Abstract
Background
Biomarkers for accurate diagnosis of early hepatocellular carcinoma (HCC) are limited in number and clinical validation. We applied SELDI-TOF-MS ProteinChip technology to identify serum profile for distinguishing HCC and liver cirrhosis (LC) and to compare the accuracy of SELDI-TOF-MS profile and alpha-fetoprotein (AFP) level in HCC diagnosis.
Patients and Methods
Serum samples were obtained from 120 HCC and 120 LC patients for biomarker discovery and validation studies. ProteinChip technology was employed for generating SELDI-TOF proteomic features and analyzing serum proteins/peptides.
Results
A diagnostic model was established by CART algorithm, which is based on 5 proteomic peaks with m/z values at 3324, 3994, 4665, 4795, and 5152. In the training set, the CART algorithm could differentiate HCC from LC subjects with a sensitivity and specificity of 98% and 95%, respectively. The results were assessed in blind validation using separate cohorts of 60 HCC and 60 LC patients, with an accuracy of 83% for HCC and 92% for LC patients. The diagnostic odd ratio (DOR) indicated that SELDI-TOF proteomic signature could achieve better diagnostic performance than serum AFP level at a cutoff of 20 ng/mL (AFP20) (92.72 vs 9.11), particularly superior for early-stage HCC (87% vs 54%). Importantly, a combined use of both tests could enhance the detection of HCC (sensitivity, 95%; specificity, 98%; DOR, 931).
Conclusion
Serum SELDI-TOF proteomic signature, alone or in combination with AFP marker, promises to be a good tool for early diagnosis and/screening of HCC in at-risk population with liver cirrhosis.
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide with China and North America showing a continuous increase in incidence and mortality rate.1 The prognosis of HCC remains poor, and most patients have a 5-year survival rate of less than 5% mainly because of the late diagnosis.2 Evidence suggests that surgical resection, liver transplantation, and other curative therapies, such as ablative treatments, can significantly improve the survival for those HCC patients with tumor detected at an early stage.3,4 Early diagnosis, therefore, is of paramount importance for prolonging the survival of HCC patients. Chronic hepatitis-related liver cirrhosis (LC) is one of the strongest predisposing factor, and ~60 to 80% of cirrhotic patients inevitably develop HCC. Therefore, early detection of HCC by incorporation of surveillance program for high-risk populations, such as those with LC or hepatitis carriers, can improve outcomes of HCC patients.5,6
Detection of serum alpha-fetoprotein (AFP) level (at a cutoff of 20 ng/mL, AFP20) and ultrasonography of intrahepatic lesions are the 2 most common clinical approaches to detect HCC.7 However, these two methods suffer from certain limitations. For instance, ultrasound imaging is highly operator dependent and is not sensitive enough to detect liver tumors in patients with cirrhotic background.8 Serum AFP level is insensitive, such that about one-third of early-staged HCC patients with small tumors (<3 cm) show same level of AFP as those in healthy subjects, making it useful only to identify late-staged HCC patients. Besides, high false-positive rate is found in ~20% of the patients with chronic hepatitis and in 20–50% of those with LC.2,9 In this regard, there is an urgent need to identify alternative serum biomarkers for early detection of HCC.10–12
For serum proteomics, surface-enhanced desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) ProteinChip technology provides an efficient and sensitive means for biomarker discovery. It enables quantitative measurement of specific peptide/protein in a high-throughput way, and small amount of materials are required for the analysis. All these features make SELDI-TOF-MS an attractive tool for clinical application. This technology has been successfully applied to facilitate biomarker discovery for various malignancies, such as breast cancer, ovarian cancer, lung cancer, nasopharyngeal cancer, and colorectal cancer.13–19 In addition, this approach has been used to study diagnostic biomarkers and/or proteomic patterns for HCC.20–22 However, these earlier SELDI-TOF-MS-based HCC studies had small sample sizes, and the patients involved were mainly in advanced stages. Therefore, we intended to investigate the serum fingerprint specifically associated with hepatitis B-related LC and early-stage HCC. In addition, we compared the accuracy of using this serum patterns with the conventional AFP biomarkers for differentiating early-staged HCC and LC patients.
Materials and Methods
Patients and Sample Collection
A total of 240 patient subjects (120 HCC patients and 120 LC patients) were included in this study, and these patients were presented to the Queen Mary Hospital (Pokfulam, Hong Kong) between 2003 and 2005. The demographics and clinicopathological data are summarized in Table 1. Informed consent was obtained from these patients. This study was approved by the Joint University/Hospital Authority’s Institutional Review Board. All patients were age- and gender-matched Chinese having the same HBsAg profile. Preliminary diagnosis of HCC was based on preoperative investigations, which included blood biochemistry, AFP assay, chest x-ray, percutaneous ultrasonography, computed tomography, and hepatic angiography in selected patients. Liver function was assessed using Child-Pugh classification. Histopathological examination of the resected specimens was conducted by board-certified pathologists to confirm the diagnosis. Tumor stages were based on the AJCC staging system. Early HCC was defined as stage I (T1N0M0). Diagnosis of cirrhosis was based on liver histology as well as laboratory and imaging evidence of hepatic decompensation or portal hypertension. The median follow-up period of cirrhotic patients was 12 months with no sign of HCC development.
Table 1.
Clinical features of patient cohort
| Group | HCC (n = 120) | Cirrhosis (n = 120) |
|---|---|---|
| Sex (M/F) | 96/24 | 88/32 |
| Age (year) | 55.4 ± 11.4 | 50.9 ± 12.6 |
| HBsAg (+) | 100 | 106 |
| AFP | 17580.8 ± 100941.3 | 24.5 ± 90.8 |
| <20 ng/mL (%) | 37 (30.8%) | 94 (78.3%) |
| 20–400 ng/mL (%) | 28 (23.3%) | 25 (20.8%) |
| ≥400 ng/mL (%) | 55 (45.8%) | 1 (0.8%) |
| Tumor stage (I/II/III/IV) | 46/36/38/0 | NA |
n number, HBsAg (+) hepatitis B virus antigen positive, AFP alpha-fetoprotein, NA not applicable
Note: All data are presented as mean ± SD
The training set consisted of 60 subjects with pathologically validated LC but without HCC and 60 HCC patients who were mainly at AJCC stage I and II (70%). Another 60 LC and 60 HCC patients were included in the blinded validation set. Pooled serum samples from healthy subjects were used as interassay quality control. Blood samples were taken prior to surgery and 3 months after surgery. Extracted serum was stored at −80°C. A commercial chemiluminescent immunoassay kit (ACS 180SE, Bayer, Leverkuesen, Germany) was used to determine the serum AFP level. In clinical practice, the upper normal range of AFP was 8–10 ng/mL and patients having >20 ng AFP/mL were considered to have liver abnormality. Therefore, this same AFP cutoff level was used in the current study to discriminate HCC in high-risk populations.23
SELDI-TOF-MS and Data Analysis
Serum samples were thawed completely and centrifuged to remove debris at 9600 g for 10 min at 4°C. SELDI-TOF-MS profiling using the IMAC-3 chip (Ciphergen Biosystems, Fremont, CA) was done as previously described.18,22 In brief, 5 μL of the cleared serum was mixed with 5 μL phosphate-buffered saline (PBS) containing 8 M urea and 2% CHAPS and then diluted with 108 μL binding buffer (0.25 M sodium chloride and 1% CHAPS in PBS). After that, 100 μL of diluted serum samples were loaded on each array spot with a robotic bioprocessor and incubated for 1 h at room temperature. After washing off unbound molecules, each array was spotted twice with 0.5 μL of freshly prepared EAM matrix solution (sinapinic acid saturated with 0.5% trifluoroacetic acid and 50% acetonitrile).
Mass spectrometric analysis was performed by SELDI-TOF-MS in a PBS-IIc ProteinChip reader (Ciphergen Biosystems) according to an automated data collection protocol. Serum spectrum was generated by averaging the results of 65 laser shots at a laser intensity of 240, a detector sensitivity of 8, in which the qualified mass peaks (signal-to-noise ratio >5) with mass-to-charge ratios (m/z) in an optimized range of 3000–12,500 Da were detected. Spectral analysis was carried out by the Biomarker Wizard program (version 3.1; Ciphergen Biosystems). After the total ion current normalization, the spectra data of the training set was analyzed using the Biomarker Pattern software (version 4.1, Ciphergen Biosystems) for model building. A classification and regression tree (CART) analysis was performed, as previously described, based on the differentially expressed protein peaks in sera of LC and HCC patients.24
The construction of CART included 2 steps, namely the tree construction step and the tree pruning step. For the tree construction process, the BPS was used to search for the best peak with defined cutoff level so that the training dataset was split into two daughter nodes. The splitting decision was based on the peak intensity of a sample. Samples went to the left daughter node if their peak intensities were equal to or less than the cutoff intensity value; otherwise, the samples would go to the right daughter node. The software continued to repeat this splitting process on each daughter node in this manner until no further gain in the classification was achieved and terminal nodes were produced. Classification of the terminal nodes was decided by a group of samples (i.e., HCC and LC), which represented the majority of samples in that group. In the second step of CART, the classification tree was cut down to a desired size that yielded the least classification error. In this study, a decision tree was generated using the Gini method with nonlinear combinations. A 10-fold cross-validation test was used for the evaluation of the classification error. The performance of the classification algorithm was then challenged in a blind study in which the test set consisted of a new batch of 120 serum samples (60 samples from each group).
Statistical Analysis
Sensitivity was calculated as the ratio of the number of HCC samples that were correctly classified as HCC (true positive) to the total number of HCC samples. Specificity was calculated as the ratio of the number of LC samples that were correctly classified as LC (true negative) to the total number of LC samples. The diagnostic odds ratio of a test was the ratio of the odds of positivity in HCC relative to the odds of positivity in the LC. The correlation analysis compared with clinical features, relative peak intensity levels between two groups was calculated using independent samples t test. For comparison of sensitivity and specificity of CART and AFP level, 2 independent sample chi-square tests were used. The correlation analysis between clinical features and peak intensity levels was calculated using bivariate correlation analysis. All the calculations were completed with the SPSS statistical package (SPSS Inc., Chicago, IL).
Results
Stability and Reproducibility of the SELDI-TOF-MS Spectra
Pooled normal sera, which were spotted randomly on different chips, were used to test for the consistency of the SELDI-TOF-MS spectra among different protein chips. The coefficients of variance (CV) for the three highest peaks with relative intensities above 25 were <20%, and their m/z ratios were <0.1%, akin to that of other studies.25,26 This suggested that little variation in the protein spectra was present for the same sample among different chips, implicating the stability and reproducibility of the SELDI-TOF-MS spectra.
CART and Its Performance
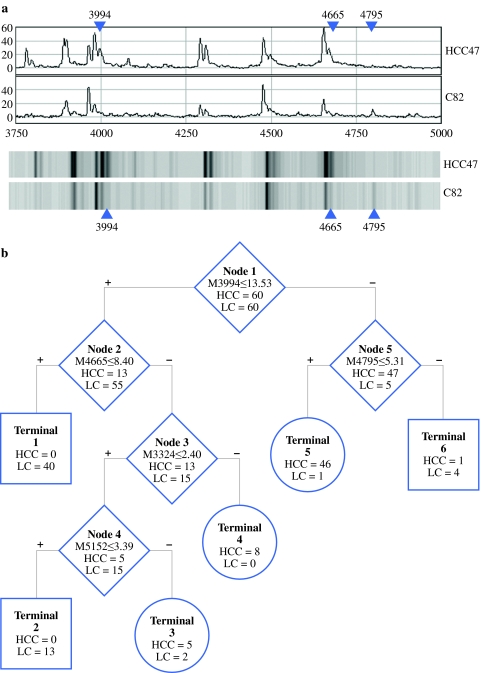
The serum spectra from 60 HCC and 60 LC subjects were used as the training set. After intensity normalization, 80 peaks were found to be statistically significant between HCC and LC patients (P < .05). Five protein peaks were regarded as classifiers to construct the CART. Figure 1a represents spectral views showing some of these protein peaks in the HCC sera compared with the LC sera. The spreadsheets of normalized intensities of these peaks were exported into the Biomarker Patterns software for CART analysis. Of the many classification trees generated by using different settings of Gini, advance, cost of BPS software, the most optimal classification tree with lowest error cost was eventually established. A CART with 6 terminal nodes was established, based on the 5 discriminative peaks with m/z of 3324, 3994, 4665, 4795, and 5152 that could separate HCC patients from LC patients (Table 2) (Fig. 1b). When the 5 proteomic peaks were used as classifiers, this CART algorithm would yield an accuracy rate of 96.7% (116 of 120), a sensitivity of 98.33%, and a specificity of 95% for predicting HCC in the training set. To validate the above finding, a blinded set consisting of a separate cohort of 60 HCC patients and 60 LC patients was used. Serum protein profiles of these samples were collected and treated the same as those in the training set. The normalized peak intensities in the blinded set were subjected to analysis. A sensitivity of 83% (50 of 60), a specificity of 92% (55 of 60), and an accuracy rate of 87.5% were yielded in the blind set. Also, 87% (40 of 46) of the HCC patients in stage I and 89% of patients in stage II were detected by the classification algorithm (Table 3).
Fig. 1.
a Trace view (above) and gel view (under) of representative SELDI-TOF-MS spectra of serum samples from HCC and cirrhosis patients in the range from m/z 3750 to 5000. The three major discriminative peaks at m/z 3994, 4665, and 4795 in the classification tree are pointed out by arrows, respectively. b The optimal classification tree generated by CART. The binary classification tree composed of 5 classifiers (open diamond node). The decision-making process involves the evaluation of if–then rules of each node from top to bottom, which eventually reaches a terminal with designed class outcome—(dark circle), HCC, or (gray square), LC
Table 2.
Intensities of five discriminative peaks
| m/z | P value | HCC group | Cirrhosis group |
|---|---|---|---|
| 3994 | <.000001 | 16.98 ± 6.17 | 8.36 ± 4.50 |
| 4665 | <.000001 | 19.57 ± 9.84 | 7.88 ± 5.22 |
| 4795 | .0000568 | 2.24 ± 1.35 | 4.17 ± 3.22 |
| 3324 | .007 | 3.13 ± 5.78 | 1.80 ± 1.44 |
| 5152 | .0000036 | 4.94 ± 2.42 | 2.99 ± 1.50 |
m/z mass-to-charge ratio
Note: All data are presented as mean ± SD
Table 3.
Number of HCC patients at different stages identified by serum SELDI-TOF-MS pattern and AFP20
| Tumor stage | Number of patients | SELDI-TOF-MS pattern (n/%) | AFP20 (n/%) |
|---|---|---|---|
| I | 46 | 40/87% | 25/54%* |
| II | 36 | 32/89% | 26/72% |
| III | 38 | 37/97% | 32/84% |
| Total | 120 | 109/91% | 83/69% |
Note: AFP20, at a cutoff value >20 ng/mL
* Compared with SELDI-TOF-MS, P < .05
Changes in Serum Proteomic Features Before and After Surgery
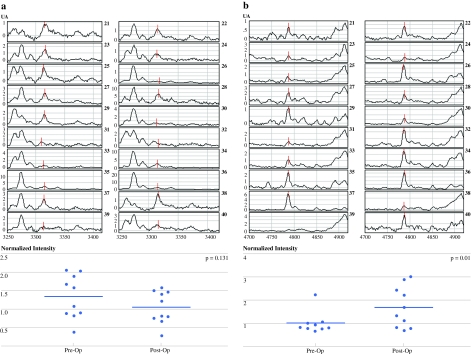
To strengthen these findings, we performed additional experiments to determine the change of proteomic profiles in 10 pairs of serum samples collected from newly recruited HCC patients, before and 3 months after surgery. As shown in Fig. 2a, there is a decrease of the serum level of m/z 3324 peak after removal of the tumors. Although the difference is not significant (P = .131), there is a trend of declination in the serum from the postsurgery group. Interestingly, the serum level of m/z 4795 peak, which was initially lower in the HCC group than the LC group, was significantly increased after surgery (P = .01) (Fig. 2b).
Fig. 2.
Changes in serum proteomic features before and after surgery by SELDI-TOF-MS. Zoom-in spectrum of m/z 3324 a and m/z 4795 b in 10 pairs of HCC serum samples before (left panel) and 3 months (right panel) after surgery. The specific proteomic features were indicated by red lines. The lower panel was the normalized intensities of the specific peak in the preoperative group plotted against the postoperative group, and the statistical significance was calculated by t test
Comparison of SELDI-TOF-MS Pattern and AFP Level for Early-Stage HCC Diagnosis
The sensitivity and specificity of SELDI-TOF-MS pattern and AFP20 to predict different stages of HCC were examined in 120 HCC patients from training and validation set. A sensitivity of 87% (40 of 46) for detecting stage I HCC patients, 89% (32 of 36) for detecting stage II and 97% (37 of 38) for detecting late-staged HCC patients (stage III) were obtained by CART analysis (Table 3). When AFP20 was used to predict HCC stages, a sensitivity of 54% (25 of 46) for stage I HCC patients, 72% (26 of 36) for stage II HCC, and 84% (32 of 38) for late-staged HCC patients (stage III) were obtained, which was less favorable than CART, especially for detecting stage I HCC (P < .05). The diagnostic odds ratios (DOR) were also calculated in this study to compare the CART analysis and AFP20 (Table 4) for predicting HCC in the blinded samples. DOR is a single indicator for diagnostic performance, such that higher DOR value means better discriminative power.27 The calculated DOR of CART algorithm was 92.72, implying that the odds for positivity among patients with HCC was 92.72 times higher than the odds for positivity among patients without HCC. The calculated DOR for AFP20 was 9.11 only. Based on these values, CART algorithm performed 10 times better than AFP20 in detecting HCC from LC patients (92.72 vs 9.11) (Table 4). Interestingly, a combined use of both CART and AFP20 increased the sensitivity to 95%, with at least 1 of the 2 tests being positive, and the specificity to 98%, when both tests were negative. The estimated DOR was 931 for the combined tests, which was better than the individual test alone (931 vs 92.72 and 9.11) (Table 4). Despite that, no significant correlation was found between the intensity values of each discriminative peak and some of the clinicopathological data of HCC patients—age, sex, AFP level, tumor size, and tumor stage (data not shown).
Table 4.
Sensitivity, specificity, and DOR of individual and/or combined use of SELDI-TOF-MS pattern and AFP20 for HCC diagnosis
| Test | Sensitivity (%) | Specificity (%) | DOR |
|---|---|---|---|
| SELDI-TOF-MS | 83 | 95 | 92.72 |
| AFP20 | 72 | 78 | 9.11 |
| SELDI-TOF-MS and/or AFP20 | 95 | 98 | 931 |
AFP 20 AFP at a cutoff value >20 ng/mL. DOR diagnostic odd ratios
DOR = sensitivity/(1-sensitivity)/(1-specificity)/specificity
Discussion
In this study, we profiled 240 serum proteomes (120 HCC and 120 LC) to identify distinctive patterns as a fingerprint for predicting HCC. Most of the clinical samples were obtained from early-staged HCC patients, implicating the potential application of the derived results in clinical settings to identify HCC at its early stage. Diagnosis of HCC at early stage is beneficial for patients as early diagnosis means better prognosis. Importantly, the experimental procedure described in this study did not involve prefractionation and albumin depletion, which were currently adopted for studying serum and inevitably might eliminate low abundance and low molecular weight disease markers. Use of serum without pretreatment also enables the process to be high throughput and cost effective.
Serum profiling is an attractive approach for finding markers or patterns for cancer diagnosis. HCC progression associates with proteomic changes, which may be partially reflected based on the changes in the blood composition of patients. Sometimes, proteins are secreted by tumor cells or by other organs in response to the presence of cancer or the hallmark of predisposing factors to HCC occurrence. Our initial efforts to identify the 5 proteomic peaks revealed the candidate markers as (m/z 3324) relaxin B chain (RLN2, P04090), (m/z 3994) neuropeptide K (TAC1, P20366), (m/z 4665) unidentified, and (m/z 4795) PACAP-related peptide or PRP-48 (ADCYAP, P18509), and (m/z 5152) Complement C3 g fragment (CPAMD1, P01024). Subsequently, we further investigated the gene expression levels of RLN2 (m/z 3324), TAC1 (m/z 3994), and PACA (m/z 4795) in 10 pairs of HCC and adjacent nontumor tissues by microarray analysis (Wong et al., unpublished data). We found RLN2 was overexpressed in the tumor tissues, whereas the ADCYAP1 level was downregulated. However, there was no significant difference in TAC1 expression between tumor and nontumor tissues. We suggest that neuropeptide K may not be directly released from the tumors, and its level may reflect the host response to the tumor development and progression.
Because of the noninvasive nature of obtaining serum when compared with other means, such as fine-needle biopsy, the information derived from serum analysis can probably have clinical implications and applications. Recent studies suggest that a single biomarker is not effective in detecting cancers. Therefore, current biomarker research is shifting from analyzing 1 marker to the simultaneous measurement of several biomarkers.28 SELDI-TOF-MS is one of the profiling approaches enabling the simultaneous analysis of several markers or peaks and is particularly useful for analyzing serum proteomes and potentially applicable for intraoperative biomarkers assays during surgery.29 For instance, Paradis et al. demonstrated a fragmented form of vitronectin as a novel serum marker for HCC using SELDI-TOF-MS.30 In addition, Cui et al. used WCX2 protein chips to generate the serum protein profiles to discriminate patients with hepatitis B, LC, or HCC.21
In accordance with other studies, we also found distinct serum profiles generated by SELDI-TOF-MS between hepatitis B-related HCC and LC that can be used to differentiate these 2 patient groups. Using the same patient cohort, we found that SELDI-TOF-MS indeed performed better than clinically used AFP20 to diagnose HCC, especially for early-staged HCC. Zinkin et al. performed a similar study to compare detection accuracy of using SELDI-TOF-MS and AFP20 to predict HCC using patients with hepatitis C background. They also demonstrated a higher sensitivity and specificity for HCC detection associated with the use of SELDI-TOF-MS when compared with AFP20, such that the sensitivity/specificity for AFP20 and SELDI-TOF-MS were 73%/71% and 79%/86%, respectively.31 Though their study differed from ours in the etiology of HCC patients (HCV related vs HBV related in this study), both studies demonstrated that serum SELDI-TOF-MS pattern employing a combination of several biomarkers was superior in detecting HCC than the known serum biomarker. Importantly, the best result for detecting HCC patients from LC patients was achieved when serum SELDI-TOF-MS pattern was combined with serum AFP detection.
There were previous concerns about the reproducibility of SELDI-TOF-MS technology.32 Because of the different experimental setups, the SELDI-TOF-MS patterns might be different for different laboratories. Still, we believe that SELDI-TOF-MS pattern is a viable means for clinical diagnosis of HCC if the assays are done in high-standard reference laboratories following stringent optimized standard operation procedures and integrated with robotic automation. The next critical step is clinical validation in multiple centers. Importantly, Kanmura et al. also demonstrated the capability of using SELDI-TOF-MS patterns to diagnose 6 of 7 HCC patients, for which these patients were later confirmed to have HCC by ultrasonography.33 In conclusion, our study shows that serum SELDI-TOF-MS profile can be used to differentiate HCC patients, even the early-staged, from cirrhotic subjects with higher sensitivity and specificity than the clinically used AFP level.
Acknowledgment
This study was jointly supported by the Scheme of National Natural Science Foundation of China (30331160411), the Hong Kong Research Grants Council (N_HKU718/03), and the Natural Science Foundation of Beijing (7042032). We acknowledge Dr. CK Hui for his clinical expertise and Professor QY He for his expert opinions and advices on protein identification in serum samples.
Conflict of Interest
The authors declare no conflict of any interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Lei Chen, David W.Y. Ho, and Nikki P.Y. Lee contributed equally.
Contributor Information
Lei Chen, Email: chentiger@medmail.com.cn.
John M. Luk, Email: jmluk@nus.edu.sg.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Hao K, Luk JM, Lee NP, Mao M, Zhang C, Ferguson MD, et al. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer. 2009;9:389. doi: 10.1186/1471-2407-9-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 4.Pang RW, Joh JW, Johnson PJ, Monden M, Pawlik TM, Poon RT. Biology of hepatocellular carcinoma. Ann Surg Oncol. 2008;15:962–971. doi: 10.1245/s10434-007-9730-z. [DOI] [PubMed] [Google Scholar]
- 5.Poon RT, Fan ST, Lo CM, Liu CL, Ng IO, Wong J. Long-term prognosis after resection of hepatocellular carcinoma associated with hepatitis B-related cirrhosis. J Clin Oncol. 2000;18:1094–1101. doi: 10.1200/JCO.2000.18.5.1094. [DOI] [PubMed] [Google Scholar]
- 6.Bruix J, Llovet JM. Prognostic prediction and treatment strategy in hepatocellular carcinoma. Hepatology. 2002;35:519–524. doi: 10.1053/jhep.2002.32089. [DOI] [PubMed] [Google Scholar]
- 7.Wun YT, Dickinson JA. Alpha-fetoprotein and/or liver ultrasonography for liver cancer screening in patients with chronic hepatitis B. Cochrane Database Syst Rev. 2003:CD002799. [DOI] [PubMed]
- 8.Colli A, Fraquelli M, Casazza G, Massironi S, Colucci A, Conte D, et al. Accuracy of ultrasonography, spiral CT, magnetic resonance, and alpha-fetoprotein in diagnosing hepatocellular carcinoma: a systematic review. Am J Gastroenterol. 2006;101:513–523. doi: 10.1111/j.1572-0241.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- 9.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 10.Luk JM, Lam CT, Siu AF, Lam BY, Ng IO, Hu MY, et al. Proteomic profiling of hepatocellular carcinoma in Chinese cohort reveals heat-shock proteins (Hsp27, Hsp70, GRP78) up-regulation and their associated prognostic values. Proteomics. 2006;6:1049–1057. doi: 10.1002/pmic.200500306. [DOI] [PubMed] [Google Scholar]
- 11.Yi X, Luk JM, Lee NP, Peng J, Leng X, Guan XY, et al. Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol Cell Proteomics. 2008;7:315–325. doi: 10.1074/mcp.M700116-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Lee NP, Leung KW, Cheung N, Lam BY, Xu MZ, Sham PC, et al. Comparative proteomic analysis of mouse livers from embryo to adult reveals an association with progression of hepatocellular carcinoma. Proteomics. 2008;8:2136–2149. doi: 10.1002/pmic.200700590. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 14.Paweletz CP, Trock B, Pennanen M, Tsangaris T, Magnant C, Liotta LA, et al. Proteomic patterns of nipple aspirate fluids obtained by SELDI-TOF: potential for new biomarkers to aid in the diagnosis of breast cancer. Dis Markers. 2001;17:301–307. doi: 10.1155/2001/674959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombes KR, Fritsche HA, Jr, Clarke C, Chen JN, Baggerly KA, Morris JS, et al. Quality control and peak finding for proteomics data collected from nipple aspirate fluid by surface-enhanced laser desorption and ionization. Clin Chem. 2003;49:1615–1623. doi: 10.1373/49.10.1615. [DOI] [PubMed] [Google Scholar]
- 16.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 17.Xiao XY, Tang Y, Wei XP, He DC. A preliminary analysis of non-small cell lung cancer biomarkers in serum. Biomed Environ Sci. 2003;16:140–148. [PubMed] [Google Scholar]
- 18.Ho DW, Yang ZF, Wong BY, Kwong DL, Sham JS, Wei WI, et al. Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry serum protein profiling to identify nasopharyngeal carcinoma. Cancer. 2006;107:99–107. doi: 10.1002/cncr.21970. [DOI] [PubMed] [Google Scholar]
- 19.Chen YD, Zheng S, Yu JK, Hu X. Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clin Cancer Res. 2004;10:8380–8385. doi: 10.1158/1078-0432.CCR-1162-03. [DOI] [PubMed] [Google Scholar]
- 20.Schwegler EE, Cazares L, Steel LF, Adam BL, Johnson DA, Semmes OJ, et al. SELDI-TOF MS profiling of serum for detection of the progression of chronic hepatitis C to hepatocellular carcinoma. Hepatology. 2005;41:634–642. doi: 10.1002/hep.20577. [DOI] [PubMed] [Google Scholar]
- 21.Cui J, Kang X, Dai Z, Huang C, Zhou H, Guo K, et al. Prediction of chronic hepatitis B, liver cirrhosis and hepatocellular carcinoma by SELDI-based serum decision tree classification. J Cancer Res Clin Oncol. 2007;133:825–834. doi: 10.1007/s00432-007-0224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He QY, Zhu R, Lei T, Ng MY, Luk JM, Sham P, et al. Toward the proteomic identification of biomarkers for the prediction of HBV related hepatocellular carcinoma. J Cell Biochem. 2008;103:740–752. doi: 10.1002/jcb.21443. [DOI] [PubMed] [Google Scholar]
- 23.Gambarin-Gelwan M, Wolf DC, Shapiro R, Schwartz ME, Min AD. Sensitivity of commonly available screening tests in detecting hepatocellular carcinoma in cirrhotic patients undergoing liver transplantation. Am J Gastroenterol. 2000;95:1535–1538. doi: 10.1111/j.1572-0241.2000.02091.x. [DOI] [PubMed] [Google Scholar]
- 24.Luk JM, Lam BY, Lee NP, Ho DW, Sham PC, Chen L, et al. Artificial neural networks and decision tree model analysis of liver cancer proteomes. Biochem Biophys Res Commun. 2007;361:68–73. doi: 10.1016/j.bbrc.2007.06.172. [DOI] [PubMed] [Google Scholar]
- 25.He M, Qin J, Zhai R, Wei X, Want Q, Rong M, et al. Detection and identification of NAP-2 as a biomarker in hepatitis B-related hepatocellular carcinoma by proteomic approach. Proteome Sci. 2008;6:10. doi: 10.1186/1477-5956-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward DG, Cheng Y, N’Kontchou G, Thar TT, Barget N, Wei W, et al. Changes in the serum proteome associated with the development of hepatocellular carcinoma in hepatitis C-related cirrhosis. Br J Cancer. 2006;94:287–292. doi: 10.1038/sj.bjc.6602923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56:1129–1135. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 28.Sun S, Lee NP, Poon RT, Fan ST, He QY, Lau GK, et al. Oncoproteomics of hepatocellular carcinoma: from cancer markers’ discovery to functional pathways. Liver Int. 2007;27:1021–1038. doi: 10.1111/j.1478-3231.2007.01533.x. [DOI] [PubMed] [Google Scholar]
- 29.Lo CY, Luk JM, Tam SC. Applicability of intraoperative parathyroid hormone assay during thyroidectomy. Ann Surg. 2002;236:564–569. doi: 10.1097/00000658-200211000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paradis V, Degos F, Dargere D, Pham N, Belghiti J, Degott C, et al. Identification of a new marker of hepatocellular carcinoma by serum protein profiling of patients with chronic liver diseases. Hepatology. 2005;41:40–47. doi: 10.1002/hep.20505. [DOI] [PubMed] [Google Scholar]
- 31.Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 32.Diamandis EP. Point: Proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clin Chem. 2003;49:1272–1275. doi: 10.1373/49.8.1272. [DOI] [PubMed] [Google Scholar]
- 33.Kanmura S, Uto H, Kusumoto K, Ishida Y, Hasuike S, Nagata K, et al. Early diagnostic potential for hepatocellular carcinoma using the SELDI ProteinChip system. Hepatology. 2007;45:948–956. doi: 10.1002/hep.21598. [DOI] [PubMed] [Google Scholar]