Abstract
Instability of (CTG)•(CAG) microsatellite trinucleotide repeat (TNR) sequences is responsible for more than one dozen neurological or neuromuscular diseases. TNR instability during DNA synthesis is thought to involve slipped strand or hairpin structures in template or nascent DNA strands, although direct evidence for hairpin formation in human cells is lacking. We have used targeted recombination to create a series of isogenic HeLa cell lines in which (CTG)•(CAG) repeats are replicated from an ectopic copy of the c-myc replication origin. In this system the tendency of chromosomal (CTG)•(CAG) tracts to expand or contract was affected by origin location and the leading or lagging strand replication orientation of the repeats, and instability was enhanced by prolonged cell culture, increasing TNR length, and replication inhibition. Hairpin cleavage by synthetic zinc finger nucleases in these cells has provided the first direct evidence for the formation of hairpin structures during replication in vivo.
Introduction
Expansion of (CTG)•(CAG) trinucleotide repeat microsatellite sequences is responsible for several neuromuscular and neurodegenerative diseases including Huntington’s disease (HD) and myotonic dystrophy type 1 (DM1). In DM1 patients instability of the (CTG)•(CAG) repeats at the DMPK locus is evident during germ cell development and in somatic cells 1. Instability increases with increasing repeat tract length, with a significant bias towards expansions. (CTG)•(CAG) expansion generally correlates with disease severity and earlier age of onset, a phenomenon termed genetic anticipation. Transgenic mouse models of DM1 also display TNR instability, with the degree of expansion bias strongly dependent on repeat length, the site of chromosomal integration and tissue type 2,3.
The tendency of (CTG)•(CAG) repeats to promote DNA instability correlates directly with their stabilities of hairpin formation 4–8. Current models of TNR instability invoke hairpin formation during DNA synthesis, either in mitotic replication or as a stage of DNA repair 1,9,10, although hairpin structures have not been shown directly to occur in mammalian cells. Agents that induce DNA damage or inhibit replication increase TNR instability 11,12. Likewise, the effects of replication polarity and origin location on TNR instability provide evidence of a role for DNA replication in TNR instability 4,13, but aside from the single stranded nature of the Okazaki fragment initiation zone, the properties of the replication fork that may account for differential hairpin formation on leading and lagging strand DNA are not known.
TNR hairpin formation in bacteria preferentially leads to contraction of direct repeat sequences in the lagging strand template and expansion of repeated sequences in the leading strand nascent DNA 14. In yeast, decreased expression of the Rad27 flap endonuclease (the ortholog of human Fen1) that catalyzes lagging strand Okazaki fragment maturation increases both contractions and expansions of the (CTG)•(CAG) TNR, with greater enhancement of expansions 15. By contrast, somatic cells of Fen1 haploinsufficient mice stably maintain a (CTG)120•(CAG)120 tract at an ectopic HD locus 16, and a (CTG)110•(CAG)110 repeat at an ectopic DMPK locus 17, although the direction of replication through the (CTG)•(CAG) microsatellites was not determined in these models.
Several models have been developed to explain how a lagging strand (CTG) hairpin might contribute to TNR contraction 1,18, while an alternative “template-push” model has been proposed in which (CAG) sequences promote hairpin formation in the leading strand template as a consequence of maintaining contact between the replicative polymerase and helicase 19. Here we have used targeted recombination by Flp recombinase to construct cell lines in which to examine the influence of replication on (CTG)•(CAG) instability and the formation of DNA hairpins. Differences in chromatin environment or genetic background were avoided by integrating cassettes containing (CTG)•(CAG) TNRs in either orientation alongside the c-myc replicator 20,21 at the same ectopic chromosomal site.
These cell lines revealed that the instability of (CTG)•(CAG) repeats is dependent on the length of the repeat, the age of the cell line, the replication polarity of the TNR, and the activity of the proximal replication origin. Instability was markedly enhanced by inhibition of replication with emetine, Fen1 siRNA or aphidicolin. The pattern of instability showed significant similarities and differences after each of these treatments. To test directly whether the (CTG)•(CAG) TNR could form hairpins in vivo, synthetic zinc finger nucleases were designed which specifically targeted these structures. Expression of these nucleases in the Flp targeted cell lines demonstrated that both leading and lagging strand DNAs form replication-dependent hairpin structures.
Results
Effectors of (CTG)•(CAG) instability
Hairpin formation in the template or nascent strands during replication can generate genomic instability by contraction or expansion, respectively, of the number of microsatellite sequence repeats (Figure 1). To investigate the influence of DNA replication on TNR stability in a human chromosomal context, we used Flp recombinase to generate clonal cell lines containing 12 or 102 (CTG)•(CAG) repeats alongside the wild type c-myc replication origin, or alongside a mutant c-myc origin inactivated by deletion of the DNA unwinding element (ΔDUE) 20–22 (Figure 2a). Flp recombinase targeted cells are collectively referred to as (CTG)n•(CAG)n cells, while individual cell lines are denoted according to the trinucleotide sequence in the lagging strand template when replicated from the flanking c-myc origin 20–22. Thus, (CTG)102 cells contain 102 (CTG) repeats in the lagging strand template for replication from the proximal c-myc origin. The single copy integration of each (CTG)•(CAG) cassette at the Flp recombinase target (FRT) site was confirmed by PCR and Southern hybridization (Supplementary Figure 1).
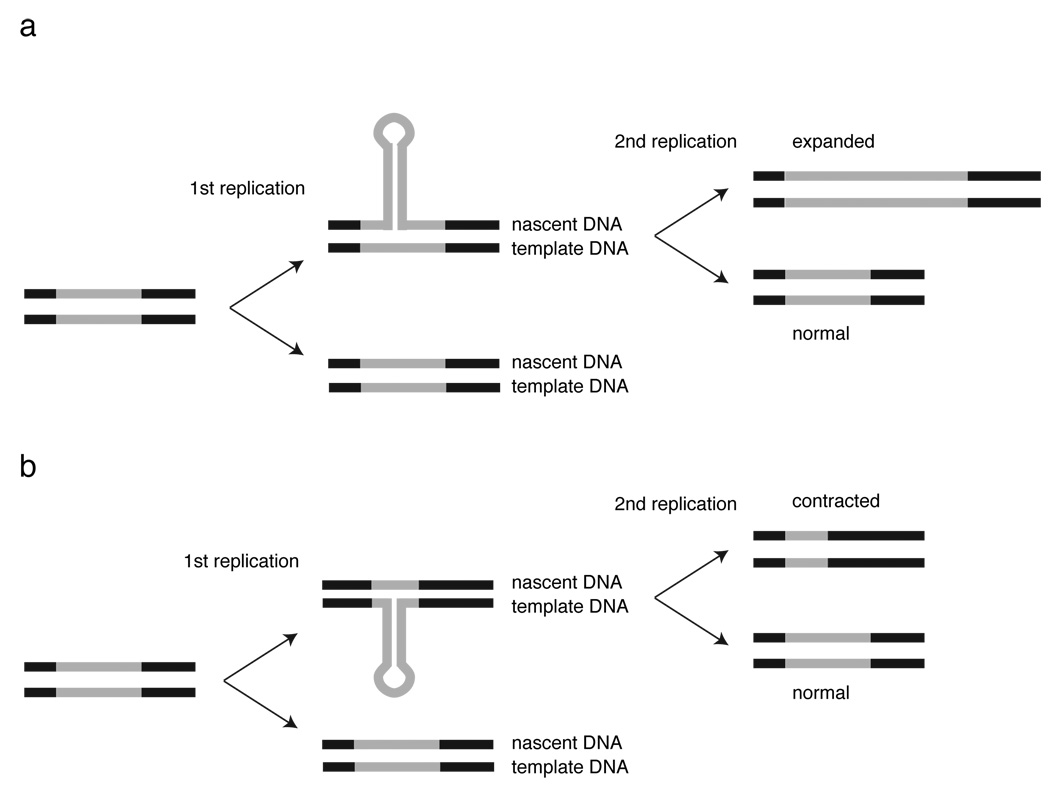
Figure 1.
Hairpin induced trinucleotide repeat instability. The TNR is indicated by grey lines, flanking DNA by black lines. (a) Nascent strand hairpin formation results in overreplication of a segment of the TNR in one chromatid. A second round of replication of the hairpin strand fixes the expanded allelle in the genome. (b) Template strand hairpin formation results in underreplication of a segment of the TNR in one chromatid. A second round of replication of the non-hairpin strand fixes the contracted allelle in the genome. (Adapted from 9).
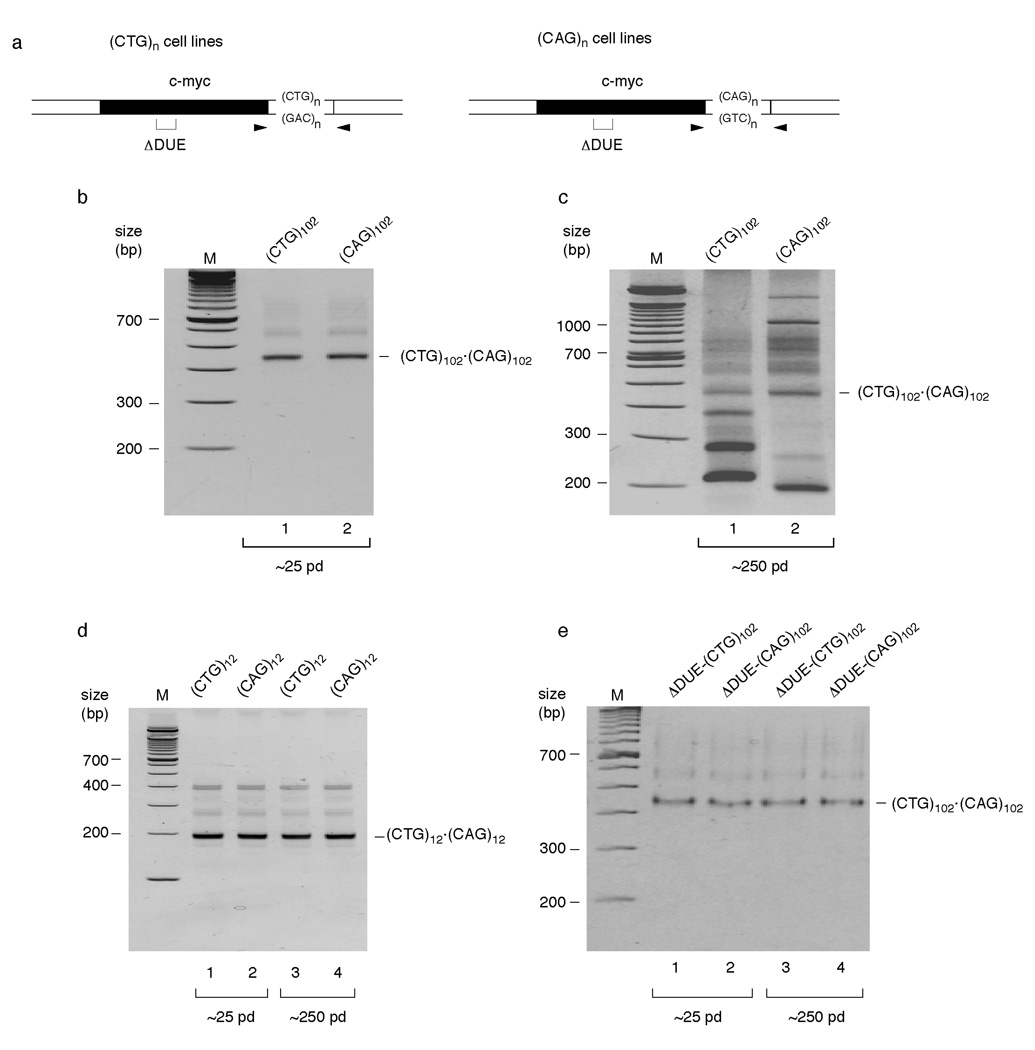
Figure 2.
DNA replication affects TNR instability. (a) Diagram of ectopic origin sites. Arrowheads, PCR primer positions; bracket, DUE deletion (ΔDUE). (b) PCR analysis of (CTG)102•(CAG)102 cells cultured for 25 population doublings. Lane 1, (CTG)102 cells; lane 2, (CAG)102 cells; M, molecular weight marker. The progenitor (CTG)102•(CAG)102 band is indicated. (c) PCR analysis of (CTG)102•(CAG)102 cells cultured for 250 population doublings. Lane 1, (CTG)102 cells; lane 2, (CAG)102 cells. (d) PCR analysis of (CTG)12•(CAG)12 cells cultured for 25 or 250 population doublings. Lanes 1, 3, (CTG)12 cells; lanes 2, 4, (CAG)12 cells. The progenitor (CTG)12•(CAG)12 band is indicated. (e) PCR analysis of ΔDUE-(CTG)102•(CAG)102 cells cultured for 25 or 250 population doublings. Lanes 1, 3, ΔDUE-(CTG)102 cells; lanes 2, 4, ΔDUE-(CAG)102 cells.
Approximately 25 population doublings after clonal selection, PCR amplification with primers flanking the repeat tracts detected no instability at the (CTG)102•(CAG)102 TNR (Figure 2b); the lower mobility shadow bands observed above the ~450 bp amplification product of the (CTG)102•(CAG)102 progenitor sequence are slipped strand artifacts of reannealing during the PCR 6, as demonstrated by re-amplification of the linear ~450 bp product band (Supplementary Figure 2). Strikingly, after about 250 population doublings, the (CTG)102 and (CAG)102 cells showed clear patterns of instability that differed for the two TNR orientations (Figure 2c). The Flp targeted cell lines are clonally derived thus, as confirmed by PCR of early passage cells, the variation in PCR product size following prolonged culture did not arise from length heterogeneity of the progenitor allele.
The (CTG)102 cells displayed predominantly contractions, while the (CAG)102 cells displayed both expansions and contractions (Figure 2c). Instability was not detected in (CTG)12 or (CAG)12 cells after 25 or 250 population doublings (Figure 2d), or when (CTG)102 or (CAG)102 TNRs were integrated next to the inactivated ΔDUE c-myc origin and grown for 250 population doublings (Figure 2e). These results indicate that (CTG)•(CAG) TNR instability depends on repeat tract length at this locus. The dependence of microsatellite expansion and contraction on proximal origin activity, and the effect of TNR orientation relative to the origin argue that the pattern of instability in this model system is also replication dependent.
Replication inhibition enhances TNR instability
Treatment of DM1 patient cells in culture with the replication inhibitors emetine or aphidicolin increased (CTG)•(CAG) instability at the DMPK locus 12. To test the effect of replication inhibitors on (CTG)•(CAG) stability at the ectopic c-myc origin, (CAG)102 or (CTG)102 cell lines at approximately 25 population doublings were treated with aphidicolin, emetine or Fen1 siRNA. When cells were treated with low dose aphidicolin to slow replication fork movement without inducing cell cycle arrest there was a clear orientation-dependent induction of TNR instability, with (CTG)102 cells showing primarily contractions and (CAG)102 cells showing both expansions and contractions (Supplementary Figure 3a). The patterns of instability in (CTG)102 cells and (CAG)102 cells treated with aphidicolin at ~25 population doublings were similar to the patterns of instability observed when these cells were maintained in long term (~250 pd) culture, suggesting that the effects of aphidicolin are qualitatively similar to those occurring in the absence of drug treatment.
Aphidicolin inhibits both leading and lagging strand DNA polymerases, whereas emetine preferentially inhibits Okazaki fragment synthesis 12,23. Similar to the patterns of instability induced by aphidicolin treatment, emetine treatment of (CTG)102 cells led primarily to contractions of the (CTG)102•(CAG)102 TNR whereas (CAG)102 cells showed a strong tendency towards expansions (Supplementary Figure 4). These results suggest that emetine-induced instability is due to hairpins formed by the lagging strand template (CTG) repeats or leading strand template (CAG) repeats. Following this reasoning, (CTG) hairpins in the leading strand nascent DNA may account for the distinct pattern of expansions seen in emetine treated (CAG)102 cells. Sequencing of expanded or contracted DNA bands after emetine treatment (Supplementary Figure 4) confirmed that these bands contain uninterrupted (CTG)•(CAG) sequences, indicating that emetine-induced instability occurs by an error-free mechanism. The differential effects of emetine on (CTG)102 and (CAG)102 cells were also evident in flow cytometry, where the drug induced a sizeable increase in the sub-G1 population of (CTG)102 cells (Supplementary Figure 5).
The effects of emetine on TNR stability are apparently not due to downregulation of Fen1 nuclease (Supplementary Figure 5), which acts during Okazaki fragment maturation and DNA repair synthesis to remove 5’ single-stranded flaps before ligation. Similar to the effects of emetine treatment however, siRNA knockdown of Fen1 led to a large increase in contractions in (CTG)102 cells, but fewer large expansions in (CTG)102 and (CAG)102 cells than aphidicolin or emetine (Supplementary Figure 6). Collectively, the results of replication inhibition suggest that slowing of replication fork movement promotes TNR instability, with (CTG)102 repeats in the lagging strand template favoring contraction, and (CTG)102 repeats in the leading strand nascent DNA favoring expansion.
The patterns of instability induced by these replication inhibitors are consistent with the predicted stability of (CTG) hairpins vs. (CAG) hairpins 24 resulting in a greater frequency of (CTG) hairpins in the lagging strand template. Nevertheless, the stronger tendency towards contractions in (CTG)102 cells vs. (CAG)102 cells could be a result of hairpin formation in the (CAG) leading strand template 19. To determine whether leading or lagging strand templates could form hairpins in vivo, we decided to interrogate these proposed structures by the expression of synthetic nucleases.
Hairpin formation in vivo
We designed zinc finger nucleases (ZFNs) that could be expressed in human cells to test directly for replication-dependent formation of hairpins in vivo. The engineered ZFNs contain the sequence nonspecific Fok 1 restriction endonuclease catalytic cleavage domain (Fok 1CD) fused to three tandem zinc finger protein domains that each recognize the (GCT) (ZFGCT zinc finger) or (AGC) (ZFAGC zinc finger) trinucleotides to produce, respectively, nucleases that target (CTG) hairpins (termed ZFNCTG), or (CAG) hairpins (termed ZFNCAG). Like Fok 1, ZFNs dimerize through their Fok 1 cleavage domains only when bound to DNA, and this dimerization is necessary for catalytic activity 25,26. Whereas co-expression of ZFNCTG and ZFNCAG is predicted to be necessary for cleavage of B-form (CTG)•(CAG) DNA (Figure 3a), we reasoned that the requirement for cleavage domain dimerization would preclude hydrolysis of B-form DNA by a single ZFN but allow digestion of (CTG) and (CAG) hairpin structures, respectively, by the individual ZFNCTG and ZFNCAG nucleases (Figure 3b).
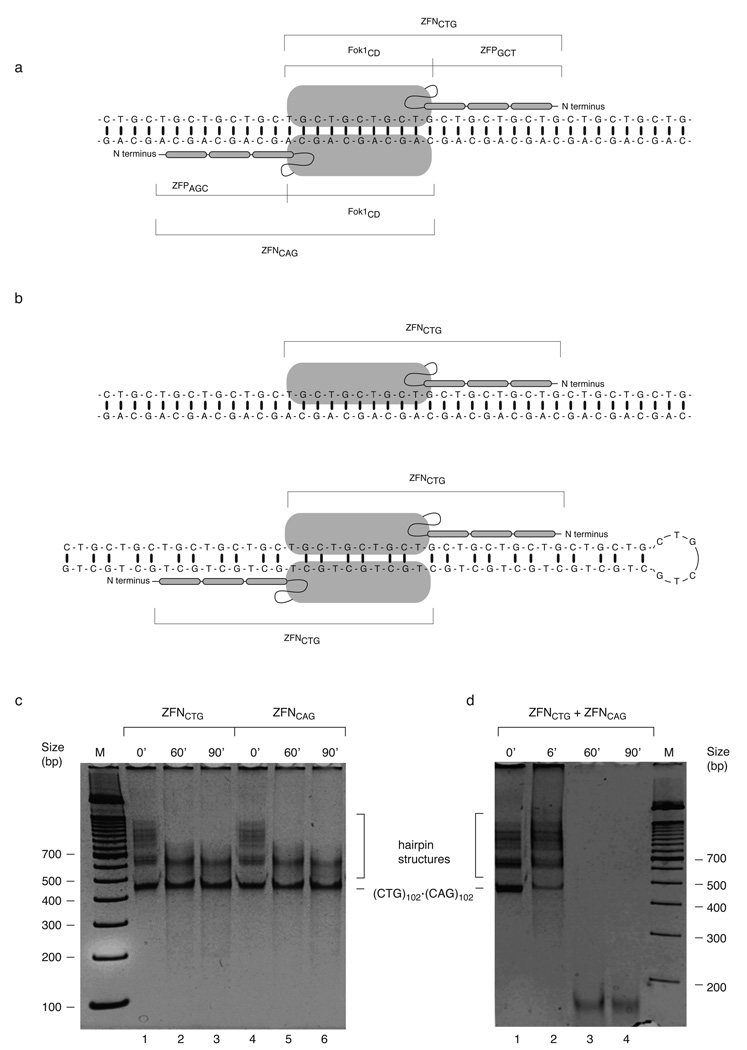
Figure 3.
ZFN cleave specifically in vitro. (a) Binding of a ZFNCTG and ZFNCAG heterodimer capable of cleaving heteroduplex DNA. Fok ICD, Fok I catalytic domain; ZFPGCT, GCT recognition zinc finger protein; ZFAGC, AGC recognition zinc finger protein. (b) Predicted modes of ZFNCTG monomer binding to heteroduplex DNA (upper) or homodimeric ZFNCTG capable of cleaving (CTG)n hairpin DNA (lower). (c) The linear (CTG)102•(CAG)102 PCR product was gel purified and reamplified. Time course of cleavage of the reamplified (CTG)102•(CAG)102 PCR products with ZFNCTG (10% of immunoprecipitate, lanes 1–3) or ZFNCAG (10% of immunoprecipitate, lanes 4–6). (d) Time course of cleavage of the reamplified (CTG)102•(CAG)102 PCR products with a mixture of ZFNCTG and ZFNCAG (5% of each immunoprecipitate).
Previous in vitro characterization of binding by individual zinc finger domains indicates that certain (GCT) zinc fingers may recognize (GCA) triplets and that some (GCA) zinc fingers may recognize (GCT) triplets 27,28. To assess the specificity of the ZFNs these nucleases were assayed in the in vitro system shown to be more permissive for nuclease digestion than in vivo 28,29. ZFNCTG and ZFNCAG were expressed individually in HeLa cells and immunoprecipitated through their FLAG epitope tags (Supplementary Figure 7). The linear (CTG)102•(CAG)102 PCR product band was reamplified and used as a substrate for cleavage by the immunoprecipitated nucleases. Consistent with the predicted specificity of these enzymes, the individual ZFNs cleaved the slow mobility, slipped strand PCR product structures 6 and a fraction of the intermediate mobility slipped strand structures, but did not cleave the (CTG)102•(CAG)102 B-form DNA during extended incubation (Figure 3c). Shorter time course digestions revealed intermediates in the slipped-strand digestion process (Supplementary Figure 8). In contrast, the mixture of ZFNCTG and ZFNCAG quantitatively cleaved the (CTG)102•(CAG)102 B-form DNA in addition to digesting the slipped strand structures (Figure 3d).
The resistance of a significant fraction of the template to digestion by the individual nucleases argued that the ZFNs were specific and that the extracts did not display nonspecific nuclease activity. To confirm this, extracts were prepared from HeLa cells expressing the same zinc finger proteins (ZFPs) omitting the Fok I nuclease domain. We observed that the ZFP containing extracts did not digest the (CTG)102•(CAG)102 DNA substrate (Supplementary Figure 8) or (CNG)46 synthetic hairpin substrates (Supplementary Figure 9). Furthermore, the ZFNCTG immunoprecipitate digested the (CTG)46 hairpin substrate but not the (CAG)46 hairpin substrate, while the ZFNCAG immunoprecipitate digested the (CAG)46 substrate but not the (CTG)46 hairpin substrate.
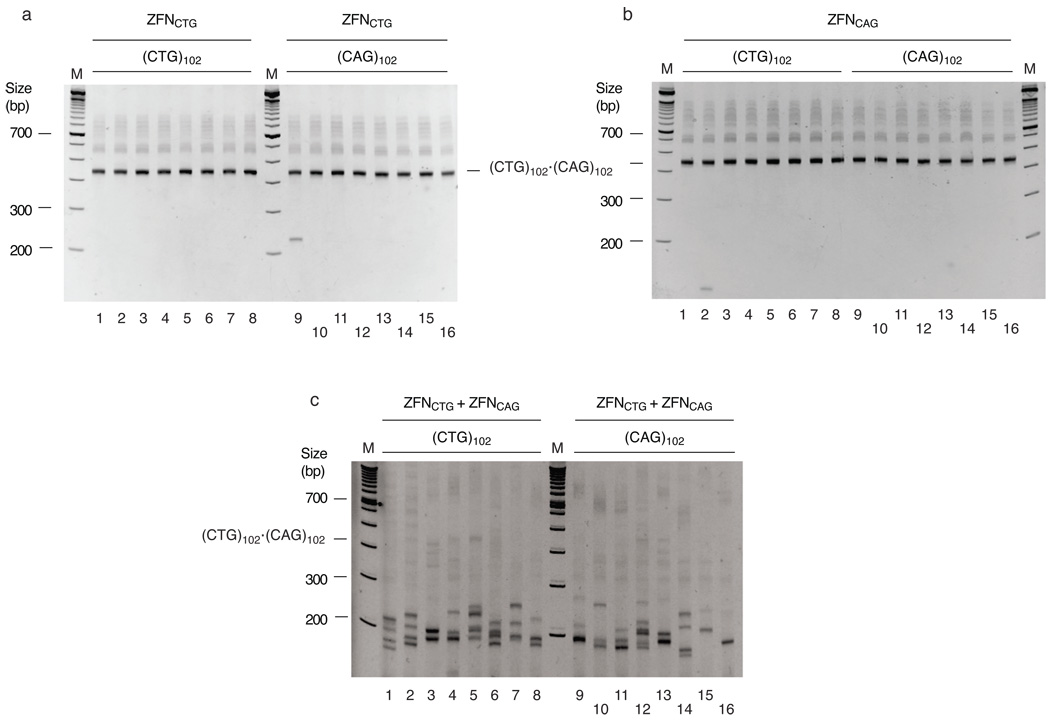
ZFNCTG or ZFNCAG were expressed in (CTG)102 or (CAG)102 cells and genomic DNA was isolated for spPCR amplification 48 hours later. As shown in Figure 4, expression of either ZFNCTG (Figure 4a) or ZFNCAG (Figure 4b) in (CTG)102 cells led to a decrease in the length of the TNR tracts. These results indicate that both the lagging strand (CTG) template and the leading strand (CAG) template strand can form hairpin structures that produce contractions in vivo. Treatment of (CAG)102 cells with ZFNCTG or ZFNCAG also resulted in shortening of the TNR tracts, reinforcing the view that either template strand can form hairpins in vivo.
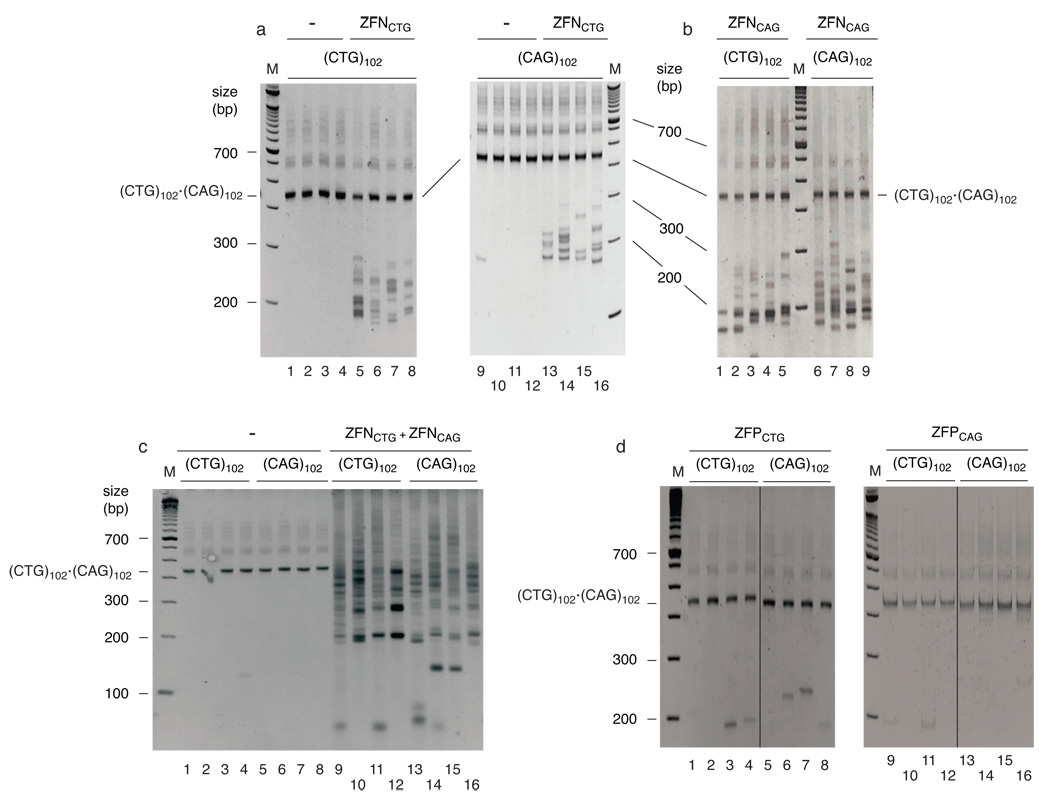
Figure 4.
(CTG)102•(CAG)102 TNRs form hairpins in vivo. (CTG)102•(CAG)102 cells were cultured for 25 population doublings. (a) spPCR of DNA from (CTG)102 cells or (CAG)102 cells transfected, respectively, with empty vector (lanes 1–4, 9–12) or ZFNCTG expression plasmid (lanes 5–8, 13–16). (b) spPCR of DNA from (CTG)102 cells (lanes 1–5) or (CAG)102 cells (lanes 6–9) transfected with ZFNCAG expression plasmid. (c) spPCR of DNA from (CTG)102 cells or (CAG)102 cells transfected, respectively, with empty vector (lanes 1–4, 5–8) or a 0.5:0.5 mixture of ZFNCTG and ZFNCAG expression plasmids (lanes 9–12, 13–16). (d) spPCR of DNA from (CTG)102 cells or (CAG)102 cells transfected with ZFPCTG (lanes 1–8) or ZFPCAG (lanes 9–16) expression plasmids. Lanes 1–4 and 5–8 were merged from nonadjacent lanes of the same gel, as were lanes 9–12 and lanes 13–16. (See Supplementary Figure 11 for full gel images).
It is formally possible that the decrease in TNR repeat length following ZFN expression is due to ZFN binding to (CTG)•(CAG) hairpins that subsequently induces cleavage by cellular nucleases. We addressed this question further by expressing the zinc finger proteins ZFPCTG and ZFPCAG in (CTG)102 or (CAG)102 cells. Omission of the Fok I nuclease domain in ZFPs resulted in significantly decreased frequencies of TNR contraction (compare Figures 4a, b, d). Based on the Fok I nuclease domain-dependent difference in TNR PCR patterns between cells expressing ZFPs and ZFNs, we conclude that the ZFNs directly cleave hairpin structures in vivo.
The progenitor (CTG)102•(CAG)102 band was preserved when cells expressed either ZFN individually, while the (CTG)102•(CAG)102 band was completely digested when either (CTG)102 or (CAG)102 cells were co-transfected with a mixture of ZFNCTG and ZFNCAG expression plasmids (Figure 4c). In contrast to the deletions that were detected by spPCR after single ZFN expression, expansions were detected after combined expression of ZFNCTG and ZFNCAG (although these are underrepresented by the spPCR). Significantly, this suggests that digestion by a homodimeric ZFN is repaired differently than digestion by a heterodimeric ZFN. Inasmuch as there is only one copy of the ectopic origin/TNR integrant per genome, we postulate that the expansions that occur after heterodimeric ZFN digestion arise by homology directed repair of double strand breaks using the misaligned sister chromatid TNR during S or G2 phase, whereas single strand gaps are formed by homodimeric ZFN hairpin excision, and are repaired by primer extension.
The resistance of the (CTG)102•(CAG)102 band to digestion by a single ZFN supports the view that the ZFNs selectively target hairpin structures. To test whether the sensitivity of the (CTG)102•(CAG)102 TNR to ZFN digestion is dependent on cell division, ZFNCTG or ZFNCAG was expressed in (CTG)102 or (CAG)102 cells during serum starvation. Digestion by ZFNCTG or ZFNCAG was markedly reduced in serum deprived cells, whereas expression of the heteromeric pair of ZFNs eliminated the (CTG)102•(CAG)102 TNR band almost completely, and generated expansion products upon spPCR (Figure 5). These data underscore the conclusion that the individual ZFNs selectively target non-B DNA structures, and that hairpin formation is replication-dependent. We also constructed (CTG)45•(CAG)45 FRT targeted cell lines, and found that these cells showed similar sensitivity to the individual ZFNs and insensitivity to ZFPs as the (CTG)102•(CAG)102 cells (Supplementary Figure 10), indicating that premutation length TNRs can form hairpin structures in vivo.
Figure 5.
(CTG)102•(CAG)102 hairpin formation is suppressed by serum starvation. (CTG)102•(CAG)102 cells were grown for 25 population doublings and transferred to medium containing 0.5% serum for 48 hr. (a) spPCR of DNA from (CTG)102 cells (lanes 1–8) or (CAG)102 cells (lanes 9–16) transfected with the ZFNCTG expression plasmid. (b) spPCR of DNA from (CTG)102 cells (lanes 1–8) or (CAG)102 cells (lanes 9–16) transfected with the ZFNCAG expression plasmid. (c) spPCR of DNA from (CTG)102 cells (lanes 1–8) or (CAG)102 cells (lanes 9–16) transfected with the ZFNCAG and ZFNCAG expression plasmids.
(CTG)12•(CAG)12 cells did not show time-dependent instability, and did not show induction of instability when treated with aphidicolin, emetine, or Fen1 siRNA (Figure 6). (CTG)12•(CAG)12 cells were also tested for their sensitivity to ZFNCTG and ZFNCAG. Because the predicted minimum binding site for a dimeric ZFN is eight trinucleotide repeats, the stems of the TNR hairpins in these cells should be too short to allow nuclease binding and dimerization, while the B-form (CTG)12•(CAG)12 TNR could theoretically bind dimeric ZFNs. Indeed, the individual ZFNs did not cleave the (CTG)12•(CAG)12 cell TNRs, but the co-expressed ZFN pair could digest the TNRs. Presumably because there are fewer possible binding configurations that would accommodate the dimeric nuclease, digestion was less efficient on the shorter repeat tracts. Considered together these data corroborate the conclusion that spontaneous or drug-induced hairpin formation and TNR instability are length-dependent phenomena.
Figure 6.
(CTG)12•(CAG)12 TNRs are stable in vivo. (CTG)12 or (CAG)12 cells were cultured for 25 population doublings. (a) PCR of DNA from (CTG)12 (lane 1, 3) or (CAG)12 cells (lane 2, 4) treated with aphidicolin, or mock treated, as indicated. (b) PCR of DNA from (CTG)12 (lane 1, 3, 5, 7) or (CAG)12 cells (lane 2, 4, 6, 8) treated with emetine, Fen1 siRNA, or mock treated, as indicated. (c) spPCR of DNA from untreated (CTG)12 (lane 1–8) or (CAG)12 cells (lane 9–16). (d) spPCR of DNA from (CTG)12 or (CAG)12 cells expressing ZFNCTG (lanes 1–8) or ZFNCAG (lanes 9–16), as indicated. (e) spPCR of DNA from (CTG)12 (Lanes 1–6) or (CAG)12 cells (lanes 7–12) expressing a 0.5:0.5 mixture of ZFNCTG and ZFNCAG.
Discussion
The site-specific integration of specific repeat length (CTG)•(CAG) tracts alongside an ectopic copy of the human c-myc replication origin has allowed analysis of the effect of repeat length, replication polarity and origin location on TNR instability. The clonal origin of each of the cell lines used in this work eliminated progenitor cell heterogeneity and differences in patient age, tissue type, genetic background, and chromosomal location that may have complicated the interpretation of previous studies. In this model we observed that (CTG)•(CAG) microsatellite instability is enhanced by prolonged cell growth, increased repeat length, origin proximity, and the inhibition of replication. Furthermore, the pattern of instability was orientation dependent. When (CTG)102 comprised the lagging template strand, the repeat showed a strong bias towards contraction in vivo, whereas both contractions and expansions were observed when (CAG)102 was present in the lagging strand template.
In bacteria and yeast (CTG)•(CAG) repeat contractions occur more often when the (CTG) sequence, which exhibits greater structure-forming potential 5,24,30, is present in the lagging strand template 31; this effect was observed in the current study as well. It has been argued alternatively that (CAG) hairpins in the leading strand template are responsible for this phenomenon 19. Our ZFN digestion data indicate that (CTG) or (CAG) sequences can both form hairpins in vivo in either the leading strand or lagging strand templates. On the other hand, since both (CTG) and (CAG) template sequences can form hairpins, the effect of replication orientation on the pattern of instability suggests that leading and lagging strands do not have the same tendency to expand or contract. Two or three major deletion products were identified in (CTG)102 cell DNA after extended culture or after replication inhibition. If both template strands are prone to contraction, it would not be expected that reversal of the TNR (i.e. in (CAG)102 cells) would lead to disappearance of these contraction products. Therefore, it seems likely that both contracted products come predominantly from one of the template strands. Since the ZFN digestions indicate that either template strand can form hairpins, we presume that a step after hairpin formation is responsible for the selective contraction on just one template strand.
Slowing of leading and lagging strand synthesis with aphidicolin accelerated the (CTG)102 TNR contraction and (CAG)102 TNR expansion in a manner similar to that seen after long term culture. Emetine inhibition of lagging strand DNA synthesis in (CTG)102 cells also resulted in the appearance of a small number of prominent contraction products, as did inhibition of Okazaki fragment maturation by Fen1 knockdown. Each of these inhibitors can also cause fork stalling and reversal 32, which may explain the resemblance of the contraction products following these alternative forms of replication inhibition 33,34. While variation in the sizes of the contraction products may be due to unique structures of the replicative intermediates induced by each inhibitor, the overall similarity in the pattern of contraction products after aphidicolin, emetine or Fen1 siRNA treatment suggests that the physical properties of the TNR are more significant in determining the results of instability than is the mechanism of replication inhibition. That TNR orientation relative to the c-myc origin affects the observed patterns of (CTG)•(CAG) instability suggests that the (CTG) and (CAG) repeats respond differently as template vs. nascent strands or leading vs. lagging strands during replication or sister chromatid homology-based repair after fork stalling.
Yang et al. have reported that aphidicolin or emetine treatment of primary fibroblasts from a fetus with DM1 strongly enhanced expansion of the (CTG)216 allele but did not affect the normal (CTG)12 allele 12. These results are in good agreement with the marked expansion induced by aphidicolin or emetine in (CAG)102 cells and the stability of (CTG)12•(CAG)12 tracts to these drug treatments. While the direction of replication through the DMPK locus was not mapped by Yang et al., the observation that (CTG)102•(CAG)102 tracts expand in (CAG)102 cells treated with emetine predicts that the (CTG)216 allele studied by Yang et al. was preferentially replicated from a downstream origin, such that the (CAG) sequence was replicated as the lagging strand template.
Replication based models posit that expansion occurs because of hairpin formation in lagging strand Okazaki fragments, or in leading strand nascent DNA during fork reversal 32,35. Several laboratories have attempted to address the role of Okazaki fragment maturation in TNR instability by inhibiting Fen1 nuclease, yet the consequences of Fen1 knockdown appear to be cell type specific 15–17,31,36–38. Variability in the chromosomal location, repeat length, and amount of natural flanking sequence 39 may also complicate interpretation of these results. In this regard, the fact that the (CTG)•(CAG) sequences used here contained less than 20 bp of 5’ or 3’ DMPK flanking sequences implies that the present results reflect the inherent characteristics of the (CTG)•(CAG) TNR with respect to length and replication polarity separate from significant shielding effects of DMPK flanking DNA.
Arguing against a major a role for Okazaki fragment maturation in (CTG)•(CAG) expansion, the most dramatic increases in expansions occurred when Okazaki fragment synthesis was inhibited by emetine, which induced two major expanded products in (CAG)102 cells and a complex array of expansion products in (CTG)102 cells. Under these conditions an extended single strand lagging template may potentiate fork reversal and hairpin formation in the displaced leading strand nascent DNA 32,35. The extruded nascent strand could then form a variety of stable (CTG) hairpin forms in (CTG)102 cells but a more limited array of stable (CAG) hairpins in (CAG)102 cells. The presence of even fewer contractions in (CAG)102 cells following emetine treatment compared to Fen1 siRNA or aphidicolin treatment supports the notion that the (CAG) repeats rarely contract in the lagging strand template and (CTG) repeats rarely contract in the leading strand template. The absence of contractions in (CAG)102 cells following emetine treatment may also point to a role for Okazaki fragment synthesis in stabilizing lagging strand (CAG) hairpins.
In the ectopic c-myc origin system, removal of the DUE eliminates origin activity 22 and (CTG)•(CAG) TNR instability. The reasons for the correlation between origin location and TNR instability may not be related to proximal origin activity, but may be due to a change in the replication polarity of the repeats (ori-switch hypothesis 9), a result of the origin presenting a distinctive chromatin or DNA structure, or a more circumscribed zone for the initiation of Okazaki fragment synthesis relative to the TNR (ori-shift hypothesis 13). Alternatively, the structure of the replication fork or the composition of the replisome may change as it moves from the origin.
HD or DM1 (CTG)•(CAG) microsatellites in humans and transgenic mice show varying degrees of expansion bias 15,17,40,41, while analyses in bacteria, yeast, and human cultured cells reveal preferential contractions 15,42,43. The ectopic (CTG)102 and (CAG)102 cell lines used here showed primarily contractions, although occasional expansions were observed upon long-term culture of (CAG)102 cells. Similarly, DM1 lymphoblastoid cell lines (LBCLs) demonstrate preferential contractions of the progenitor repeat length 42. Upon continued culture, LBCLs containing the contracted alleles disappear from the population, and are replaced by more rapidly proliferating mutants with large repeat expansions 42. These results evoked the hypothesis that mitotic drive may contribute to the expansion bias observed at the tissue level. During unperturbed cell division (CTG)102 and (CAG)102 cells most often experience TNR contractions upon extended culture, while three different means of replication inhibition rapidly induced repeat contraction in (CTG)102 cells and expansion in (CAG)102 cells. These observations, and the report that the aphidicolin and emetine regimens used here do not select for drug resistant cell lines 12, suggest that the induced instability results from aberrant DNA synthesis rather than growth selection.
Extended passage (CTG)102 cells accumulate primarily contractions in the (CTG)•(CAG) TNR. The replication-dependent sensitivity of the (CTG)•(CAG) TNR to cleavage by either ZFNCTG or ZFNCAG in short term passage (CTG)102 cells implies that both the leading and lagging template strands can form hairpins during unperturbed replication 35. ZFN excision of a hairpin is predicted to generate gapped linear DNA. In our assay, excision of nascent strand hairpins would not be detectable after gap filling, whereas excision of template strand hairpins would result in contractions. Therefore, the contractions induced by either ZFNCTG or ZFNCAG treatment of (CTG)102 or (CAG)102 cells (and (CTG)45 or (CAG)45 cells) support the conclusion that both the leading and lagging template strands can form hairpins, and it is also plausible that the ZFNs cleave leading strand nascent DNA hairpins. Furthermore, since instability is not detected in early passage cells in the absence of ZFN treatment, we presume that hairpin structures do not ordinarily persist long enough to be fixed as repeat length changes by a second round of replication.
Shishkin et al. have recently proposed a template switching mechanism for (GAA)•(TTC) expansion that predicts nascent strand hairpin formation as a consequence of leading strand polymerase copying of Okazaki fragments 44. While the strong enhancement of expansions in (CAG)102 cells treated with emetine might seem at odds with this model, it is possible that these expansions occur as the (CAG)102 cells recover from the drug treatment. It has also been proposed that a common property of expandable repeats is their tendency to form noncanonical DNA structures, although the structures formed by different repeats may vary 9,45–47. Hence, different repeats such as (GAA)•(TTC) 44 or (ATTCT)•(AGAAT) 22 may expand through structural intermediates that reflect their particular physical tendencies.
In the yeast model of large (GAA)•(TTC) expansion, instability is not dependent on fork stalling 44. However, replication inhibition by aphidicolin, emetine, or Fen1 siRNA may potentiate expansion or contraction if the replication fork is destabilized. The present system offers several opportunities to test the contribution of the replication fork stabilization complex 48, helicases and recombination proteins to TNR stability in human chromosomes.
Methods
Plasmid construction
(CTG)•(CAG) trinucleotide repeats were amplified from genomic DNA or plasmid clones by PCR and inserted into the Not I restriction site at the 3’ edge of the c-myc replicator plasmids pFRT.myc or pFRT.myc. ΔDUE 21. DMPK flanking DNA in the TNR insert comprise the 5' sequence TTGTAGCCGGGAATGCTG and the 3' sequence TGGTCTGTGATCCCCC. All primer sequences used in this work are available from the authors. For cloning of TNR PCR products, bands were separated by electrophoresis on a 2% agarose gel and extracted. The purified PCR products were subcloned into the Not I site of pFRT.myc and sequenced.
Cell culture
HeLa/406 acceptor cells containing a single chromosomal Flp recombinase target were constructed, grown, and transfected as described 20. Transfectants were subjected to drug selection using G418 and ganciclovir 21. Cells were grown at 37 °C and 5% CO2 in DMEM supplemented with 10% newborn calf serum. (CTG)n•(CAG)n cells were treated with emetine or aphidicolin as described 12. Approximately 5×105 cells were grown in DMEM with 1 µM emetine (18 hr) or 0.2 µM aphidicolin (18 hr), and allowed to recover in drug-free medium for two population doublings. One cycle of treatment includes drug exposure and recovery. This treatment was repeated for a total of 5 cycles. Control experiments were performed using drug free medium. (CTG)102•(CAG)102 cells (30% confluent) were transfected with 50 nM Fen1 siRNA in 6-well plates every 48 hr for a total of 3 transfections. Mock experiments were carried out using non-specific siRNA. Whole-cell lysates were prepared from treated or untreated HeLa cells and probed using antibodies against human Fen1 (Santa Cruz) or Mcm7 (a gift from Dr. Aloys Schepers, GSF-Haematologikum).
Zinc finger plasmids
The single zinc finger plasmids pc3XB-ZF72, pcXB-ZF83, and a mammalian expression vector, pST1374, were purchased from Addgene Inc. (Cambridge, MA). Proteins with three zinc fingers (ZFPs) were assembled using standard molecular cloning methods. ZFPCTG, containing three ZF72 zinc fingers (Addgene plasmid 13206), and ZFPCAG, containing three ZF83 zinc fingers (Addgene plasmid 13217), were designed with Zinc Finger Targeter (ZiFiT) Version 2.0 49. ZFPs cDNAs were subcloned into the mammalian expression vector pST1374 (Addgene plasmid 13426) between the coding sequences of the FLAG tag and Fok I DNA cleavage domain to generate zinc finger nucleases (ZFNs). Plasmids expressing ZFPs were constructed by introducing a stop codon at the Bam HI restriction site between zinc finger domain and Fok I cleavage domain in plasmid pST1374ZFNCTG or pST1374ZFNCAG to generate pST1374ZFNCTG or pST1374ZFPCAG respectively.
For in vitro digestion, ZFP and ZFN proteins were expressed in HeLa cells and immunoprecipitated using EZview Red Anti-FLAG M2 affinity gel (Sigma). ZFN and ZFP proteins were expressed from the same vectors at similar levels (Supplementary Figure 4). The in vitro digestion 29 substrates were produced by PCR amplification of linear (CTG) 102•(CAG)102 DNA or by annealing of TNR oligonucleotide 5’-TCCTTGTAGCCGGGAATG(CNG)46GGGGGATCACAGACCATT-3’ to 5’-AATGGTCTGTGATCCCCCAAAACATTCCCGGCTACAAGGA-3’ (where (CNG)46 stands for (CTG)46 or (CAG)46) to form hairpin structures. The digestion products were analyzed by electrophoresis in 7.5% polyacrylamide gels. For in vivo digestion ZFN or ZFP plasmid DNAs were transfected into the indicated cell lines at 50% confluence in 6-well plates twice every 72 hr. Cells were harvested 72 hr after the second transfection, and genomic DNA was isolated for PCR.
PCR
Standard PCR used 40 ng genomic DNA per reaction. Amplification conditions were 94 °C (5’); 35 cycles of 94 °C (30”), 55 °C (30”), 72 °C (2’); and 72 °C (7’). spPCR used 130 pg genomic DNA per reaction. Amplification conditions were 94 °C (5’); 35 cycles of 94 °C (30”), 55 °C (30”), 72 °C (30”); and 72 °C (7’). Under the short extension conditions of spPCR, the expanded templates seen by standard PCR of (CTG)102 and (CAG)102 genomic DNA were not detected. PCR products were resolved in 7.5% polyacrylamide gels. Images were obtained on a Fuji LAS-3000 using ImageReader and Adobe Photoshop software.
Flow cytometry
Cells were trypsinized, washed in PBS and fixed overnight at −20 °C in 70% ethanol. Cells were resuspended in PBS (pH 7.4), treated with 100 U of RNase A for 20 min at 37 °C, and stained with 50 µg of propidium iodide/ml. DNA content was detected using a Becton Dickinson FACScan flow cytometer.
Supplementary Material
Acknowledgments
The authors thank Drs. C. Pearson and D. Monckton for their comments on this work. This work was supported by a grant from the WSU Boonshoft School of Medicine to GL and by a grant from the NIH (GM53819) to ML.
Footnotes
Author contributions. Experiments were conceived and designed by GL, ML, JJB and RRS, and performed by GL, XC and ML. The manuscript was drafted by ML and revised by all authors.
Competing interests. The authors declare that they have no competing interests as defined by the Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
References
- 1.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 2.Lia AS, et al. Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum Mol Genet. 1998;7:1285–1291. doi: 10.1093/hmg/7.8.1285. [DOI] [PubMed] [Google Scholar]
- 3.Fortune MT, Vassilopoulos C, Coolbaugh MI, Siciliano MJ, Monckton DG. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum Mol Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 4.Miret JJ, Pessoa-Brandao L, Lahue RS. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson CE, Wang YH, Griffith JD, Sinden RR. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM. Replication and expansion of trinucleotide repeats in yeast. Mol Cell Biol. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc Natl Acad Sci U S A. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 10.Sinden RR, Pytlos MJ, Potaman V. Mechanisms of DNA repeat expansion. In: Fry M, Usdin K, editors. Human nucleotide expansion disorders. Vol. xvii. Berlin; New York: Springer; 2006. pp. 3–53. [Google Scholar]
- 11.Hashem VI, et al. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 2004;32:6334–6346. doi: 10.1093/nar/gkh976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Lau R, Marcadier JL, Chitayat D, Pearson CE. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am J Hum Genet. 2003;73:1092–1105. doi: 10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleary JD, Pearson CE. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 2005;21:272–280. doi: 10.1016/j.tig.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Trinh TQ, Sinden RR. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature. 1991;352:544–547. doi: 10.1038/352544a0. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, Freudenreich CH. Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene. 2007;393:110–115. doi: 10.1016/j.gene.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiro C, McMurray CT. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol Cell Biol. 2003;23:6063–6074. doi: 10.1128/MCB.23.17.6063-6074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Broek WJ, Nelen MR, van der Heijden GW, Wansink DG, Wieringa B. Fen1 does not control somatic hypermutability of the (CTG)(n)*(CAG)(n) repeat in a knock-in mouse model for DM1. FEBS Lett. 2006;580:5208–5214. doi: 10.1016/j.febslet.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 18.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delagoutte E, Goellner GM, Guo J, Baldacci G, McMurray CT. Single-stranded DNA-binding protein in vitro eliminates the orientation-dependent impediment to polymerase passage on CAG/CTG repeats. J Biol Chem. 2008;283:13341–13356. doi: 10.1074/jbc.M800153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malott M, Leffak M. Activity of the c-myc replicator at an ectopic chromosomal location. Mol Cell Biol. 1999;19:5685–5695. doi: 10.1128/mcb.19.8.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G, Malott M, Leffak M. Multiple functional elements comprise a mammalian chromosomal replicator. Mol Cell Biol. 2003;23:1832–1842. doi: 10.1128/MCB.23.5.1832-1842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu G, Bissler JJ, Sinden RR, Leffak M. Unstable Spinocerebellar Ataxia Type 10 (ATTCT)*(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol Cell Biol. 2007;27:7828–7838. doi: 10.1128/MCB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burhans WC, et al. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO Journal. 1991;10:4351–4360. doi: 10.1002/j.1460-2075.1991.tb05013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gacy AM, McMurray CT. Influence of hairpins on template reannealing at trinucleotide repeat duplexes: a model for slipped DNA. Biochemistry. 1998;37:9426–9434. doi: 10.1021/bi980157s. [DOI] [PubMed] [Google Scholar]
- 25.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci U S A. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mani M, Smith J, Kandavelou K, Berg JM, Chandrasegaran S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem Biophys Res Commun. 2005;334:1191–1197. doi: 10.1016/j.bbrc.2005.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segal DJ, Dreier B, Beerli RR, Barbas CF., 3rd Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5'-GNN-3' DNA target sequences. Proc Natl Acad Sci U S A. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittelman D, et al. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0902420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll D, Morton JJ, Beumer KJ, Segal DJ. Design, construction and in vitro testing of zinc finger nucleases. Nat Protoc. 2006;1:1329–1341. doi: 10.1038/nprot.2006.231. [DOI] [PubMed] [Google Scholar]
- 30.Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 32.Carbajal Rivera ML, Ordaz Tellez MG, Castaneda Sortibran A, Rodriguez-Arnaiz R. Emetine and/or its metabolites are genotoxic in somatic cells of Drosophila melanogaster. J Toxicol Environ Health A. 2007;70:1713–1716. doi: 10.1080/10937400601171241. [DOI] [PubMed] [Google Scholar]
- 33.Hashem VI, Rosche WA, Sinden RR. Genetic recombination destabilizes (CTG)n.(CAG)n repeats in E. coli. Mutat Res. 2004;554:95–109. doi: 10.1016/j.mrfmmm.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr Opin Struct Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Sinden RR, Pytlos MJ, Potaman VN. Mechanisms of DNA repeat expansion. In: Fry M, U K, editors. Human Nucleotide Expansion Disorders. Springer; 2006. pp. 3–53. [Google Scholar]
- 36.Gordenin DA, Kunkel TA, Resnick MA. Repeat expansion--all in a flap? Nat Genet. 1997;16:116–118. doi: 10.1038/ng0697-116. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 38.Jackson SM, et al. SCA7 CAG/CTG repeat expansion is stable in Drosophila melanogaster despite modulation of genomic context and gene dosage. Gene. 2005;347:35–41. doi: 10.1016/j.gene.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Libby RT, et al. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum Mol Genet. 2003;12:41–50. doi: 10.1093/hmg/ddg006. [DOI] [PubMed] [Google Scholar]
- 40.Monckton DG, Wong LJ, Ashizawa T, Caskey CT. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum Mol Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- 41.Seznec H, et al. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum Mol Genet. 2000;9:1185–1194. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- 42.Khajavi M, et al. "Mitotic drive" of expanded CTG repeats in myotonic dystrophy type 1 (DM1) Hum Mol Genet. 2001;10:855–863. doi: 10.1093/hmg/10.8.855. [DOI] [PubMed] [Google Scholar]
- 43.Hashem VI, Rosche WA, Sinden RR. Genetic assays for measuring rates of (CAG).(CTG) repeat instability in Escherichia coli. Mutat Res. 2002;502:25–37. doi: 10.1016/s0027-5107(02)00026-x. [DOI] [PubMed] [Google Scholar]
- 44.Shishkin AA, et al. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potaman VN, et al. Unpaired Structures in SCA10 (ATTCT)(n).(AGAAT)(n) Repeats. J Mol Biol. 2003;326:1095–1111. doi: 10.1016/s0022-2836(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 47.Sinden RR, Pytlos-Sinden MJ, Potaman VN. Slipped strand DNA structures. Front Biosci. 2007;12:4788–4799. doi: 10.2741/2427. [DOI] [PubMed] [Google Scholar]
- 48.Voineagu I, Surka CF, Shishkin AA, Krasilnikova MM, Mirkin SM. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat Struct Mol Biol. 2009;16:226–228. doi: 10.1038/nsmb.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.