Abstract
Mechanistic studies of O-GlcNAc glycosylation have been limited by an inability to monitor the glycosylation stoichiometries of proteins obtained from cells. Here, we describe a powerful method to visualize the O-GlcNAc-modified protein subpopulation using resolvable polyethylene glycol mass tags. This approach enables rapid quantification of in vivo glycosylation levels on endogenous proteins without the need for protein purification, advanced instrumentation, or expensive radiolabels. In addition, the glycosylation state (e.g., mono-, di-, tri-) of proteins is established, providing information regarding overall O-GlcNAc site occupancy that cannot be obtained using mass spectrometry. Finally, we apply this strategy to rapidly assess the complex interplay between glycosylation and phosphorylation, and discover an unexpected reverse yin-yang relationship on the transcriptional repressor MeCP2, which was undetectable by traditional methods. We anticipate that this mass-tagging strategy will advance our understanding of O-GlcNAc glycosylation, as well as other post-translational modifications and poorly understood glycosylation motifs.
Protein post-translational modifications represent an important molecular mechanism for the control of complex biological systems. One example is O-GlcNAc glycosylation, the dynamic addition of N-acetyl-D-glucosamine to serine or threonine residues of intracellular proteins1–3. O-GlcNAc plays key roles in diverse processes such as glucose homeostasis1,4, cardiac muscle survival5, cell cycle progression6, and synaptic transmission7. However, like many post-translational modifications, O-GlcNAc glycosylation has intrinsic characteristics (e.g., labile, present in low cellular abundance, ubiquitous), which have hindered both its detection and mechanistic studies2.
Hundreds of O-GlcNAc-modified proteins have been identified by mass spectrometry8–10, yet in vivo glycosylation levels are known for only a few11–15, and there are currently no methods to assess whether proteins exist in singly, doubly or multiply glycosylated states in vivo. Knowledge of glycosylation stoichiometries is essential for understanding the function and regulation of O-GlcNAc. Moreover, the ability to quantify O-GlcNAc stoichiometries should help overcome a major bottleneck in the field – namely, the prioritization of proteins in the O-GlcNAc proteome for in-depth characterization and functional analysis.
Current methods to assess glycosylation stoichiometries are time-consuming and require large amounts of purified protein. GlcNAc levels are typically quantified by radioactivity or high pH anion exchange chromatography-pulsed amperometric detection (HPAEC-PAD)11–15. However, there are no general methods for measuring O-GlcNAc glycosylation levels on unpurified, endogenous proteins. Furthermore, monitoring the dynamics of O-GlcNAc glycosylation on specific proteins is challenging because O-GlcNAc site-specific antibodies are notoriously difficult to generate16. As such, rapid, parallel, and low-cost methods are needed to study O-GlcNAc glycosylation stoichiometries and to understand their dynamic regulation across different tissues and (patho)physiological states.
In addition to quantifying stoichiometries, a related challenge is to understand the interplay between different post-translational modifications. For instance, there is an intriguing potential for O-GlcNAc and phosphorylation to act reciprocally or engage in combinatorial crosstalk at the level of signaling cascades1,7,17–20. To establish ‘yin-yang’ reciprocity, the addition of a phosphate group must affect the glycosylation state of the same molecule and vice versa. However, until now, there have been no general methods to study the kinetics of glycosylation on only the phosphorylated protein subpopulation or vice versa.
We describe a novel strategy that overcomes these challenges, unraveling the stoichiometry and dynamics of O-GlcNAc glycosylation in vivo and revealing new insights into the intricate interplay between glycosylation and phosphorylation. Using this strategy, we show that O-GlcNAc stoichiometries occur over a wide range in vivo and are subject to tight regulatory control. In addition, we identify a complex, reverse yin-yang relationship on the transcriptional repressor MeCP2 that would be missed using traditional approaches.
RESULTS
Mass-tagging strategy to quantify O-GlcNAc stoichiometry
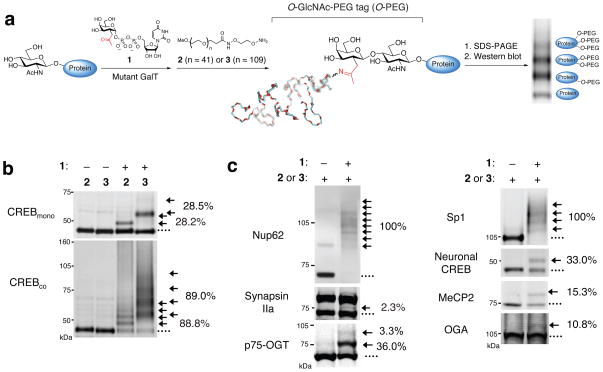
We reasoned that the selective attachment of a defined molecular mass tag to terminal GlcNAc sugars would enable rapid visualization of O-GlcNAc glycosylated proteins of interest (Fig. 1a). Specifically, O-GlcNAc-modified proteins from cell lysates could be labeled with the mass tag, resolved by SDS-PAGE, and visualized in parallel by immunoblotting with antibodies against proteins of interest. Simple inspection of the mass-shifted bands would establish whether a protein was mono-, di- or multiply glycosylated, and in vivo glycosylation stoichiometries could be determined by quantifying the relative intensities of each band. We chose polyethylene glycol (PEG) for the mass tag because it is aqueous-soluble, highly flexible, chemically inert, and available in various well-defined molecular weight ranges. Although PEG has been used extensively to modulate the pharmacokinetics and other properties of proteins21, it has not been exploited as a tool to advance an understanding of post-translational modifications. Aminooxy-functionalized 2 and 3 were readily synthesized in one chemical step from commercially available PEG 2K and 5K derivatives, respectively (Supplementary Scheme 1 and Supplementary Figs. 1–4).
Figure 1.
Mass-tagging strategy for quantifying O-GlcNAc glycosylation levels on specific proteins. (a) Schematic of the approach. O-GlcNAc-modified proteins are chemoenzymatically labeled using the UDP-ketogalactose analogue 1 and an engineered GalT enzyme, followed by reaction with an aminooxy-functionalized PEG mass tag (2 or 3). This approach enables facile visualization of O-GlcNAc-glycosylated species upon SDS-PAGE and immunoblotting with antibodies against proteins of interest. (b) Validation of the approach using CREBmono and CREBco. CREBmono was 28.2% glycosylated and existed primarily in the mono-glycosylated state, whereas CREBco was 88.8% glycosylated and present in multiply glycosylated forms. (c) In vivo O-GlcNAc stoichiometries vary significantly even among proteins with similar functions. Cell lysates (100 μg) from 293T cells (source of Sp1), adult rat brain (source of MeCP2, Nup62, and synapsin Ia (upper band) and IIa (lower band)), embryonic neuronal cultures (source of CREB, and OGA) or purified p75-OGT (0.43 μg) from Sf9 cells were subjected to chemoenzymatic labeling, SDS-PAGE, and immunoblotting with antibodies against the indicated proteins. See Methods for details. In all cases, 1 was excluded as a control for selectivity. The indicated glycosylation stoichiometries were determined by measuring the relative intensities of the glycosylated and nonglycosylated bands. (….) denotes the nonglycosylated protein fraction, (→) denotes O-GlcNAc glycosylated protein fraction shifted with 2 or 3. Full-length blots are presented in Supplementary Figure 9.
To attach the mass tag onto O-GlcNAc proteins, we used our previously reported chemoenzymatic labeling strategy8,9,22. This approach allows for selective and complete labeling of terminal GlcNAc sugars using an engineered β-1,4-galactosyltransferase22,23 (Y289L GalT). Structural and mechanistic studies have shown that GalT selectively labels terminal GlcNAc sugars23,24 without regard for the surrounding peptide sequence23, indicating that it should not discriminate between specific glycosylation sites. Moreover, the GalT labeling method has been the gold standard for quantitative radiolabeling of purified O-GlcNAc glycosylated proteins in vitro12.
We first investigated the approach on two preparations of purified cAMP-response element binding protein (CREB), which differed only in their extent of glycosylation25. CREB expressed alone in Sf9 cells (CREBmono) and CREB co-expressed with O-GlcNAc transferase (OGT; CREBco) were incubated with the Y289L GalT and UDP-ketogalactose analogue 122, after which the transferred ketone moiety was reacted with the aminooxy-functionalized PEG derivative 2 or 3 (Fig. 1a). The labeled proteins were resolved by SDS-PAGE and analyzed by Western blotting using an anti-CREB antibody. Reaction of CREBmono with 2 or 3 produced an expected shift of molecular mass (2 kDa or 5 kDa, respectively) consistent with labeling of a single GlcNAc moiety (Fig. 1b). Omission of the UDP-ketone substrate 1 prevented the mass shift and confirmed the specificity of the reaction. By measuring the relative intensities of the glycosylated and nonglycosylated protein bands, the stoichiometry of glycosylation of CREBmono was calculated to be 28.2% (24.1% monoglycosylated, 4.1% diglycosylated). As expected, CREBco exhibited a higher extent of glycosylation (88.8%) as well as the presence of multiple mass-shifted bands. Inspection of the blot revealed four 2 kDa- or 5 kDa-incremental shifts in molecular mass, indicating that CREBco contains at least four glycosylation sites and exists as a mixture of mono-, di-, tri- and tetraglycosylated forms (31.8% monoglycosylated, 29.2% diglycosylated, 18.2% triglycosylated and 9.6% tetraglycosylated). Accounting for the number of O-GlcNAc units in each glycoform (e.g., 1 for the monoglycosylated form, 2 for the diglycosylated form, etc.) and the percentage of each glycoform, we calculated a ratio of 183:32 GlcNAc groups for CREBco:CREBmono. This corresponds to a 5.7-fold increase in glycosylation, similar to that obtained previously by radiolabeling25.
Approach Validation
For accurate quantification of O-GlcNAc stoichiometries, the following criteria must be satisfied: 1) the enzymatic labeling and PEGylation reaction must proceed to completion, 2) the PEG tag must not interfere significantly with Western blotting, and 3) detection of the labeled glycoprotein must be linear across a wide range of stoichiometries. Completion of both the enzymatic labeling and oxime formation steps has been demonstrated by mass spectrometry of O-GlcNAc-modified peptides22. As further verification, we incubated PEG-labeled CREBmono with an aminooxybiotin derivative to cap any unreacted ketone groups. No biotin incorporation was detected by streptavidin-IR800, whereas the CREBmono control lacking 3 gave a strong streptavidin signal (Supplementary Fig. 5). Furthermore, we tested the labeling of two proteins known to be highly glycosylated, Nup62 and Sp113,26,27. Rat brain or 293T cell lysates were labeled using an optimized procedure and immunoblotted for Nup62 or Sp1, respectively. Importantly, we detected complete labeling of both proteins and the addition of 10 PEG units to Nup62, consistent with labeling of the 10 reported Nup62 sites27 (Fig. 1c).
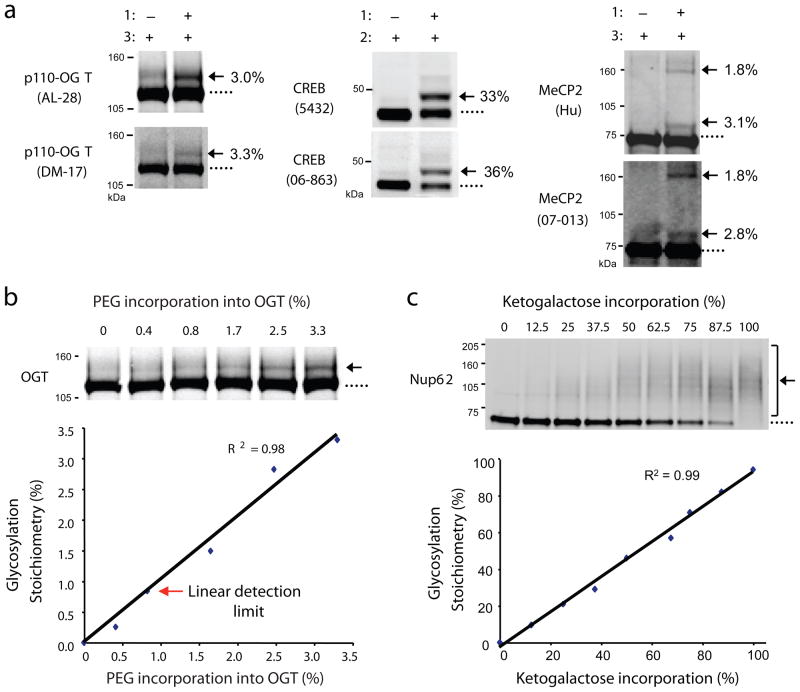
To confirm that the PEG tag does not interfere significantly with immunoblotting, we compared several antibodies. The anti-OGT antibodies AL-28 and DM-17 detected the same extent of glycosylation on OGT (Fig. 2a). Similarly, no differences in the stoichiometries of CREB or the transcriptional repressor MeCP2 were calculated using two distinct antibodies against each protein. Given the low stoichiometry of OGT glycosylation (3.3%), we used OGT to test the detection sensitivity of the mass-tagging method. We measured the glycosylation stoichiometries of six OGT standards, which were generated by serial dilution of labeled and unlabeled lysates (Fig. 2b). As little as 0.8% of glycosylated OGT could be readily detected, highlighting the sensitivity of the approach.
Figure 2.
Validation of the approach. (a) Distinct antibodies detect the same glycosylation stoichiometry on OGT (AL-28 and DM-17), CREB (Chemicon 5432 and Upstate 06-863), and MeCP2 (Hu and Upstate 07-013). p110-OGT was expressed in Sf9 cells, endogenous CREB was from rat liver, and MeCP2 was co-expressed with OGT in Sf9 cells. (b) As little as 0.8% of glycosylated OGT is readily detected. Sf9 cell lysate containing over-expressed p110-OGT was chemoezymatically labeled with PEG derivative 3 and diluted with unlabeled lysate to generate standards with varying percentages of label incorporation. The lysate was resolved by SDS-PAGE and immunoblotted with the anti-OGT antibody DM-17. The limit of detection was defined as the lowest stoichiometry value within 10% of the linear fit. See Supplementary Methods for details. (c) Detection of PEG incorporation into ketogalactose-labeled Nup62 is linear across a wide range of stoichiometries (0–100%). 293T cell lysate was labeled with UDP-ketogalactose 1 and then diluted with varying amounts of unlabeled lysate to simulate different levels of glycosylation. Each mixture was reacted with 2, resolved by SDS-PAGE, and immunoblotted for Nup62. Full-length blots are presented in Supplementary Figure 9.
Lastly, we confirmed that PEGylation of O-GlcNAc glycosylated proteins was linearly related to the glycosylation stoichiometry, particularly at high stoichiometries where a large number of PEG molecules might preclude linear detection. A standard curve was generated to simulate a wide range of glycosylation (0–100%) by treating lysates containing varying concentrations of ketogalactose-labeled proteins with 2 (Fig. 2c). Importantly, the intensity of the mass-shifted bands (i.e. the extent of label incorporation) was linearly proportional to the extent of Nup62 glycosylation over the entire stoichiometry range.
Profiling in vivo glycosylation stoichiometries
An important implication of the approach is that labeling O-GlcNAc proteins with resolvable mass tags should enable the first comprehensive profiling of in vivo glycosylation levels. To investigate the generality of the approach, we compared the glycosylation levels of a diverse set of proteins from different biological samples. We found that there exists a broad range of in vivo O-GlcNAc stoichiometries, even among proteins with similar functions (Fig. 1c). Whereas synapsin IIa was glycosylated at a very low level (2.3%) in the adult rat brain cortex, the entire population of Nup62 was modified in the same tissue sample. The transcription factor Sp1 was highly glycosylated at multiple sites in accordance with previous reports13, while a smaller (33.0%) subpopulation of CREB was singly modified in embryonic cortical neurons. MeCP2, a transcriptional repressor whose loss underlies mental retardation in Rett’s Syndrome, was 15.3% mono-glycosylated in the rat hypothalamus, a small brain structure where MeCP2 regulates stress and metabolism28. Interestingly, low glycosylation levels were observed on OGA and the long form of OGT in unstimulated, cortical neurons (10.8% and undetectable, respectively). The long form of OGT also exhibited low levels of mono-glycosylation in Sf9 cells (3.3%; Fig. 2a), although the short form of OGT showed higher levels of glycosylation (36.5% mono-glycosylated, 3.3% di-glycosylated; Fig. 1c). Five distinct glycosylation sites have been identified on OGT by mass spectrometry29, yet OGT exists predominantly in the mono-glycosylated state in vivo. These results highlight the complementary information obtained using the mass-tagging approach and mass spectrometry analyses.
Quantitative studies of O-GlcNAc levels and dynamics
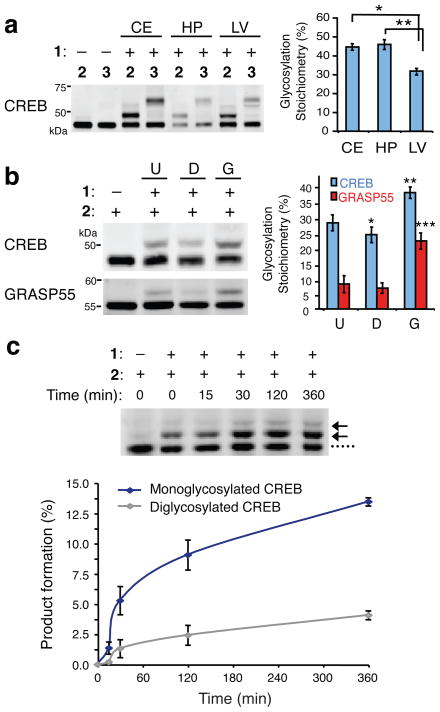
The ability to monitor the glycosylation levels of proteins in vivo also permits comparisons across different tissues, organs, or disease states. Endogenous CREB exhibited similar glycosylation levels in the adult rat hippocampus and cerebellum (44.5 ± 1.6% and 45.8 ± 2.6%, respectively; Fig. 3a) but was consistently glycosylated at lower levels in the adult rat liver (31.7 ± 1.0%). The strong reproducibility of the measurements across multiple different animals is striking and suggests that physiological glycosylation levels are under tight regulatory control.
Figure 3.
Monitoring O-GlcNAc glycosylation levels on proteins across various tissues and after cellular stimulation. (a) CREB is O-GlcNAc glycosylated at higher levels in the rat cerebellum (CE) and hippocampus (HP) than in the liver (LV). Cell lysates from the indicated tissues were chemoenzymatically labeled, resolved by SDS-PAGE, and immunoblotted for CREB. A representative immunoblot from one animal is shown. Plotted are the glycosylation stoichiometries of CREB averaged across multiple animals (n = 4–6). * P = 0.0003, ** P = 0.03. (b) O-GlcNAc glycosylation levels of CREB and GRASP55 decreased in rat embryonic neuronal cultures upon DON treatment (D) and increased upon GlcN treatment (G) relative to untreated neurons (U). Cell lysates were chemoenzymatically labeled, resolved by SDS-PAGE, and immunoblotted for the indicated proteins. A representative immunoblot is shown. Plotted are the glycosylation stoichiometries averaged across n = 3 experiments. * P = 0.002, ** P = 0.006, *** P = 0.001. (c) The rate constant for monoglycosylation of CREB is approximately threefold higher than that for diglycosylation of CREB. Neuro2a cells were treated with 10 mM GlcN for the indicated times. Crude nuclear protein lysates were chemoezymatically labeled, resolved by SDS-PAGE, and immunoblotted for CREB. Formation of monoglycosylated or diglycosylated CREB was plotted as a function of time for n = 3 experiments. See Supplementary Methods for details. Data represent mean ± s.e.m. Statistical analyses were performed using the Student’s t-test (two-tailed, paired). Full-length blots are presented in Supplementary Figure 9.
In addition to comparisons across different tissue samples, the mass-tagging approach also facilitates studies of the dynamic cycling of O-GlcNAc. Cortical neurons were treated either with deoxynorleucine (DON) to block UDP-GlcNAc synthesis30 or with glucosamine (GlcN) to enhance UDP-GlcNAc synthesis30. DON decreased overall protein O-GlcNAc levels by 0.70 ± 0.11–fold, while GlcN increased them by 2.4 ± 0.1–fold, as measured with a general O-GlcNAc antibody (Supplementary Fig. 6). Using our mass-tagging approach, we were able to quantify changes in glycosylation on specific proteins, including CREB and the Golgi stacking protein GRASP55 (1.4 ± 0.1–fold and 3.1 ± 0.6–fold, respectively for GlcN; 0.87 ± 0.05–fold and 0.86 ± 0.02–fold, respectively for DON; Fig. 3b). Notably, while the fold-increase in glycosylation differed considerably for CREB and GRASP55, both proteins underwent a similar change in glycosylation stoichiometry in response to GlcN (13.1 ± 0.2% and 10.2 ± 0.5%, respectively), consistent with the notion that UDP-GlcNAc acts as a global modulator of OGT activity. Furthermore, we observed differences in the rates of glycosylation for the mono- and diglycosylated forms of CREB. In response to GlcN, the relative rate of monoglycosylation was approximately 3-fold higher than that of diglycosylation (Fig. 3c), suggesting that once CREB becomes O-GlcNAc glycosylated, it is less likely to be glycosylated a second time. Together, these studies underscore the power of the mass-tagging approach to provide new insights into the kinetics of O-GlcNAc glycosylation on specific modified subpopulations in vivo, information that would be impossible to obtain using existing methods.
Direct interrogation of the interplay between PTMs
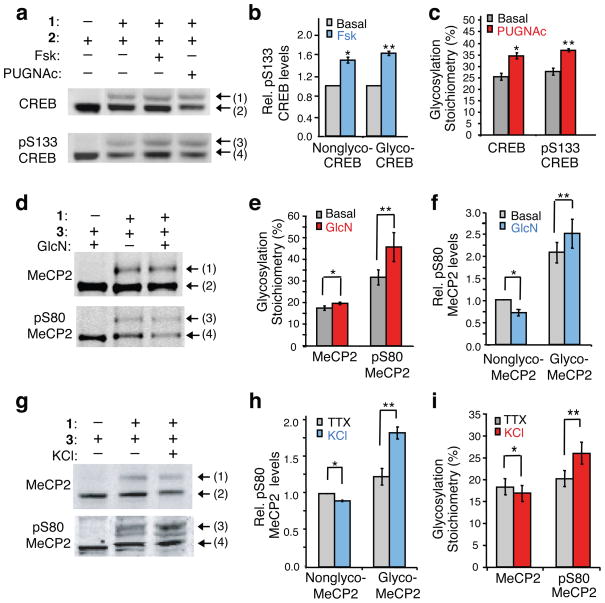
Establishing the presence or absence of a reciprocal ‘yin-yang’ relationship between O-GlcNAc and O-phosphate is challenging because it typically requires knowledge of the glycosylation sites, extensive mutagenesis, and cellular analyses31–33. Thus, we investigated the potential for our mass-tagging strategy to provide rapid, new insights into the interplay between O-GlcNAc and phosphorylation. Many stimuli induce phosphorylation of CREB at Ser-133 (pS133), thereby activating CREB-dependent transcription and regulating key processes such as glucose homeostasis, drug addiction, learning and memory34,35. To determine whether O-GlcNAc glycosylation affects pS133 phosphorylation of CREB and vice versa, we stimulated HEK cells with the cAMP agonist forskolin (Fsk) to induce pS133 phosphorylation or with the OGA inhibitor O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenyl carbamate (PUGNAc) to induce CREB glycosylation. After PEG labeling, the cell lysates were immunoblotted with a pS133-specific antibody or total CREB antibody. This approach enabled rapid visualization of four distinct subpopulations of CREB: (1) glycosylated, (2) nonglycosylated, (3) glycosylated and phosphorylated, and (4) nonglycosylated and phosphorylated CREB (Fig. 4a). Simple inspection of the blot revealed that a significant fraction of the CREB subpopulation was simultaneously phosphorylated and glycosylated, indicating that the two modifications can reside on the same molecule and are not mutually exclusive (i.e. not yin-yang).
Figure 4.
Dissecting the interplay between O-GlcNAc and phosphorylation. Detection of four subpopulations of CREB or MeCP2: glycosylated (1), nonglycosylated (2), glycosylated and phosphorylated (3), and nonglycosylated and phosphorylated (4). (a) CREB can be simultaneously glycosylated and phosphorylated. Mass-tagged lysates from 293T cells treated with Fsk, PUGNAc or vehicle were immunoblotted with a pS133-specific or general CREB antibody. (b) Fsk stimulates pS133 phosphorylation of glycosylated and non-glycosylated CREB similarly (n = 6). (c) Inhibition of OGA increases CREB glycosylation levels on phosphorylated and total CREB similarly (n = 4). Neither Fsk nor PUGNAc affected overall CREB glycosylation or pS133 phosphorylation levels, respectively (Supplementary Fig. 7). (d) Complex interplay between MeCP2 glycosylation and phosphorylation. Mass-tagged lysates from neurons treated with GlcN or vehicle were immunoblotted with a pS80-specific or general MeCP2 antibody (n = 6). (e) GlcN increases MeCP2 glycosylation levels predominantly on the pS80 subpopulation. (f) GlcN decreases pS80 levels on the nonglycosylated subpopulation and increases pS80 levels on the glycosylated subpopulation. (g) Reverse yin-yang relationship on MeCP2 under physiological conditions. Mass-tagged nuclear extracts from synchronously depolarized or non-depolarized neurons were immunoblotted with a pS80-specific or general MeCP2 antibody (n = 8). (h) Neuronal depolarization induces pS80 levels on glycosylated MeCP2. (i) Depolarization increases the glycosylation level of pS80 MeCP2, while decreasing that of total MeCP2. UDP ketogalactose 1 was excluded as a control. Data represent mean ± s.e.m. Statistics were analyzed by Student’s t-test. * P = 0.0005, ** P = 0.00007 for (b). * P = 0.02, ** P = 0.0007 for (c). * P = 0.04, ** P = 0.008 for (e). * P = 0.01, ** P = 0.04 for (f). * P = 0.01, ** P = 0.001 for (h). * P = 0.04, ** P = 0.001 for (i). Full-length blots are presented in Supplementary Figure 9.
Comparison of the levels of pS133 induction on the glycosylated and nonglycosylated subpopulations revealed that forskolin stimulated CREB phosphorylation to a similar extent in both fractions (nonglyco-CREB 1.47 ± 0.05–fold, glyco-CREB 1.55 ± 0.04–fold; Fig. 4a,b). Similarly, the level of PUGNAc-induced CREB glycosylation was unaffected by the presence of phosphorylation at S133 (Fig. 4a,c). Thus, pS133 phosphorylation can be stimulated independently of glycosylation and vice versa, and we observe no evidence of interplay between the cAMP signaling pathway and OGA activity toward CREB.
We next investigated the potential for interplay between phosphorylation and glycosylation on MeCP2. MeCP2 phosphorylation at Ser-421 and dephosphorylation at Ser-80 have been shown to de-repress specific genes and may play an important role in coordinating activity-dependent gene expression36,37. Using the traditional method of immunoblotting with phospho-specific antibodies, we observed intriguing evidence for a potential yin-yang relationship at Ser-80 on MeCP2. Specifically, increasing O-GlcNAc levels with GlcN led to a 30.1 ± 6.9% decrease in phosphorylation at Ser-80 (pS80) in rat cortical neurons, whereas phosphorylation at Ser-421 was unaffected (Supplementary Fig. 8a,b). To characterize further the potential yin-yang relationship at Ser-80, we exploited our mass-tagging strategy on neuronal lysates and immunoblotted with a pS80-specific antibody or total MeCP2 antibody (Fig. 4d). As before, the approach resulted in rapid visualization of four distinct post-translationally modified subpopulations. Notably, a significant fraction of MeCP2 was found to be simultaneously phosphorylated and glycosylated, definitively establishing the absence of a simple yin-yang relationship.
We quantified the GlcN-induced change in glycosylation on the pS80 subpopulation relative to the overall MeCP2 population. Surprisingly, glycosylation was induced to a greater extent on the phosphorylated subpopulation of MeCP2 (1.42 ± 0.08–fold for pS80 MeCP2; 1.14 ± 0.06–fold for total MeCP2; Fig. 4d,e), indicative of a reverse yin-yang relationship. We also examined the effects of GlcN on MeCP2 phosphorylation levels. Consistent with our earlier result, GlcN induced an overall decrease in pS80 phosphorylation. However, the glycosylated subpopulation underwent an unexpected increase in phosphorylation at Ser-80 (1.20 ± 0.06–fold), and pS80 levels decreased selectively on the nonglycosylated population (0.71 ± 0.07–fold, Fig. 4d,f), again the opposite of a yin-yang relationship.
To examine whether this reverse yin-yang relationship occurred in response to physiological stimuli, we induced membrane depolarization of neurons with KCl. Synchronous activation of embryonic neurons with depolarizing amounts of KCl reduced the overall pS80 levels on MeCP2, as reported37 (Supplementary Fig. 8c). Despite an overall decrease in global pS80 levels, membrane depolarization enhanced Ser-80 phosphorylation selectively on the glycosylated MeCP2 subpopulation (1.56 ± 0.15– fold) and decreased Ser-80 phosphorylation only on the nonglycosylated MeCP2 subpopulation (0.90 ± 0.03–fold; Fig. 4g,h). Whereas KCl treatment induced a modest overall decrease in O-GlcNAc glycosylation on total MeCP2 (0.89 ± 0.03–fold), the pS80 subpopulation underwent an increase in glycosylation (1.26 ± 0.03–fold; Fig. 4g,i). Together, these results provide strong evidence for a reverse yin-yang relationship on MeCP2.
Discussion
In this study, we demonstrate a powerful new approach for visualizing the O-GlcNAc-glycosylated protein subpopulation in complex biological samples. Through the selective, chemoenzymatic attachment of PEG mass tags to O-GlcNAc modification sites, in vivo glycosylation stoichiometries can be readily quantified on endogenous proteins, without the need for protein purification, advanced instrumentation, or expensive radiolabels. In addition, the approach allows for direct interrogation of proteins of interest by immunoblotting, without requiring O-GlcNAc site identification or O-GlcNAc site-specific antibodies. This feature is critical given that glycosylation sites have been mapped for only a small fraction of the O-GlcNAc proteome, and comprehensive analyses of all glycosylation sites within a given protein are lacking, even for many well-studied proteins.
Our mass-tagging approach yields information that cannot be obtained using current methods. Because glycosylation levels can be rapidly monitored by immunoblotting in parallel, O-GlcNAc glycosylation stoichiometries can be readily profiled across the proteome. We quantified the stoichiometries of a broad range of proteins, including proteins that eluded detection by the O-GlcNAc antibodies RL-2 and CTD110.6. Even among proteins with similar functions, we observed a wide range of O-GlcNAc stoichiometries in vivo. For example, both the transcriptional repressor MeCP2 and transcription factor CREB were mono-glycosylated at moderate levels in neurons (15.3% and 33.0%, respectively), while the transcription factor Sp1 was 100% glycosylated at multiple sites in vivo. Similarly, the long form of OGT exhibited very low levels of glycosylation in Sf9 cells (3.3%), whereas the short form of OGT had a significantly higher extent of glycosylation in these cells (39.7%). Unexpectedly, OGT was primarily mono-glycosylated despite having multiple known glycosylation sites, which suggests a high potential for the transient regulation of OGT activity by O-GlcNAc in response to cellular stimuli. These findings underscore the complementarity between our mass-tagging strategy and mass spectrometry approaches. Although mass spectrometry provides key information about the identity and number of O-GlcNAc sites, it cannot readily determine their relative occupancy or interrelationship within the same molecule of protein. In contrast, our mass-tagging approach provides a direct read-out of glycosylation stoichiometry and state (e.g., mono-, di-, tri-, etc.), and when applied in conjunction with site-directed mutagenesis, it may reveal O-GlcNAc stoichiometries at specific amino acid sites.
The ability to monitor glycosylation stoichiometries and states provides unique insights into the cellular functions and regulation of O-GlcNAc glycosylation. For example, we probed the glycosylation level of CREB across different tissues, cells and organisms in response to a variety of perturbations. Our results revealed tissue-specific differences and tight regulatory control over the levels of CREB glycosylation in vivo. Moreover, we found that the glycosylation state of CREB influenced its kinetics of glycosylation: upon activation of the hexosamine biosynthesis pathway, unglycosylated CREB was glycosylated three-times faster than mono-glycosylated CREB, suggesting that the presence of one GlcNAc sugar on CREB decreases the rate of deposition of another sugar. Although the presence of only one OGT gene may suggest uniform regulation of O-GlcNAc substrates, evidence from quantitative mass spectrometry analyses indicates that OGT regulates its substrates more discretely29. Our approach allows this phenomenon to be explored in detail across and within proteins of interest in response to specific stimuli. Such information would be difficult, if not impossible, to obtain using current methods.
Elucidating the interplay of O-GlcNAc with other post-translational modifications is another powerful application of the mass-tagging approach. For example, we demonstrated that the mass tag enables rapid visualization of distinct post-translationally modified subpopulations, which are distinguished by their glycosylation or phosphorylation status. As such, the approach provides a direct read-out of whether the two modifications are mutually exclusive on proteins of interest (i.e. yin-yang) or whether they can co-exist on the same molecule. We found that glycosylation and phosphorylation occur independently in the case of CREB under the stimuli tested. In addition, we discovered a surprising reverse yin-yang relationship on MeCP2, which was undetectable by traditional methods and revealed only by the mass-tagging approach. A yin-yang relationship on MeCP2 was observed by stimulating O-GlcNAc levels with GlcN and monitoring phosphorylation levels on the total MeCP2 population. However, our mass-tagging strategy enabled changes in glycosylation to be monitored specifically on the phosphorylated subpopulation and vice versa. We found that neuronal activity or stimulation of the hexosamine biosynthesis pathway induced O-GlcNAc glycosylation selectively on the S80-phosphorylated subpopulation of MeCP2 – the opposite of a yin-yang relationship. One possibility is that glycosylation may mark a specific subset of MeCP2-regulated genes and render them less susceptible to activity-dependent derepression. Our results provide strong evidence for the close coupling of glycosylation and phosphorylation on MeCP2 and, more broadly, we find that the net change in glycosylation or phosphorylation on the global protein population can be the opposite of changes occurring on specific modified subpopulations, an observation with significant implications for studying the interplay between modifications. The complexities observed with MeCP2 underscore the importance of carefully dissecting the intricate interplay between post-translational modifications on a molecular level and of developing new methods to address these questions.
In conclusion, we have developed a new mass-tagging strategy to advance our understanding of the stoichiometry, complex regulation, and cellular dynamics of O-GlcNAc glycosylation. In the future, we anticipate extending this approach to explore the complex ‘codes’ or networks of post-translational modifications on O-GlcNAc-modified proteins through the use of antibodies against other modifications. Moreover, we envision that this general strategy of tagging modifications with resolvable PEG mass tags will prove valuable for the study of other post-translational modifications and poorly understood glycosylation motifs.
METHODS
General procedure for chemoenzymatic labeling with mass tags
Protein lysates (100 μg at 3.3 μg μl−1 in 1% SDS) were diluted 5-fold to a final buffer concentration of 10 mM HEPES pH 7.9, 10 mM MgCl2, 100 mM NaCl and 1.8% Triton-X100, and 0.2% SDS. The lysates (100 μg at 0.67 μg μl−1) were incubated with Y289L GalT23 (7 μg; 0.1 mg ml−1 final concentration) and UDP-ketogalactose derivative 1 (500 μM) for 16 h at 4 °C. The samples were then diluted to 200 μl and precipitated by sequential mixing with 600 μl of MeOH, 200 μl of CHCl3, and 450 μl H2O, followed by centrifugation at 23,000 ×g for 15 min. Precipitated protein was washed with 450 μl of MeOH and centrifuged at 23,000 ×g for 10 min. After allowing the protein pellet to dry, the pellet was resuspended in labeling buffer (43.6 μl at 2.3 μg μl−1; 7 M urea, 10 mM HEPES pH 7.9, 2% CHAPS, 1 mM DTT, Complete™ protease inhibitor cocktail (Roche) and 1 mM PMSF). The samples were then acidified to pH 4.5 by the addition of 1.4 μl of 1.8 M NaOAc (final concentration 50 mM) and incubated with 6 mM aminooxy-functionalized PEG (2 or 3; 5 μl of a 60 mM aqueous stock) for 20–24 h at RT. To stop the reaction, the samples were neutralized with 1M HEPES pH 7.9 (2 μl), precipitated using the MeOH/CHCl3/H2O method described above, and re-suspended in 1% SDS. The UDP-ketogalactose derivative 1 was omitted as a control in each case. If desired, complex N-glycans can be removed prior to (or during) labeling using PNGaseF as previously reported25,38. Membrane-associated glycoproteins can be removed readily by subcellular fractionation (see Supplementary Methods). However, such procedures are not necessary as the O-GlcNAc proteins of interest are selectively visualized by immunoblotting.
Quantification of O-GlcNAc stoichiometries
The following samples were used to detect glycosylation of the indicated proteins in Fig. 1c: 293T whole cell lysate (Sp1), rat hypothalamus crude nuclear pellet (MeCP2), rat brain detergent-soluble fraction (synapsin IIa, Nup62), whole cell lysate from cultured embryonic neurons (CREB), cytosolic fraction from PUGNAc-treated cultured embryonic neurons (OGA), and p75-OGT purified from Sf9 cells. For Fig. 3a, the liver, hippocampus or cerebral cortex was harvested from adult Sprague Dawley rats, and crude nuclear pellets were processed. Animal protocols were approved by the Institutional Animal Care and Use Committee at Caltech, and the procedures were performed in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. Cell lysates were prepared as described in the Supplementary Methods. Each sample and its corresponding negative control (lacking ketogalactose probe 1 incorporation) was subjected to chemoenzymatic labeling with PEG mass tags, resolved on 4–12% Bis-Tris NuPAGE gels (Invitrogen), and transferred to nitrocellulose or PVDF membranes. The membranes were immunoblotted with antibodies against each protein of interest (see Supplementary Methods). After incubation with secondary antibodies (IRDye 800 goat anti-rabbit or Alexa Fluor 680 goat anti-mouse), proteins were visualized and quantified using an Odyssey infrared imaging system (LI-COR Biosciences). To quantify O-GlcNAc stoichiometries, the intensities of the PEG-shifted band (glycosylated protein fraction) and the unshifted band (non-glycosylated protein fraction) were measured using Odyssey imaging software (Version 2.1). The resulting values of the PEG-shifted bands were corrected for non-specific background by subtracting the background intensity from negative control reactions. For data and statistical analyses, mean values, standard error of the mean, and P-values (paired, two-tailed, Student’s T-tests, α-value = 0.05) were calculated using the program Excel.
Monitoring O-GlcNAc dynamics
Rat cortical neurons were treated with DON (5 μM, 6 h), GlcN (10 mM, 6 h) or vehicle (2 mM HEPES pH 7.5) and harvested by rapid lysis in boiling 1% SDS containing Complete ™ protease inhibitor cocktail and 1 mM PMSF. Lysates were sonicated, centrifuged, and measured for protein concentration using the BCA assay. Each sample (100 μg) was subjected to chemoenzymatic labeling with the PEG derivatives 2 or 3. Following resolution by SDS-PAGE and gel transfer, the membranes were blotted sequentially with anti-CREB and anti-GRASP55 antibodies. Glycosylation stoichiometries were calculated using both PEG derivatives.
Studying the interplay between phosphorylation and glycosylation
For CREB, HEK 293T cells were treated with Fsk (10 μM, 25 min), PUGNAc (100 μM, 6 h), or vehicle only (DMSO for Fsk, water for PUGNAc). Following treatment, cells were harvested by rapid lysis in boiling 1% SDS containing Complete™ protease inhibitor cocktail, 1 mM PMSF, 50 mM NaF and 1 mM Na3VO4, and the lysates were sonicated, centrifuged, and measured for protein concentration. The lysates (100 μg) were then subjected to chemoenzymatic labeling as described above, resolved by SDS-PAGE (30 μg), and immunoblotted in parallel for pS133 and total CREB. pS133 CREB levels and glycosylation stoichiometries were calculated. The pS133 signals were corrected for total CREB levels in each fraction and then normalized with respect to the basal pS133 phosphorylation level in each case.
To assess the interplay between MeCP2 phosphorylation at Ser-80 (pS80) or Ser-421 (pS421) and glycosylation, embryonic day 18 cortical neurons were incubated after 4 days in vitro in the presence or absence of GlcN (10 mM, 6 h). pS421 levels were induced by neuronal depolarization (55 mM KCl, 2 h). pS421-phosphorylated MeCP2 was detected from whole cell lysate as the slower migrating MeCP2 band upon 8% SDS-PAGE and MeCP2 immunoblotting36,37. pS80 levels were assessed by parallel immunoblotting of nuclear extracts with pS80-specific and total MeCP2 antibodies.
For glycosylation assays of pS80 and total MeCP2, nuclear lysates of neuronal samples were subjected to chemoenzymatic labeling with PEG derivative 3 as described above, resolved in parallel on 4–12% Bis-Tris NuPAGE gels, and immunoblotted with either a pS80-specific or total MeCP2 antibody. To quantify pS80 levels, the pS80 signal was corrected for the total MeCP2 signal in each fraction. pS80 levels were plotted relative to the levels of basal nonglycosylated MeCP2. Glycosylation stoichiometries of pS80 and total MeCP2 were calculated as described above.
Supplementary Material
Acknowledgments
We thank the following people for their generous gifts: G. Hart for the OGT antibody, S. Whiteheart for the OGA antibody, W. Wang and K. Hu for the MeCP2 antibodies, P. Greengard and A. Nairn for the synapsin antibody, and P. Qasba for the Y289L GalT construct. We thank P. Clark and T. Wilson for a critical reading of the manuscript. We also thank P. Clark for preparing the Y289L GalT used in these studies and for contributing to the aminooxy-linker synthesis. This work was supported by grants from the National Institutes of Health (R01 GM084724 to L.H.W. and F31 NS056525-02 to J.E.R.), and Rett Syndrome Research Foundation (Y.E.S.).
Footnotes
AUTHOR CONTRIBUTIONS
J.E.R. and L.H.W. conceived of and designed the experiments. J.E.R. performed the biochemistry and labeling experiments, J.F. and Y.E.S. contributed to the development and characterization of the phospho-specific Ab, C.J.R. synthesized and characterized the first set of aminooxy-functionalized PEG probes, and S.Y. synthesized the UDP-ketogalactose substrate and additional aminooxy-functionalized PEG probes. J.E.R. and L.H.W. wrote the manuscript, and all authors participated in editing the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Hanover JA, Krause MW, Love DC. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagen.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rexach JE, Clark PM, Hsieh-Wilson LC. Chemical approaches to understanding O-GlcNAc glycosylation in the brain. Nat Chem Biol. 2008;4:97–106. doi: 10.1038/nchembio.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 4.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 5.Ngoh GA, et al. Unique hexosaminidase reduces metabolic survival signal and sensitizes cardiac myocytes to hypoxia/reoxygenation injury. Circ Res. 2009;104:41–49. doi: 10.1161/CIRCRESAHA.108.189431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slawson C, et al. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- 7.Tallent MK, et al. In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J Biol Chem. 2009;284:174–181. doi: 10.1074/jbc.M807431200. [DOI] [PubMed] [Google Scholar]
- 8.Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc Natl Acad Sci U S A. 2004;101:13132–13137. doi: 10.1073/pnas.0403471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark PM, et al. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Enrichment and site-mapping of O-Linked N-Acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation (ETD) mass spectrometry. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold CS, et al. The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J Biol Chem. 1996;271:28741–28744. doi: 10.1074/jbc.271.46.28741. [DOI] [PubMed] [Google Scholar]
- 12.Dong DL, Xu ZS, Hart GW, Cleveland DW. Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H. J Biol Chem. 1996;271:20845–20852. doi: 10.1074/jbc.271.34.20845. [DOI] [PubMed] [Google Scholar]
- 13.Jackson SP, Tjian R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 14.Luthi T, Haltiwanger RS, Greengard P, Bahler M. Synapsins contain O-linked N-acetylglucosamine. J Neurochem. 1991;56:1493–1498. doi: 10.1111/j.1471-4159.1991.tb02043.x. [DOI] [PubMed] [Google Scholar]
- 15.Roquemore EP, et al. Vertebrate lens alpha-crystallins are modified by O-linked N-acetylglucosamine. J Biol Chem. 1992;267:555–563. [PubMed] [Google Scholar]
- 16.Teo CF, et al. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Gucek M, Hart GW. Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc Natl Acad Sci U S A. 2008;105:13793–13798. doi: 10.1073/pnas.0806216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dias WB, Cheung WD, Wang Z, Hart GW. Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. J Biol Chem. 2009;284:21327–21337. doi: 10.1074/jbc.M109.007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- 20.Wells L, Kreppel LK, Comer FI, Wadzinski BE, Hart GW. O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J Biol Chem. 2004;279:38466–38470. doi: 10.1074/jbc.M406481200. [DOI] [PubMed] [Google Scholar]
- 21.Veronese FM, Pasut G. PEGylation, successful approach to drug delivery. Drug Discov Today. 2005;10:1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 22.Khidekel N, et al. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 23.Ramakrishnan B, Qasba PK. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: point mutation broadens beta 4Gal-T1 donor specificity. J Biol Chem. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 24.Holt GD, Hart GW. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem. 1986;261:8049–8057. [PubMed] [Google Scholar]
- 25.Lamarre-Vincent N, Hsieh-Wilson LC. Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J Am Chem Soc. 2003;125:6612–6613. doi: 10.1021/ja028200t. [DOI] [PubMed] [Google Scholar]
- 26.Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci U S A. 1987;84:7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holt GD, et al. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol. 1987;104:1157–1164. doi: 10.1083/jcb.104.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fyffe SL, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59:947–958. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khidekel N, et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 30.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- 31.Medina L, Grove K, Haltiwanger RS. SV40 large T antigen is modified with O-linked N-acetylglucosamine but not with other forms of glycosylation. Glycobiology. 1998;8:383–391. doi: 10.1093/glycob/8.4.383. [DOI] [PubMed] [Google Scholar]
- 32.Cheng X, Cole RN, Zaia J, Hart GW. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry. 2000;39:11609–11620. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- 33.Yang WH, et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 34.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 35.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Z, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao J, et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tai HC, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J Am Chem Soc. 2004;126:10500–10501. doi: 10.1021/ja047872b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.