Abstract
Patients with hepatitis C virus (HCV) and human immunodeficiency virus (HIV) coinfection for whom prior treatment of HCV with interferon-ribavirin has failed may require subsequent treatment with new HCV protease inhibitors (PIs). We evaluated the diversity of HCV nonstructural protein 3 (NS3) in 26 HCV- and HIV-coinfected patients receiving stable antiretroviral therapy (ART) who were treated with interferon-ribavirin. Plasma HCV RNA clonal analysis was performed. There was greater baseline NS3 diversity in patients with nonresponse or relapse than in those with sustained virologic response. Interferon-ribavirin treatment did not result in significant changes in HCV protease gene diversity or significant HCV PI resistance mutations. The effect of prior interferon-ribavirin treatment on HCV NS3 will likely not impact HCV PI efficacy in HIV-coinfected patients receiving ART.
The management of hepatitis C virus (HCV) infection has become increasingly important among patients with human immunodeficiency virus (HIV) coinfection, in order to prevent HCV-related liver disease, hepatocellular carcinoma, and death. To date, the treatment of HCV infection has comprised therapy based on pegylated interferon alpha and ribavirin for at least 24–48 weeks. Important factors associated with achieving sustained virologic response (SVR) after therapy for HCVinfection are baseline HCV load and genotype. In addition, patients with HIV coinfection, particularly those infected withHCVgenotype 1, experience significantly lower rates of treatment response compared with HCV-monoinfected patients (SVR rate in patients with HCV genotype 1 monoinfection, 40%–50%; SVR rate in patients with HCV genotype 1 and HIV coinfection, 15%–30%) [1].
The N-terminal domain of nonstructural protein 3 (NS3) encodes a serine protease that catalyzes posttranslational processing of the HCV polyprotein in a step crucial to the assembly and replication of viral particles. Several drugs that target the HCV protease gene are currently in development. HCV protease inhibitors (PIs) block the cleavage of nonstructural proteins and restore innate, interferon-mediated host cellular responses to HCV replication [2], rapidly reducing HCV loads during PI exposure [3]. Boceprevir and telaprevir are 2 PIs that have demonstrated efficacy in treating both treatment-naive and treatment-experienced patients with HCV genotype 1 infection. When combined with pegylated interferon and ribavirin, these PIs resulted in significantly higher rates of SVR (60%–75%) among treatment-naive patients with HCV genotype 1 infection and among patients with HCV genotype 1 infection for whom interferon-based therapy had previously failed [4–6]. Unfortunately, rapid emergence of PI resistance in the NS3 domain was found with PI monotherapy, which necessitated combination treatment with HCV PIs, pegylated interferon, and ribavirin.
Because the HCV RNA polymerase is prone to errors and because HCV has high replicative capacity, HCV circulates in individuals as closely related yet distinct viruses, which are termed quasispecies. Factors such as immune pressure, viral kinetics, genotype, and treatment impact genetic diversity. The NS3 region of HCV genotype 1 has demonstrated significant variability at the amino acid level (up to 10%–11% variability for genotypes 1a and 1b) and nucleotide sequence level (up to 26%–30% variability) in monoinfected patients [7]. What impact such NS3 genetic diversity may have on the use of PIs, given the emerging data on NS3 PI resistance mutations, is unknown.
Further understanding of the underlying NS3 diversity among populations of previously treated and HIV-coinfected patients is required. Elsewhere we evaluated the NS3 diversity in coinfected patients who initiate antiretroviral therapy (ART) for HIV infection and found no significant effect of ART on HCV protease diversity [8]. Here we evaluate the effect of interferon-ribavirin therapy on HCV NS3 protease gene sequence diversity and amino acid changes in HIV-coinfected patients who receive ART.
Methods
The institutional review boards of the National Institute of Allergy and Infectious Diseases and Stanford University approved this study. Patients were previously enrolled in 2 prospective studies that analyzed the HCV virologic responses to the following regimens: 1.5 µg of peginterferon alfa-2b (PegIntron; Schering-Plough) per kilogram of body weight or 180 µg of peginterferon alfa-2a (Pegasys; Roche Laboratories) subcutaneously weekly with ribavirin (Rebetrol; Schering-Plough) at a dose of 400 mg every morning and 600 mg every evening for patients with <75 kg of body weight and at a dose of 600 mg twice per day for patients with >75 kg of body weight; or 900 or 1200 µg of albinterferon alfa-2b (Zalbin; Human Genome Sciences) every 2 weeks with 1000–1200 mg/day of ribavirin twice daily, at doses based on body weight. All patients in our analysis were coinfected with HCV genotype 1 and HIV, and all were receiving ART.
Patients were evaluated for HCV quasispecies diversity at baseline (prior to initiation of interferon-based therapy) and at a follow-up time point no more than 4 weeks after completion of HCV therapy. Outcomes were defined as SVR (undetectable viral load 24 weeks after completion of therapy), virologic nonresponse (detectable viral load throughout treatment or a reduction of <2 log{in10} IU/mL in viral load at week 12 of treatment), or virologic relapse (decrease in viral load to undetectable levels during treatment but subsequent increase to detectable levels after treatment discontinuation). All patients with SVR attained an undetectable HCV load at the follow-up time point and thus were compared with the patients with nonresponse or relapse only at baseline. Within the nonresponse/relapse group, the variability of NS3 quasispecies at follow-up was compared with that at baseline to assess the change in genetic diversity with therapy.
HCV genotyping was performed using the Versant HCV genotype line probe assay (version 2.0; Siemens Healthcare Diagnostics). Plasma HIV and HCV RNA loads were quantified using the Versant HIV RNA assay (version 3.0; Siemens Healthcare Diagnostics; lower limit of detection, 50 copies/mL) and the Abbott HCV assay (Abbott; lower limit of detection, 20 copies/mL), respectively. CD4+ T cell counts were measured by means of flow cytometry.
HCV NS3 sequences from each patient were generated; ∼20 clones per sample per time point were sequenced. The reverse-transcription polymerase chain reactions generated amplicons spanning nucleotides 3315–4298 of the HCV genome (National Center for Biotechnology Information Reference Sequence NC_004102). Diversity, including ratios between synonymous and nonsynonymous nucleotide substitutions (dS/dN) and normalized Shannon entropy, was analyzed using Molecular Evolutionary Genetics Analysis software (version 4) [9], as described in detail elsewhere [8].
Comparisons within groups (paired comparisons) or between groups (unpaired comparisons) were determined using Wilcoxon signed rank tests and Mann-Whitney U tests, respectively. Categorical measures between groups were determined using the Fisher exact probability test. All reported P values are 2-tailed, and results for which P < .05 are considered statistically significant.
Results
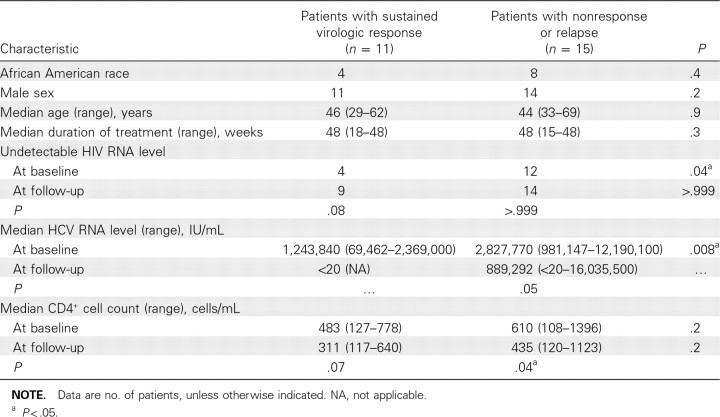
Twenty-six coinfected patients were evaluated; baseline demographic characteristics are presented in Table 1. The patients were predominantly male (25 [96%] of 26 patients); 12 patients [46%] were African American and 11 (42%) were white (non-Hispanic). The mean age was 45 years. Eleven patients attained SVR, and 15 were characterized as having nonresponse or relapse. The mean time from baseline to the end of treatment was 43 weeks for patients with SVR and 38.3 weeks for patients with nonresponse or relapse; the median duration of treatment was 48 weeks in both groups. Two of 11 patients with SVR received 18–24 weeks of therapy because of adverse events, and 7 of 15 patients with nonresponse or relapse received 15–36 weeks of therapy because of virologic nonresponse or adverse events. The patients in the SVR and nonresponse/relapse groups were similar with respect to age, race, and baseline and follow-up CD4+ cell counts. Patients with nonresponse or relapse had higher baseline HCV loads (median, 2.83 × 106 IU/mL) compared with those of patients with SVR (median, 1.24 × 106 IU/mL; P = .008). In the nonresponse/relapse group, there was a nonsignificant decrease in the median HCV load after treatment (from 2.83 × 106 to 8.89 × 105 IU/mL); in addition, whereas decreases in CD4+ cell count were observed in both groups between baseline and follow-up, only the decrease in the nonresponse/relapse group was significant (median at baseline vs follow-up, 610 vs 435 cells/mL; P = .04).
Table 1.
Characteristics of Patients with Human Immunodeficiency Virus (HIV) and Hepatitis C Virus (HCV) Coinfection
Genetic diversity and complexity measures were calculated from nucleotide sequences of 903 NS3 gene clones (median, 23 clones per patient per time point; range, 13–30 clones). There was no difference in the number of clones analyzed per patient per time point between the 2 groups or between time points within the nonresponse/relapse group. All clones phylogenetically clustered appropriately by patient, and all patients' sequences segregated appropriately by genotype (data not shown).
Differences in the genetic variability and complexity between patients at the nucleotide and amino acid levels are shown in Table 2. At the nucleotide level, there was no significant difference between baseline and follow-up in the NS3 gene genetic distances in the nonresponse/relapse group (median genetic distance at baseline vs follow-up, 0.012 vs 0.007 nucleotides; P = .21). Similarly, there was no significant change in dS/dN between baseline and the post-HCV treatment time points.
Table 2.
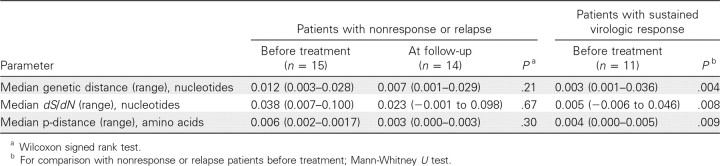
Comparison of HCV Nonstructural Protein 3 Gene Quasispecies Parameters in Patients with Human Immunodeficiency Virus (HIV) and Hepatitis C Virus (HCV) Coinfection
Although no change in diversity was noted over time among the patients with nonresponse or relapse, significant differences between the 2 outcome groups were observed with respect to baseline NS3 diversity. Greater baseline NS3 diversity was observed in patients who subsequently experienced virologic nonresponse or relapse, compared with those who attained SVR (median genetic distance in nonresponse/relapse group vs SVR group, 0.012 vs 0.003 nucleotides; P = .004; median dS/dN in nonresponse/relapse group vs SVR group, 0.038 vs 0.005 nucleotides; P = .008).
At the amino acid level, the diversity was evaluated by examining the p-distance between clones from each patient. Again, in patients with nonresponse or relapse, there was no significant change in p-distance between the 2 time points. However, the p-distance at baseline in the nonresponse/relapse group was significantly higher than that of the SVR group (median p-distance in nonresponse/relapse group vs SVR group, 0.006 vs 0.004 amino acids; P = .009).
NS3 amino acid changes were then examined for known resistance mutations and functional enzyme residues. No active site positions (H57, D81, and S139) or metal-binding residues (C97, C99, C145, and H149) had amino acid substitutions in any of the clones examined. In the nonresponse/relapse group, 2 patients at baseline and 2 different patients at follow-up each had a single clone with the R155G substitution. Those with the R155G substitution at baseline each had a different single, additional clone at baseline with a significant NS3 mutation (P88L and A156V). None of these mutations had persisted at both follow-up time points. In the SVR group, all of the clones from 1 patient had a V36M mutation at baseline.
Discussion
Our study is the first (to our knowledge) to evaluate HCV NS3 sequence diversity in HIV-coinfected patients and the relationship between NS3 diversity and interferon-based treatment. Our results indicate there is no significant effect of interferon-ribavirin therapy on NS3 quasispecies diversity in coinfected patients who experience nonresponse or relapse. Major NS3 PI resistance or active site mutations were rarely found. These results suggest that prior HCV treatment with interferon-ribavirin would not markedly impact the potential efficacy of subsequent HCV PI treatment in coinfected patients.
A significant association was seen between treatment failure and higher baseline NS3 diversity in this cohort of patients with HCV genotype 1 infection. Prior studies have demonstrated correlations between increased quasispecies variability in the HCV hypervariable region 1 (HVR1) at baseline and nonresponse to antiviral therapy in patients with HCV genotype 1 monoinfection [10]. Some studies have found that low baseline HVR1 genetic complexity, in addition to other factors such as baseline viral load, predict interferon response [11], whereas other studies have found that quasispecies complexity correlates with early, but not sustained, virologic response [12, 13]. Similar observations have been made with other genotypes and interferon-treated HIV-coinfected patients [14]. The results of these studies suggest that HVR1 complexity and diversity, through host selection pressure or other factors, influences interferon responsiveness. In contrast, fewer studies have evaluated the associations between NS3 quasispecies diversity and interferon response, and none have been described in a coinfected population. Donlin et al [15] evaluated full HCV open reading frame consensus sequences from patients with HCV genotype 1a or 1b monoinfection and found—in contrast to our results—that marked, versus poor, virologic response at day 28 of HCV therapy was associated with lower NS3 diversity. Of note, significantly lower baseline HCV loads were seen among responders and correlated with interferon response.
PI mutations observed in our study included V36M, P88L, R155G, and A156V. The V36M mutation confers low to moderate resistance to telaprevir and higher level resistance in conjunction with a R155K or A156T mutation; it has also been noted to occur at low frequencies (<1%) in PI treatment-naive patients. The A156V mutation is associated with resistance to multiple PIs, including telaprevir; whereas a different substitution at this site, A156T, confers high-level resistance to boceprevir and telaprevir. The R155G mutation is a minor variant of unclear significance that emerges after PI use and may confer low-level telaprevir resistance. The R155K/T mutation, however, is a common resistance mutation for multiple PIs, particularly telaprevir [16]. The occurrence of the P88L mutation has been noted after exposure to the now discontinued PI BILN-2061, and the significance of this mutation for current PIs is unclear. Thus, NS3 resistance mutations are occasionally found in PI treatment-naive patients; however, interferon-ribavirin treatment does not appear to select for these mutations or increase their prevalence.
Our results are potentially limited by the small sample size in each group. Thus, we were unable to evaluate NS3 diversity differences between genotypes 1a and 1b. Significantly more patients with nonresponse or relapse had undetectable HIV loads at baseline; however, HIV RNA level has not been found to correlate with HCV diversity. Although NS3 diversity was higher among patients with nonresponse or relapse, HCV load was also higher in this group. In accordance with the results of prior studies, we found that lower HCV load was independently associated with response. However, our primary analysis was not affected, as no significant NS3 diversity changes with treatment were noted in the nonresponse/relapse group. Of note, we acknowledge that recent use of ultradeep sequencing suggests the prevalence of minor variants in even treatmentnaive patients with HCV infection; however, the impact of such low-frequency resistance mutations on PI treatment efficacy is unstudied, and comparisons between clonal analysis and deep sequencing have not yet been made.
In summary, we found no significant effect of HCV interferon-based therapy on NS3 protease sequence diversity or sequence mutations in our cohort of patients with HCV genotype 1 and HIV coinfection. As HCV PIs are introduced in clinical practice, further understanding of HCV NS3 diversity in this therapeutically challenging coinfected population is needed. However, our results indicate that the potential efficacy of HCV PIs will be unaffected by previous HCV treatment in coinfected patients.
Footnotes
Potential conflicts of interest: none reported.
Financial support: Department of Veterans Affairs; Intramural Research Program of the National Institutes of Health (National Institute of Allergy and Infectious Diseases and National Institutes of Health Clinical Center; project Z01 AI000390-25 and ZIA AI00390-26 to S.K.).
References
- 1.Ghany M, Strader D, Thomas D, Seeff L. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49(4):1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foy E, Li K, Sumpter R, Jr, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci U S A. 2005;102(8):2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamarre D, Anderson P, Bailey M, et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426(6963):186–189. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- 4.Kwo P, Lawitz EJ, McCone J, et al. Programs and abstracts of the 44th Annual Meeting of the European Association for the Study of the Liver. Geneva, Switzerland: European Association for the Study of the Liver; 2009. HCV SPRINT-1 final results: SVR 24 from a phase 2 study of boceprevir plus PegIntron (peginterferon alfa-2b)/ribavirin in treatment-naive subjects with genotype-1 chronic hepatitis C. Abstract 4. [Google Scholar]
- 5.McHutchison JG, Everson GT, Gordon SC, et al. Telaprevir with peg interferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360(18):1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 6.McHutchison JG, Manns MP, Muir AJ, et al. Telaprevir for previously treated chronic HCV infection. N Engl JMed. 2010;362(14):1292–1303. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 7.Holland-Staley CA, Kovari LC, Golenberg EM, Pobursky KJ, Mayers DL. Genetic diversity and response to IFN of the NS3 protease gene from clinical strains of the hepatitis C virus. Arch Virol. 2002;147(7):1385–1406. doi: 10.1007/s00705-002-0807-5. [DOI] [PubMed] [Google Scholar]
- 8.Winters MA, Chary A, Eison R, Asmuth D, Holodniy M. Impact of highly active antiretroviral therapy on hepatitis C virus protease quasispecies diversity in HIV co-infected patients. J Med Virol. 2010;82(5):791–798. doi: 10.1002/jmv.21679. [DOI] [PubMed] [Google Scholar]
- 9.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 10.Morishima C, Polyak S, Ray R, et al. Hepatitis C virus-specific immune responses and quasi-species variability at baseline are associated with nonresponse to antiviral therapy during advanced hepatitis C. J Infect Dis. 2006;193(7):931–940. doi: 10.1086/500952. [DOI] [PubMed] [Google Scholar]
- 11.Salmeron J, Casado J, de Rueda P, et al. Quasispecies as predictive factor of rapid, early and sustained virological responses in chronic hepatitis C, genotype 1, treated with peginterferon-ribavirin. J Clin Virol. 2008;41(4):264–249. doi: 10.1016/j.jcv.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 12.Abbate I, Lo Iacono O, Di Stefano R, et al. HVR-1 quasispecies modifications occur early and are correlated to initial but not sustained response in HCV-infected patients treated with pegylated- or standard-interferon and ribavirin. J Hepatol. 2004;40(5):831–836. doi: 10.1016/j.jhep.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Chambers TJ, Fan X, Droll DA, et al. Quasispecies heterogeneity within the E1/E2 region as a pretreatment variable during pegylated interferon therapy of chronic hepatitis C virus infection. J Virol. 2005;79(5):3071–3083. doi: 10.1128/JVI.79.5.3071-3083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shire N, Horn P, Rouster S, Stanford S, Eyster ME, Sherman K. HCV kinetics, quasispecies, and clearance in treated HCV-infected andHCV/HIV-1-coinfected patients with hemophilia. Hepatology. 2006;44(5):1146–1157. doi: 10.1002/hep.21374. [DOI] [PubMed] [Google Scholar]
- 15.Donlin MJ, Cannon NA, Yao E, et al. Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy. J Virol. 2007;81(15):8211–8224. doi: 10.1128/JVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson AJ, McHutchison JG. Antiviral resistance and specifically targeted therapy for HCV (STAT-C) J Viral Hepat. 2009;16(6):377–387. doi: 10.1111/j.1365-2893.2009.01124.x. [DOI] [PubMed] [Google Scholar]