; the other two R group positions contain equimolar mixtures of the other functionalities. For example, mixture 126 in the N-methyl aminocyclic thiourea library (Supplemental information Table 1) contains the defined functionality N′N-dimethyl-S-propylamine at position R1, and a mixture of the 37 R2 and 80 R3 functionalities at the other two positions. Thus, this single mixture contains a total of 2960 (1*37*80) possible compounds, each present at ~337 pg/ml (~900 pM) when tested in reactions at 1 μg/ml, and this allows evaluation of the functionality N′N-dimethyl-S-propylamine at position R1 with respect to inhibition of recombination. Functionalities at R2 and R3 were screened in the same fashion. The mixtures from DCR 528 (Figure 3 in Supplemental information) averaged two-fold more HJ accumulation than DCR 527 (data not shown) and thus we chose to identify compounds from DCR 528. These top ranking mixtures are designated with an asterisk in Supplemental information, Figure 3, and each mixture was titrated more extensively in the bent-L pathway and ranked based on potency. The top defined mixtures were also shown to inhibit attL x attR (excisive12) recombination, albeit with lower potencies2, 14. In order to test the specificity of these mixtures, they were assayed in several other reactions involving DNA transactions and were found not to inhibit restriction digests by NdeI and HindIII, or plasmid relaxation by E. coli topoisomerase I at 10 μg/ml14. The mixtures were also tested (as described in ref4) at 25 μg/ml, and shown to inhibit the growth of S. aureus14. Based on these data, three R1, three R2, and one R3 functionalities were selected and the combinations resulted in nine individual compounds (Table 1) that were synthesized as described15, 16.
Table 1.
Deconvolution of positional scanning data for the N-methyl aminocyclic thiourea library
| Compound | R1 | R2 | R3 |
|---|---|---|---|
| 1530-1 | N′N-dimethyl-S-propylamine | R-hydroxy-methyl | 2-(4-nitro-phenyl)-ethyl |
| 1530-2 | R-hydroxy-methyl | ||
| 1530-3 | S-3-methyl-1-methyl-indole | ||
| 1530-4 | N′N-dimethyl-S-propylamine | S-3-methyl-1H-indole | |
| 1530-5 | R-hydroxy-methyl | ||
| 1530-6 | S-3-methyl-1-methyl-indole | ||
| 1530-7 | N′N-dimethyl-S-propylamine | S-hydroxy-methyl | |
| 1530-8 | R-hydroxy-methyl | ||
| 1530-9 | S-3-methyl-1-methyl-indole |
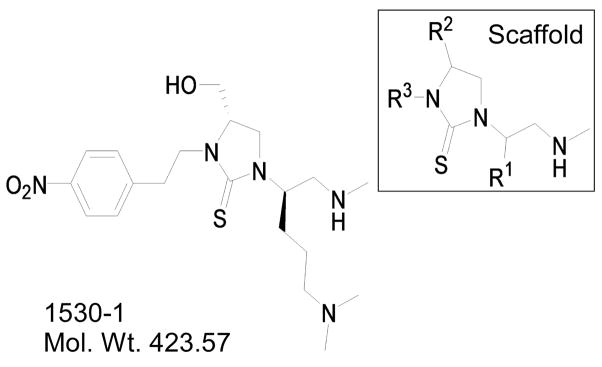
The crude compounds were characterized in several in vivo and in vitro assays similar to those used to assess the defined mixtures14. Based on these data TPI1530-1 (Figure 2) was selected for further study.
Figure 2.
1530–1 is built on a N-methyl aminocyclic thiourea scaffold with functionalities N′N-dimethyl-S-propylamine, R-hydroxy-methyl, and 2-(4-nitro-phenyl)-ethyl (R1, R2, and R3, respectively).
Compound characterization
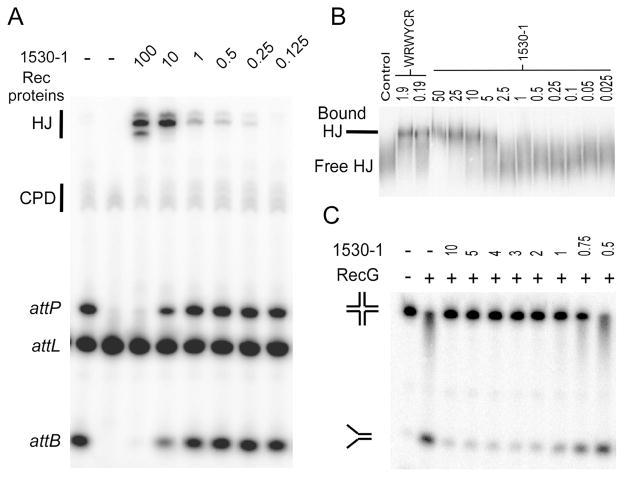
Compound 1530–1 was purified to > 95% and verified by LC-MS. We titrated 1530–1 into excision recombination reactions (Figure 3A) and determined IC50 values of 3 μg/ml for accumulation of HJs, and 5 μg/ml for inhibition of recombination (compared with IC50 values for peptide wrwycr of 0.04 and 0.12 μg/ml, respectively; Table 2). We tested for whether 1530–1 inhibits several restriction enzymes as well as for whether it binds non-specifically to double stranded DNA (an attL site12 in the absence of recombination enzymes). 1530–1 inhibited restriction digests at 100 μg/ml by 22% compared to untreated reactions (Table 2); however, no evidence of binding to double stranded DNA alone was detected at this concentration. This suggested specificity for the common HJ intermediate in the recombination assays. To test this further, 1530–1 was examined for its ability to bind to protein-free HJ substrates and shown to form a specific complex similar to that seen with peptide WRWYCR (Figure 3B). We then tested the effects of the compound on RecG-mediated unwinding of HJs. RecG is a monomeric SFX family helicase that recognizes HJs and unwinds these substrates in the presence of ATP and magnesium17. This helicase bears no structural or mechanistic similarity to the proteins in the recombination assays and has been used previously to examine the specificity of our peptides for the HJ7, 10. As seen in Figure 3C, 1530–1 does inhibit RecG activity in a dose dependent manner with an IC50 of 0.85 μg/ml (compared with 0.12 μg/ml for wrwycr). These data further corroborate a model where the inhibitor recognizes the structure of the HJ intermediate.
Figure 3.
1530–1 interacts with Holliday junctions. All concentrations in μg/ml. (A) Dose dependent inhibition of excision SSR. Rec protein refers to inclusion of Int and Xis proteins and Integration Host Factor (IHF). (B) 1530–1 causes a shift in the migration rate of HJ DNA. The specific shift seen with 1530–1 is comparable to the shift seen in control reactions with peptide WRWYCR. (C) 1530–1 inhibits RecG-mediated unwinding of HJ DNA. RecG binds to HJs and, in the presence of ATP and Mg+2 will unwind the DNA to a replication fork. This activity is inhibited by 1530–1 in a dose dependent manner.
Table 2.
Comparison of 1530–1 and wrwycr for selected activities.
| Compound | Mol. Weighta | Excision | RecG | NdeI | Growth inhibitionc | |
|---|---|---|---|---|---|---|
| IC50 (Rec) | IC50 (HJ) | IC50 | % cleavageb | Stm (galE rfaA) | ||
| 1530–1 | 423.5 | 5 | 3 | 0.85 | 78 | 100 |
| wrwycra | 1936.2 | 0.04 | 0.02 | 0.12 | 100 | 12.5 |
All concentrations in μg/ml.
Molecular weights given are for active species, which is a homodimer in the case of wrwycr.
Restriction digest data are given as a percentage of activity at 100 μg/ml compared to untreated reactions.
The concentration at which 50% growth inhibition is seen.
The ability of 1530–1 to inhibit bacterial growth was examined using several Gram (+) and Gram (−) bacteria. In contrast to the activity of the DCR 528 library, compound 1530–1 did not inhibit bacterial growth, even at the highest concentration tested (100 μg/ml, data not shown). Lack of inhibitory activity may be due poor to permeability of 1530–1 into bacterial cells. To address this possibility we tested a hyperpermeable Salmonella enterica serovar Typhimurium (Stm) strain with mutations in the galE and rfa genes; this strain has shorter lipopolysaccharide (LPS) chains in the outer membrane and increased permeability to many compounds, including our peptides4, 18–20. At 100 μg/ml, 1530–1 inhibited the growth of this mutant by 50% after 22 hours of incubation (data not shown). Alternatively, 1530–1 may be rapidly effluxed from the cell. To address this we tested an E. coli strain with a deletion of the acrAB genes, which inactivates the major drug efflux pump in the bacterium and increases susceptibility to many antibiotics21.
At 100 μg/ml of 1530–1 the growth of the efflux mutant was not inhibited (data not shown). These data suggest that 1530–1 may have antibiotic activity but that poor membrane permeability may prevent the compound from entering cells and finding its target14.
In summary, we have identified and purified an N-methyl aminocyclic thiourea that recognizes HJs and inhibits enzymes that catalyze the resolution of these intermediates of DNA recombination and repair. This compound is composed of a similar combination of R-group functionalities as the side chains of the peptide inhibitors3, 6: aromatic and basic functionalities are present in both the most potent peptides and in the other N-methyl aminocyclic thioureas that were identified (Table 1). Molecular modeling corroborated by fluorescence quenching studies on peptide WRWYCR suggested that aromatic functionalities may be involved in base stacking interactions with the HJ10, while functionalities that are positively charged could stabilize binding to the negatively charged DNA. Together, these functionalities may represent the core characteristics necessary for interactions with HJs, although not necessarily for entry into bacteria. One attribute which 1530–1 does not share with the peptides is an efficient mechanism for forming dimers, which the most effective hexapeptides do via disulfide linkage between two monomers. Future screening of other chemical libraries combining a primary screen for HJ binding activity, followed by a secondary screen for growth inhibition may yield compounds with greater antimicrobial activity.
Supplementary Material
(A) A simplified schematic of a tyrosine recombinase-mediated site-specific recombination pathway. Four Int monomers (ovals) bind and synapse attachment sites on two DNA molecules (shown as pairs of black or green strands). Two of the Int monomers are initially active (shown as light grey ovals) and each cleaves opposing strands of DNA, forming a covalent protein-DNA adduct. The free DNA strands are exchanged and then rejoined to form the Holliday junction (HJ) intermediate. Dark grey ovals are initially inactive Int monomers, and isomerization of the HJ causes them to become active for catalysis (note the transition to light grey ovals). A second round of DNA cleavage, strand exchange, and ligation on the remaining strands produces recombinant products. Accessory proteins Xis and IHF (not shown) are pathway-dependent. The HJ is the target of the peptide inhibitors, an example of the most potent of which, WRWYCR, is shown in (B) in the active form of a homodimer. Peptide binding stabilizes the HJ and prevents the 2nd round of cleavage, and product formation.
Each library is listed with the Torrey Pines Institute for Molecular Studies designation. Chemical names and structures refer to the various scaffolds upon which the small molecule libraries were built, and not to individual compounds. The number of mixtures in each library is defined by the sum of the functionalities possible for each R group (shown on each scaffold as R1 − R3 or R1 − R4). The number of compounds in each library is defined by the possible functionalities at each R group multiplied together. For example, the N-methyl aminocyclic thiourea library, DCR528, contains 157 mixtures (40 R1 + 37 R2 + 80 R3) and a total of 118,400 compounds (40 R1 * 37 R2 * 80 R3).
Defined R group mixtures were tested in the bent-L recombination pathway at a concentration of 1 μg/ml. Gels were quantitated for % of HJ accumulation and mixtures were ranked in order of this activity. The most active mixtures marked with an asterisk were chosen and the corresponding functionalities are given. Ave = average % HJ accumulation for all possible functionalities at that R group position.
Acknowledgments
This work was supported by Public Health Service grant RO1 GM052847 from the National Institute of General Medical Sciences to AMS. MCR is the recipient of an Achievement Rewards for College Scientists scholarship. We thank Matt Parker for critical reading of the manuscript.
Footnotes
The concentration of individual compounds (in μg/ml) that are present in the library or in the defined mixtures are calculated from the final concentration of the mixture in the treated reaction divided by the diversity of the mixture. For instance, treatments with library DCR 528 at 10 μg/ml gives a final compound concentration of 84 pg/ml (10 μg/ml/118,400 compounds), and treatments with a defined DCR 528 mixture (such as mixture 126) at 1 μg/ml gives a final compound concentration of 337 pg/ml (1 μg/ml/2960 compounds).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Azaro MALA, editor. λ Integrase and the λ Int family. ASM Press; Washington D.C: 2002. [Google Scholar]
- 2.Cassell G, Klemm M, Pinilla C, Segall A. J Mol Biol. 2000;299:1193. doi: 10.1006/jmbi.2000.3828. [DOI] [PubMed] [Google Scholar]
- 3.Boldt JL, Pinilla C, Segall AM. J Biol Chem. 2004;279:3472. doi: 10.1074/jbc.M309361200. [DOI] [PubMed] [Google Scholar]
- 4.Gunderson CW, Segall AM. Mol Microbiol. 2006;59:1129. doi: 10.1111/j.1365-2958.2005.05009.x. [DOI] [PubMed] [Google Scholar]
- 5.Klemm M, Cheng C, Cassell G, Shuman S, Segall AM. J Mol Biol. 2000;299:1203. doi: 10.1006/jmbi.2000.3829. [DOI] [PubMed] [Google Scholar]
- 6.Fujimoto DF, Pinilla C, Segall AM. J Mol Biol. 2006;363:891. doi: 10.1016/j.jmb.2006.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kepple KV, Boldt JL, Segall AM. Proc Natl Acad Sci U S A. 2005;102:6867. doi: 10.1073/pnas.0409496102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. DNA Repair (Amst) 2007;6:967. doi: 10.1016/j.dnarep.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson CW, Boldt JL, Authement RN, Segall AM. J Bacteriol. 2009;191:2169. doi: 10.1128/JB.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kepple KV, Patel N, Salamon P, Segall AM. Nucleic Acids Res. 2008;36:5319. doi: 10.1093/nar/gkn512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houghten RA, Pinilla C, Appel JR, Blondelle SE, Dooley CT, Eichler J, Nefzi A, Ostresh JM. J Med Chem. 1999;42:3743. doi: 10.1021/jm990174v. [DOI] [PubMed] [Google Scholar]
- 12.Segall AM, Nash HA. Genes Cells. 1996;1:453. doi: 10.1046/j.1365-2443.1996.d01-254.x. [DOI] [PubMed] [Google Scholar]
- 13.Pinilla C, Appel JR, Borras E, Houghten RA. Nat Med. 2003;9:118. doi: 10.1038/nm0103-118. [DOI] [PubMed] [Google Scholar]
- 14.Ranjit DK. San Diego State University. 2004. [Google Scholar]
- 15.Nefzi A, Ostresh JM, Giulianotti M, Houghten RA. Journal of Combinatorial Chemistry. 1999;1:195. [Google Scholar]
- 16.Nefzi A, Ostresh JM, Meyer JP, Houghten RA. Tetrahedron Letters. 1997;38:931. [Google Scholar]
- 17.Whitby MC, Lloyd RG. J Biol Chem. 1998;273:19729. doi: 10.1074/jbc.273.31.19729. [DOI] [PubMed] [Google Scholar]
- 18.Ames BN, Lee FD, Durston WE. Proc Natl Acad Sci U S A. 1973;70:782. doi: 10.1073/pnas.70.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kadam SK, Rehemtulla A, Sanderson KE. J Bacteriol. 1985;161:277. doi: 10.1128/jb.161.1.277-284.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikaido H. Microbiol Mol Biol Rev. 2003;67:593. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami S, Tamura N, Saito A, Hirata T, Yamaguchi A. J Biol Chem. 2004;279:3743. doi: 10.1074/jbc.M308893200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) A simplified schematic of a tyrosine recombinase-mediated site-specific recombination pathway. Four Int monomers (ovals) bind and synapse attachment sites on two DNA molecules (shown as pairs of black or green strands). Two of the Int monomers are initially active (shown as light grey ovals) and each cleaves opposing strands of DNA, forming a covalent protein-DNA adduct. The free DNA strands are exchanged and then rejoined to form the Holliday junction (HJ) intermediate. Dark grey ovals are initially inactive Int monomers, and isomerization of the HJ causes them to become active for catalysis (note the transition to light grey ovals). A second round of DNA cleavage, strand exchange, and ligation on the remaining strands produces recombinant products. Accessory proteins Xis and IHF (not shown) are pathway-dependent. The HJ is the target of the peptide inhibitors, an example of the most potent of which, WRWYCR, is shown in (B) in the active form of a homodimer. Peptide binding stabilizes the HJ and prevents the 2nd round of cleavage, and product formation.
Each library is listed with the Torrey Pines Institute for Molecular Studies designation. Chemical names and structures refer to the various scaffolds upon which the small molecule libraries were built, and not to individual compounds. The number of mixtures in each library is defined by the sum of the functionalities possible for each R group (shown on each scaffold as R1 − R3 or R1 − R4). The number of compounds in each library is defined by the possible functionalities at each R group multiplied together. For example, the N-methyl aminocyclic thiourea library, DCR528, contains 157 mixtures (40 R1 + 37 R2 + 80 R3) and a total of 118,400 compounds (40 R1 * 37 R2 * 80 R3).
Defined R group mixtures were tested in the bent-L recombination pathway at a concentration of 1 μg/ml. Gels were quantitated for % of HJ accumulation and mixtures were ranked in order of this activity. The most active mixtures marked with an asterisk were chosen and the corresponding functionalities are given. Ave = average % HJ accumulation for all possible functionalities at that R group position.