Abstract
Diverse and precise control is essential for eukaryotic gene expression. This is accomplished through the recruitment of a myriad of proteins to a nascent messenger RNA (mRNA) to mediate modifications, such as capping, splicing, 3′-end processing, and export. Despite being important for every cell, however, the mechanism by which the formation of diverse messenger ribonucleoprotein (mRNP) particles contributes to maintaining intricate systems in the multicellular organism remains incompletely defined. We identified and characterized a mutant gene named erecta mRNA under-expressed (emu) that leads to the defective mRNA accumulation of ERECTA, a developmental regulator in the model plant Arabidopsis thaliana. EMU encodes a protein homologous to a component of the THO complex that is required for the generation of functional mRNPs. Further analysis suggested that EMU is genetically associated with SERRATE, HYPONASTIC LEAVES1, and ARGONAUTE1, which are required for proper RNA maturation or action. Furthermore, mutations in another THO-related gene led to embryonic lethality. These findings support the presence and importance of the THO-related complex in plants as well as yeast and vertebrates.
Keywords: gene regulation, plant, THO complex
INTRODUCTION
The precise regulation of gene expression is essential throughout the life of an organism. During the transcription of primary messenger RNAs (mRNAs), concomitant processing steps take place, including 5′-capping, intron splicing, and 3′-polyadenylation, after which transport-competent mature mRNAs are exported to cytoplasmic translational machinery. A myriad of different proteins bind to the mRNA as it progresses to form mature messenger ribonucleoprotein (mRNP) particles in the nucleus, and these particles are involved in particular steps of the gene expression pathway. Individual mRNP complex components can be thought of as adaptors that allow mRNAs to interface with the numerous intracellular machineries mediating their processing, subcellular localization, translation, and decay (Hieronymus and Silver 2004; Moore 2005; Luna et al. 2008). Despite being important for every cell, however, the mechanism by which the formation of diverse mRNPs contributes to maintaining intricate systems in the multicellular organism remains incompletely defined.
MicroRNAs (miRNAs) were discovered through studies of developmental timing and morphogenesis. They are endogenous small regulatory RNAs that are loaded onto effector RNP complexes, mediating various functions at the transcriptional and/or post-transcriptional level—such as mRNA cleavage, translational repression, and regulation of chromatin structure. miRNAs show distinct regulatory functions with respect to their targets, which contain a sequence motif partially complementary to the miRNA; some miRNAs determine spatial accumulation, others play a buffering role that ensures the robustness of their expression level and pattern, and still others establish the temporal expression of target genes (Bartel 2004; Carthew and Sontheimer 2009; Voinnet 2009). Except for a few miRNAs that seem to be products of RNA polymerase III (Borchert et al. 2006), most genes encoding miRNAs are first transcribed by RNA polymerase II to produce primary miRNAs (Cai et al. 2004; Lee et al. 2004; Xie et al. 2005). Accordingly, they are subject to an intricate series of transcriptional regulation, as is the case with protein-coding mRNAs (Pawlicki and Steitz 2010). A link between mRNA and miRNA biogenesis pathways has been recently indicated in Arabidopsis, and this has been extended to RNA silencing in animals (Gregory et al. 2008; Kim et al. 2008; Laubinger et al. 2008; Gruber et al. 2009; Sabin et al. 2009; Büssing et al. 2010). Connecting these two processes permits more robust and accurate regulation of gene expression, and further inquiry into the crosstalk is expected to reveal additional roles.
Here we identified and characterized Arabidopsis genes that encode proteins homologous to the components of the THO complex that is required for generation of functional mRNPs. Our findings support the presence and importance of the THO-related complex in plants as well as yeast and vertebrates.
RESULTS
erecta mRNA under-expressed (emu) phenotypes
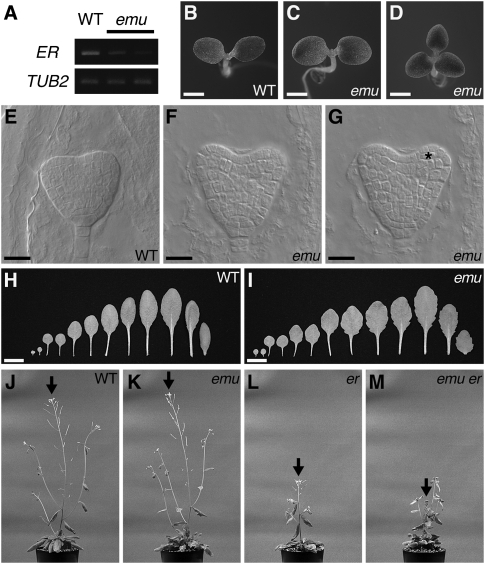
The recessive emu mutant was originally identified as a mutation that decreases the transcript accumulation of ERECTA (ER), which encodes the founding member of the ER family of receptor kinases that function as key regulators of the morphogenesis of above-ground organs in Arabidopsis (Fig. 1A; Torii et al. 1996; van Zanten et al. 2009). Apart from the effect on ER expression, the emu mutation confers pleiotropic phenotypes. For example, the number of cotyledons varies in emu (Fig. 1B–D). Whereas wild-type seedlings invariably produced two cotyledons (n = 725), 1.3% of emu seedlings had one or three cotyledons (n = 1287). During Arabidopsis embryogenesis, cotyledons grow out of the apical region of embryos at the heart stage (Barton and Poethig 1993). In wild-type embryos, two protrusions were clearly visible at the peripheral zone of the apical region, where cells in the outermost layer divided only anticlinally, developing into the future epidermis (Fig. 1E). In 2.8% of emu embryos, abnormal cell division was observed in the outermost cell layer of the apical region (n = 319) (Fig. 1F,G). Such defects in cell division were not observed in wild-type embryos and were previously described in other genetic backgrounds that alter the cotyledon number (Saiga et al. 2008; Ploense et al. 2009). Furthermore, the leaves of emu plants exhibited a more serrated margin than wild-type plants during later developmental stages (Fig. 1H,I). These phenotypes are not observed in er mutants, and emu plants did not display er phenotypes (Torii et al. 1996). On the other hand, emu er double mutants exhibited a novel phenotype, which neither emu nor er confers. In these double mutants, elongation of the primary inflorescence ceased prematurely, altering plant architecture (Fig. 1J–M). This is reminiscent of er erecta-like1 (erl1) erl2 plants, suggesting close interactions among EMU and ERL genes, which are functionally orthologous to ER (Shpak et al. 2004). These data suggest general roles for EMU in both embryonic and post-embryonic development, rather than a specific role as a regulator of ER expression. The F1 offspring from emu and wild-type plants exhibited wild-type appearances, and 23% of F2 offspring displayed emu phenotypes (n = 187), indicating that emu is a single nuclear recessive mutation.
FIGURE 1.
emu confers pleiotropic phenotypes. (A) RT-PCR analysis of ER expression in emu. RNA samples were prepared from shoots grown for 10 d after germination. The β-tubulin gene (TUB2) was amplified as a control. (B−D) Wild-type (WT) (B) and emu (C,D) seedlings. Plants were grown for 3 d after germination. (E−G) Wild-type (E) and emu (F,G) embryos. The same emu embryo was visualized at different foci to show three cotyledons (F,G). An asterisk indicates abnormal cell division. (H,I) Leaf series of wild-type (H) and emu (I) plants. For each genotype, the left two leaves are cotyledons, and the right-most leaf is the first cauline leaf. Plants were grown for 26 d after germination. (J−M) Wild-type (J), emu (K), er-105 (L), and emu er-105 (M) plants grown for 46 d after germination. emu er-105 double mutants exhibited a novel phenotype, which neither emu nor er confers. Arrows indicate the primary inflorescences. Photographs were taken at the same magnification. Scale bars: 1 mm in B−D; 20 μm in E−G; 1 cm in H,I.
EMU encodes a putative THO complex component, Hpr1
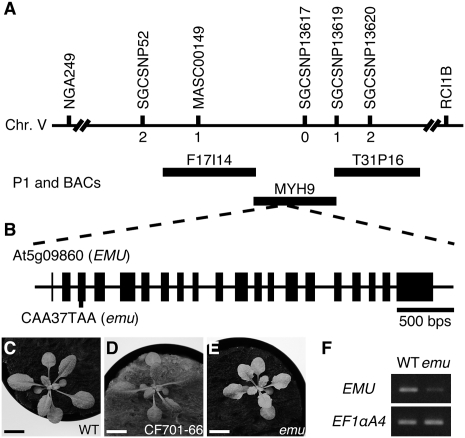
To understand the molecular basis of emu phenotypes, the EMU gene was isolated using a map-based strategy. Sequence analysis revealed a single base substitution at the At5g09860 locus, and constitutive expression of the At5g09860 cDNA in emu restored the wild-type appearance (Fig. 2). Thus, we concluded that the identified mutation accounts for the emu phenotype. This C-to-T substitution in the third exon produces a stop codon at position 37, and the resulting emu product lacks a large portion of the predicted protein. The emu mutation reduced the accumulation of the EMU transcript, which is presumably induced by nonsense-mediated decay (Fig. 2F).
FIGURE 2.
Map-based cloning of EMU. (A) Genetic mapping of EMU. The markers used for mapping are shown as well as the number of recombinants between emu and these markers. (B) Gene structure of EMU/At5g09860, with exons represented as closed boxes. The site of the lesion and nucleotide change in the emu allele is indicated. (C−E) Complementation of emu. Constitutive expression of wild-type At5g09860 cDNA in the emu mutant restored the wild-type appearance (D). Plants were grown for 17 d after germination. (F) EMU expression was analyzed by RT-PCR. Total RNA was extracted from seedlings grown for 7 d after germination. EF1αA4 was amplified as a control. Scale bars: 1 cm.
BLAST searches of nonredundant databases within GenBank revealed that the predicted EMU product is similar in amino acid sequence to the Hpr1/p84/pThoc1 protein in Drosophila melanogaster, Homo sapiens, and Mus musculus (Supplemental Fig. S1; Durfee et al. 1994; Rehwinkel et al. 2004; Wang et al. 2006). Although the similarity in primary sequence is low, the predicted EMU is also similar to the yeast Hpr1 protein, the founding member of these evolutionally conserved proteins, and stretches of conserved regions are recognizable (Supplemental Fig. S2; Aguilera and Klein 1990). Similar proteins are encoded in the genomes of various plant species, suggesting their functional importance in plants (Supplemental Figs. S1, S3).
THO complex components are conserved in Arabidopsis
The yeast Hpr1 forms a multimeric protein complex named the THO complex with the proteins Tho2, Mft1, and Thp2 (Chávez et al. 2000). The human and fruit fly THO complexes were recently characterized, and both were shown to contain homologs of yeast Hpr1 and Tho2 (Sträßer et al. 2002; Rehwinkel et al. 2004; Masuda et al. 2005). However, whether a similar complex functions in plants has not yet been established. Using BLAST searches, we identified Arabidopsis genes homologous to the fruit fly THO genes—THO2 and THOC6—named AtTHO2 (At1g24706) and AtTHOC6 (At2g19430), respectively (Rehwinkel et al. 2004). In the yeast, human, and fruit fly genomes, a single gene encodes each component of the THO complex. However, in the Arabidopsis genome, there are two genes homologous to the fruit fly THOC5 and THOC7, respectively, and these genes will be referred to as AtTHOC5A (At5g42920), AtTHOC5B (At1g45233), AtTHOC7A (At3g02950), and AtTHOC7B (At5g16790). No obvious homologs of the yeast MFT1 and THP2 genes were identified in the Arabidopsis genome. Next, we analyzed expression patterns of these genes using publicly available microarray data. No probe set is available for AtTHOC5B; however, expression data for other genes were obtained. EMU, AtTHO2, AtTHOC5A, AtTHOC6, AtTHOC7A, and AtTHOC7B are expressed throughout the plant in a similar manner (Supplemental Fig. S4). Interestingly, the expression of AtTHOC7B is highly specific to seed. This may suggest a functional divergence between AtTHOC7A and AtTHOC7B.
AtTHO2 is required for embryo development
To investigate the biological roles of the Arabidopsis homologs of the THO components, we searched for T-DNA insertion mutants of AtTHO2 and AtTHOC6, both of which were predicted to be present as a single-copy gene in the genome. Four and two T-DNA insertion mutants were identified for AtTHO2 and AtTHOC6, respectively (Supplemental Fig. S5A,B). Under our growth conditions, the atthoc6 mutants did not display discernible morphological phenotypes. Given that this gene is absent in yeast, the lack of a mutant phenotype may imply that AtTHOC6 is not an essential subunit of the putative Arabidopsis THO complex. Alternatively, AtTHOC6 may exhibit another function in Arabidopsis. Lee et al. (2008) suggested that a subset of WD40 proteins that contain a damaged DNA binding 1 (DDB1)-binding WD40 (DWD) motif, including AtTHOC6, may act as a substrate receptor for E3 ubiquitin ligase.
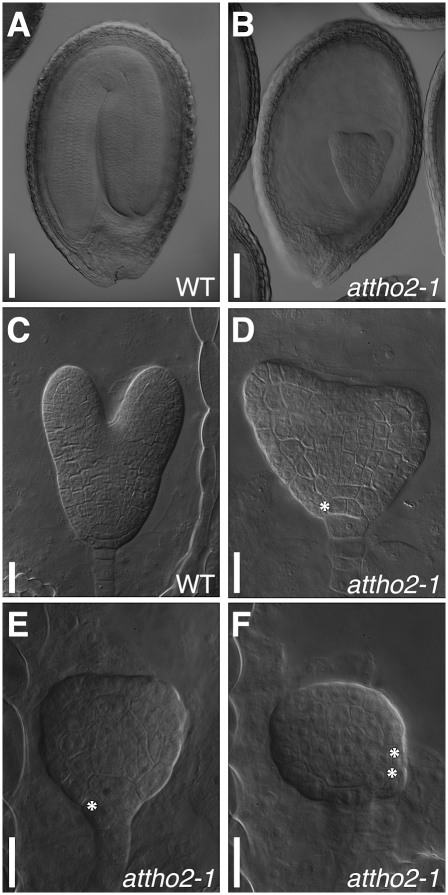
Attempts to obtain seedlings homozygous for T-DNA insertions in AtTHO2 by PCR-based genotyping did not identify homozygous plants, and pale-green ovules were segregated in fruits produced from plants heterozygous for the T-DNA insertion (Supplemental Fig. S5C,D). Therefore, it is likely that attho2 mutants result in lethality during embryo development. To address this possibility, we examined mature embryos produced from plants heterozygous for the T-DNA insertion. Approximately 25% of the embryos exhibited abnormal morphology (Fig. 3A,B). Consistent with our observation, AtTHO2 is listed in the “pending additions” page in the SeedGenes database at www.seedgenes.org, of which its goal is to establish a catalog of essential genes in Arabidopsis, and is called EMB2793 (Meinke et al. 2008). Abnormal embryos can be recognized from as early as the heart stage by their retarded development and irregular divisions compared with the corresponding wild-type embryo (Fig. 3C–F). These results indicate that AtTHO2 is indispensable for embryogenesis, suggesting a fundamental role for this gene in Arabidopsis development.
FIGURE 3.
attho2 mutations lead to embryonic lethality. (A,B) Mature embryos extracted from a plant heterozygous for the attho2-1 mutant allele, exhibiting either the wild-type (A) or mutant (B) phenotype. (C) A wild-type late-heart stage embryo. (D) An embryo segregating among wild-type late-heart stage embryos. (E,F) Embryos segregating among wild-type heart stage embryos. Asterisks denote aberrant cell divisions. Scale bars: 100 μm in A,B; 20 μm in C−F.
Splicing defects in emu
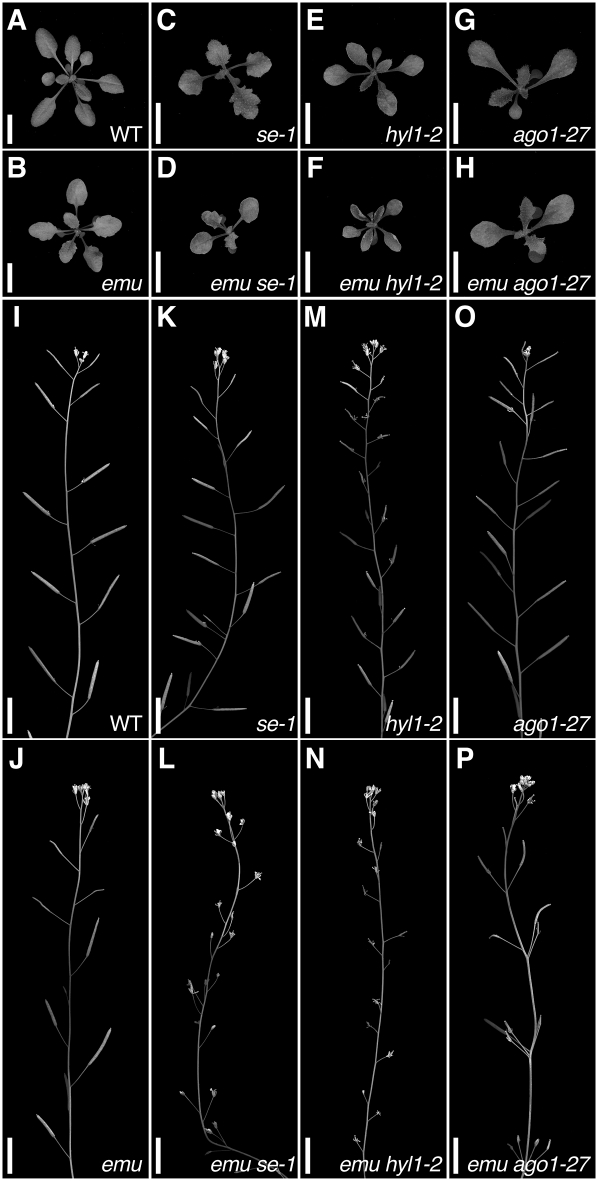
Similar to emu, a weak loss-of-function allele of SERRATE (SE), se-1, causes a serrated-leaf phenotype and alters the cotyledon number (Clarke et al. 1999; Prigge and Wagner 2001). Moreover, a null se allele results in embryonic lethality, which is reminiscent of mutations in AtTHO2 (Lobbes et al. 2006). Therefore, we generated emu se-1 double-mutant plants to examine the interaction between these two genes. The leaves of emu se-1 plants exhibited an upward-curling phenotype as seen in stronger se-2 and se-3 alleles (Fig. 4A–D; Grigg et al. 2005). In addition, the double mutant was sterile, although emu and se-1 alone have little impact on seed production (Fig. 4I–L). These results suggest a strong genetic interaction between emu and se-1.
FIGURE 4.
Double mutants between emu and miRNA mutants exhibit novel phenotypes. (A−H) Plants grown for 21 d after germination. (I−P) Primary inflorescences of plants grown for 51 d after germination. (A,I) Wild type; (B,J) emu; (C,K) se-1; (D,L) emu se-1; (E,M) hyl1-2; (F,N) emu hyl1-2; (G,O) ago1-27; and (H,P) emu ago1-27. Scale bars: 1 cm.
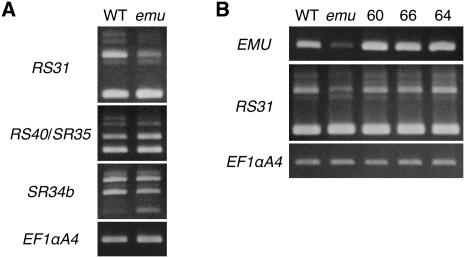
This strong genetic interaction raised the possibility that EMU and SE may have similar functions. SE encodes a zinc finger protein that has been implicated in mRNA splicing (Prigge and Wagner 2001; Laubinger et al. 2008). Interestingly, some functions of the THO complex are dependent on splicing, and studies in animals have indicated that the THO complex interacts with the spliceosome (Rappsilber et al. 2002; Zhou et al. 2002; Masuda et al. 2005; Cheng et al. 2006). To determine whether EMU as well as SE are involved in mRNA splicing, we characterized the splicing patterns in emu mutant plants by examining transcripts of genes encoding serine/arginine-rich (SR) proteins, which belong to a conserved family of splicing regulators in eukaryotes and are extensively alternatively spliced (Reddy 2007; Long and Caceres 2009). The splicing patterns of SR genes were analyzed by RT-PCR using primers described previously (Palusa et al. 2007). As a result, the alternative splicing patterns of RS31, RS40/SR35, and SR34b were found to be altered in emu mutant plants, and the expression of EMU cDNA restored the wild-type alternative splicing pattern (Fig. 5). This suggests that EMU is involved at least in the regulation of alternative splicing of mRNA.
FIGURE 5.
Alternative splicing patterns of SR genes are altered in emu. RT-PCR analysis of the expression of SR genes in seedlings grown for 7 d after germination. (A) Alternative splicing patterns of some SR genes are altered in the emu mutant. (B) Expression of EMU cDNA restored the wild-type expression pattern of RS31. Either CF701-60 (60), CF701-66 (66), or CF701-64 (64) contained a single copy of the 35S∷EMU transgene as a homozygote and was used for analysis.
Genetic interactions between emu and miRNA mutants
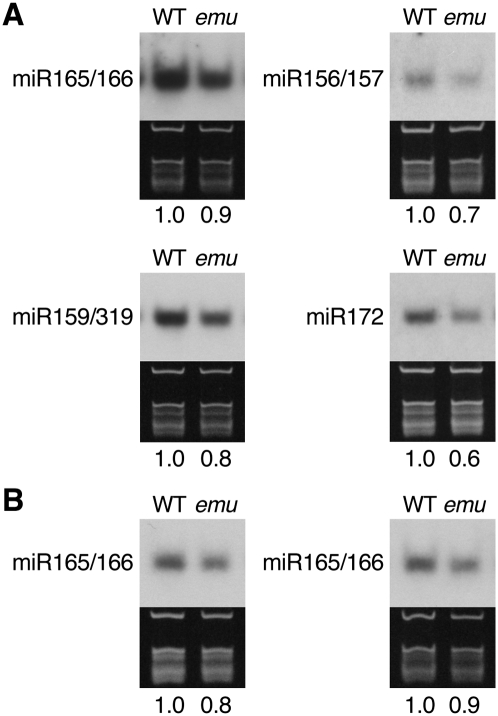
It was demonstrated that SE also functions in miRNA processing through interacting with HYL1, which acts solely in miRNA biogenesis (Lu and Fedoroff 2000; Han et al. 2004; Vazquez et al. 2004; Grigg et al. 2005; Lobbes et al. 2006; Yang et al. 2006; Fang and Spector 2007; Dong et al. 2008; Laubinger et al. 2008). Although there is no evidence that the THO complex plays a direct role in miRNA processing in metazoa, the observed genetic interaction between emu and se-1 prompted us to further examine a potential role for EMU in miRNA biogenesis. Therefore, we characterized emu hyl1-2 double-mutant plants. emu hyl1-2 plants resembled emu se-1 plants, conferring upward-curling leaves and sterility (Fig. 4E,F,M,N). Similarly, the emu mutation enhanced the phenotypes conferred by mutations in other genes involved in miRNA biogenesis, HUA ENHANCER 1 and HASTY (data not shown; Chen et al. 2002; Bollman et al. 2003; Park et al. 2005; Yu et al. 2005). To directly assess the potential role for EMU in miRNA biogenesis, miRNA levels were examined. In emu plants, various miRNAs accumulated less than in wild-type plants (Fig. 6). Considering that the mutant phenotype conferred by the emu mutation is much weaker than the embryonic lethal phenotype seen in the attho2 mutants, these mild reductions might be due to a modest effect of emu on the putative Arabidopsis THO complex. To elucidate whether the slight decrease of miRNA accumulation impacts on miRNA-mediated gene regulation in emu plants, we generated double-mutant plants between emu and plants containing a hypomorphic allele of ARGONAUTE1 (AGO1), ago1-27, because miRNAs interact with AGO1 proteins to regulate gene expression (for review, see Vaucheret 2008). Interestingly, the emu mutation enhanced the slightly altered phyllotaxy of ago1-27 inflorescences, wherein the internode length is more random than that in wild-type plants, resulting in multiple fruits emanating from the same node (Fig. 4O,P; Morel et al. 2002; Smith et al. 2009). Taken together, these data suggest that EMU may be involved in assuring proper miRNA action.
FIGURE 6.
miRNA accumulation is reduced in emu. (A) RNA samples were prepared from inflorescence apices. (B) RNA samples were prepared from rosette leaves (left) and inflorescences (right) from plants grown for 27 d after germination. Ethidium bromide-stained gels on which rRNA and tRNA bands are visible are shown as a loading control. The intensity of the hybridization signal relative to WT is indicated and was calculated after normalization to rRNA.
DISCUSSION
We identified Arabidopsis genes encoding proteins homologous to the THO complex components. Although further studies are required to validate the presence of the THO complex in Arabidopsis, this study supports the existence of the THO complex in plants as well as in yeast and metazoa and underscores the importance of THO-related proteins in the context of plant development.
The discovery that EMU encodes an Arabidopsis homolog of the yeast Hpr1 is particularly important because the in vivo functions of the THO components in multicellular organisms remain elusive. Recently, a role for mouse Thoc1/Hpr1/p84 during embryogenesis was revealed using mice containing the mutant alleles (Wang et al. 2006, 2007), and researchers have now started to analyze THO components in a physiological context in multicellular organisms. In the present study we showed that mutations in EMU and AtTHO2 compromise Arabidopsis development. Particularly, attho2 mutations lead to embryonic lethality, highlighting the essential role of this factor in Arabidopsis development.
Our genetic and molecular analyses shed some light on the putative role of EMU. As expected from the functions of its counterparts in yeast and animals, EMU appears to be required to guarantee accuracy and proper levels of RNA biosynthesis. The observed genetic interactions between emu and se-1 may provide a clue to the mechanism of action of EMU. Interestingly, mutations in the Arabidopsis genes that encode the two cap-binding complex (CBC) subunits—CAP-BINDING PROTEIN20 (CBP20) and ABSCISIC ACID HYPERSESITIVE1 (ABH1)/CBP80—cause a serrated leaf phenotype and mRNA splicing defects as observed in emu and se-1 mutants (Hugouvieux et al. 2001; Papp et al. 2004; Kuhn et al. 2007; Laubinger et al. 2008; Raczynska et al. 2010). It is also noteworthy that the abh1-8 mutation enhanced the phyllotaxy defect of ago1-38 mutant inflorescences as seen in emu ago1-27 plants (Gregory et al. 2008). Furthermore, emu was sensitive to the exogenous application of abscisic acid (Supplemental Fig. S6), as are cbp and se-1 (Hugouvieux et al. 2001; Bezerra et al. 2004; Papp et al. 2004). These observations raise the possibility that EMU is involved in connecting SE and CBC, which are involved in miRNA biogenesis but do not appear to interact directly with each other (Gregory et al. 2008; Kim et al. 2008; Laubinger et al. 2008). Although we were unable to detect the two hybrid interactions among ABH1, EMU, SE, and AtTHO2 observed in yeast (C. Furumizu and Y. Komeda, unpubl.), this hypothesis remains to be tested in planta. In addition, our speculation is supported by the finding that the human transcription/export (TREX) complex, which is comprised of the core THO complex, is recruited to a region near the 5′ end of the mRNA and that TREX recruitment requires the 5′ cap (Sträßer et al. 2002; Cheng et al. 2006).
The TREX complex has a conserved role in coupling transcription to mRNA export in yeast and metazoa and consists of two export factors, Yra1/ALY/REF and Sub2/UAP56, in conjunction with the THO complex (Sträßer et al. 2002; Masuda et al. 2005). The Arabidopsis genome contains at least one (At5g59950) and two (At5g11170 and At5g11200) genes, which are homologous to Yra1/ALY/REF and Sub2/UAP56, respectively. This suggests the possibility that a complex similar to the TREX complex could exist in Arabidopsis as well. In addition to utilizing the genetic resources provided by this and previous studies, future experimental validation and biological characterization of these predicted complexes will allow us to further explore the vast world of RNA and may contribute to a better understanding of the molecular mechanisms, whereby the intricate systems are built up in plants.
MATERIALS AND METHODS
Plant materials and growth conditions
All plants were in the Columbia (Col) background, and Col was used as the wild type. The seeds of ago1-27 were kindly provided by Hervé Vaucheret. The attho2-1 (SALK_072011), attho2-2 (SALK_130342), attho2-3 (CS848337), attho2-4 (SALK_129441), atthoc6-1 (SALK_051022C), atthoc6-2 (SALK_004825), hyl1-2 (SALK_064863), and se-1 (CS3257) seeds were obtained from the Arabidopsis Biological Resource Center (ABRC) (Alonso et al. 2003). Plants were grown on rock-wool bricks supplemented with vermiculite under cool white fluorescent light in long day conditions (16 h light/8 h dark) at an average temperature of 22°C. For RNA extraction from young seedlings, seeds were germinated on nutrient agar plates containing half-strength Murashige and Skoog Plant Salt Mixture (Wako Pure Chemical Industries), 1% sucrose, and 0.8% Bacto Agar (Difco). The pH of the medium was adjusted to 5.7 with KOH.
Mutant isolation
A genetic screen was conducted as previously described (Furumizu and Komeda 2008). The emu mutant was backcrossed six times to wild-type Col, and the individual plants that showed the mutant phenotypes in the absence of the ERECTA∷GUS transgene (Yokoyama et al. 1998) were chosen for further analyses.
Map-based cloning
The emu mutant was crossed to Ler, and F2 seedlings with the emu phenotype were examined for recombination between the mutation and PCR-based polymorphic markers. The emu mutation was initially mapped to chromosome 5 between markers NGA249 and RCI1B. Further analysis of 678 emu-like F2 plants allowed us to locate EMU to a region of 138 kb located between markers MASC00149 and SGCSNP13619. Sequencing of this interval identified a C-to-T substitution in the third exon of At5g09860. The entire coding region of At5g09860 in emu, in the parental line harboring the ERECTA∷GUS transgene, in Col, and in Ler, was sequenced to confirm that this C-to-T substitution occurred only in the emu mutant. The initial mapping used cleaved-amplified polymorphic sequence (CAPS) and simple sequence length polymorphism (SSLP) markers from The Arabidopsis Information Resource (TAIR, www.arabidopsis.org). The derived cleaved amplified polymorphic sequence (dCAPS) markers used for fine mapping of the emu mutation were generated based on information available from TAIR (Neff et al. 1998). Details regarding primer sequences and polymorphisms are available upon request.
Complementation of emu
The EMU cDNA was amplified by PCR from the Col cDNA template using primers 5g09860-FW101 (5′-CCCGGGAAATTGTTCTTCCTCCACTC-3′) and 5g09860-RV101 (5′-CCCGGGTTACGCAAAAGAACTTAATTG-3′), and was subcloned into pGEM-T (Promega) to yield pCF700. pCF700 was sequenced to exclude amplification errors. The EMU cDNA was excised from pCF700 by digestion with SmaI and cloned into the SmaI site of pCF196 (Furumizu and Komeda 2008) to yield pCF701. This complementation construct was introduced into the Agrobacterium tumefaciens strain C58C1 by electroporation and was used to transform emu plants by the floral dip method using the surfactant Silwet L-77 (Clough and Bent 1998). Transformants were selected on nutrient agar plates containing 50 μg/mL kanamycin.
PCR-based genotyping
PCR-based genotyping markers were designed for emu, attho2, and atthoc6 alleles. Two markers were designed for the emu allele. In the absence of the complementation construct, the emu allele was identified by a CAPS marker. For this, a 0.9-kb genomic fragment was amplified using the primer pair 5g09860-FW (5′-CCGGCGACTCATAGTCCGTG-3′) and 5g09860-RV3 (5′-CGGTCAAATCCTCCAGCAG-3′). After digestion with AflII, the emu allele produces two fragments (0.5 and 0.4 kb), while the wild-type allele is resistant to AflII digestion. In the presence of the complementation construct, the emu allele was identified by a dCAPS marker. In the wild type, the primer pair 12D1M-F2 (5′-TTGACAGAAACAAACAAAGTTGGAT-3′) and 5g09860-RV4 (5′-AAGTATATACGCATACCACG-3′) produces the full-length product of 0.13 kb, which becomes 0.11 kb after digestion with MboI. The emu allele is resistant to MboI. For the T-DNA insertion mutants, the presence of T-DNA insertions was confirmed by PCR using the primer pairs LB-FW (5′-GGCGTTACCCAACTTAATCGCC-3′) and 1g24706-RV (5′-TCCGCTTTGTGCCAGTCTC-3′) for attho2-1; 1g24706-FW (5′-TCTTAATCCAGTGGCACACG-3′) and RB-RV (5′-AAACGATCCAGATCCGGTGC-3′) for attho2-2; 1g24706-FW2 (5′-GTCACTGAACAGTTCGACAG-3′) and pCSA110LB-RV (5′-TACAGCAAGAACGGAATGCG-3′) for attho2-3; 1g24706-FW2 and LB-FW for attho2-4; and THOC6-FW (5′-GCAAGCTAGTAATTTGATCG-3′) and LB-FW for atthoc6-1 and atthoc6-2. The respective wild-type alleles were genotyped by PCR using the primer pairs 1g24706-FW and 1g24706-RV for attho2-1 and attho2-2; 1g24706-FW2 and 1g24706-RV2 (5′-TACAACGCTGCAGGAACTCC-3′) for attho2-3; 1g24706-FW2 and 1g24706-RV4 (5′-CGTCTGTAACCCAATGTGG-3′) for attho2-4; and THOC6-FW and THOC6-RV (5′-GCGCAAGAATCACCAGCTG-3′) for atthoc6-1 and atthoc6-2. The se-1 allele was detected by a PCR-based genotyping marker generated based on the information previously described (Prigge and Wagner 2001). PCR using the primer pair SE-FW3 (5′-AGGTAGCGGGAGAGGTGGAC-3′) and SE-RV3 (5′-ACGTTACCTGCGCAGCCGTC-3′) produces PCR products of 82 and 75 base pairs for the wild-type and se-1 alleles, respectively. PCR amplification was performed under standard conditions.
Microscopy
For examination of embryo phenotypes, tissues were cleared as previously described (Yadegari et al. 1994). Samples were visualized using Nomarsky differential interference contrast optics on a Leica DMRX E microscope (Leica Microsystems).
RNA analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions.
For RT-PCR analysis, total RNA was treated with RNase-free DNase I (Takara Bio). First-strand cDNA was synthesized from 2 μg of RNA with an oligo dT-adaptor primer using AMV Reverse Transcriptase XL (Takara Bio) according to the manufacturer's instructions. One-fortieth of the reverse transcriptase reaction was subjected to PCR amplification. The gene-specific primers used were 5g09860-FW4 (5′-AGCATATACTGGAGCGCG-3′) and 5g09860-RV2 (5′-AGCTAGCAGATTTGCACTC-3′) for EMU. Primers for SR genes, EF1αA4, and TUB2 were previously described (Lee et al. 2000; Nesi et al. 2000; Palusa et al. 2007). The PCR program started at 94°C for 2 min, followed by the optimum number of cycles of 30 sec at 94°C, 30 sec at 56°C, and 1 min at 68°C. PCR products were subjected to agarose gel electrophoresis and were visualized by staining with ethidium bromide. RT-PCRs were performed three times on two independent RNA batches, and the results were comparable in all experiments.
For miRNA gel-blot analysis, total RNA (5 μg) was loaded onto a warmed 15% polyacrylamide, 7 M urea gel and run at 100 V in 0.5X TBE buffer for 2.5–3 h. The gel was divided in two; the upper gel was stained with ethidium bromide to show 5S RNA and tRNAs, and the lower gel was used for RNA blot. Small RNAs were electroblotted to a positively charged Hybond-N+ membrane (Amersham Biosciences) at constant current (4 mA/cm2) for 45 min in 0.5X TBE buffer. After transfer, the membrane was rinsed in 20X SSC, the RNA was crosslinked to the membane with 1200 × 100 μJ of energy using a Stratalinker UV crosslinker (Stratagene), and the membrane was baked at 80°C for 30 min. The membrane was hybridized with DNA oligonucleotide probes specific for miR156 (5′-GTGCTCACTCTCTTCTGTCA-3′), miR159 (5′-TAGAGCTCCCTTCAATCCAAA-3′), miR165 (5′-GGGGGATGAAGCCTGGTCCGA-3′), miR166 (5′-GGGGAATGAAGCCTGGTCCGA-3′), and miR172 (5′-ATGCAGCATCATCAAGATTCT-3′). The probe was end-labeled with [γ-32P]-ATP using T4 Polynucleotide Kinase (Takara Bio). Unincorporated nucleotides were removed using G-25 spin columns (Amersham Biosciences) as per the manufacturer's instructions. Membranes were prehybridized for at least 3 h and hybridized overnight at 42°C using ULTRAhyb-Oligo Hybridization Buffer (Ambion). Membranes were washed four times for 30 min each with 2X SSC, 0.5% SDS at 42°C. The membranes were visualized using exposure to X-ray films for several days at −80°C with an intensifying screen. The intensities of the bands were quantified from the original figures using the ImageJ 1.43 program (http://rsb.info.nih.gov/ij/), and normalized miRNA/rRNA values are represented. RNA gel-blot analyses were performed at least twice on at least two independent RNA batches, and the results were comparable in all experiments.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to Hervé Vaucheret and ABRC for the mutant seeds. We thank John Bowman, Naoto Yabe, Mitsutomo Abe, Shunsuke Saiga, Nobutoshi Yamaguchi, and members of our laboratories for valuable discussions. Yuichiro Watanabe and Yuko Tagami gave us detailed protocols and helpful advice for miRNA analysis. This work was supported by Grants-in-Aid (to H.K. and Y.K.) and a Research Fellowship for Young Scientists (to C.F.) from the Japan Society for the Promotion of Science.
NOTE ADDED IN PROOF
Recently, Yelina et al. (2010) published a similar study on the putative Arabidopsis THO/TREX complex.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2265710.
REFERENCES
- Aguilera A, Klein HL 1990. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol 10: 1439–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Barton MK, Poethig RS 1993. Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119: 823–831 [Google Scholar]
- Bezerra IC, Michaels SD, Schomburg FM, Amasino RM 2004. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J 40: 112–119 [DOI] [PubMed] [Google Scholar]
- Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS 2003. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL 2006. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 13: 1097–1101 [DOI] [PubMed] [Google Scholar]
- Büssing I, Yang JS, Lai EC, Großhans H 2010. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J 29: 1830–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A 2000. A protein complex containing Tho2, Hpr1, Mft1, and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D 2002. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Clarke JH, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M 1999. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J 20: 493–501 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N 2008. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Mancini MA, Jones D, Elledge SJ, Lee WH 1994. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J Cell Biol 127: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL 2007. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumizu C, Komeda Y 2008. A novel mutation in KNOPF uncovers the role of α-glucosidase I during post-embryonic development in Arabidopsis thaliana. FEBS Lett 582: 2237–2241 [DOI] [PubMed] [Google Scholar]
- Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR 2008. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M 2005. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, et al. 2009. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Goud S, Song L, Fedoroff N 2004. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Silver PA 2004. A systems view of mRNP biology. Genes Dev 18: 2845–2860 [DOI] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI 2001. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477–487 [DOI] [PubMed] [Google Scholar]
- Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH 2008. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary microRNAs. Plant Cell Physiol 49: 1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Breton G, Schroeder JI 2007. mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J 50: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D 2008. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW 2008. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J 2006. SERRATE: A new player on the plant microRNA scene. EMBO Rep 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF 2009. The SR protein family of splicing factors: Master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N 2000. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R, Gaillard H, González-Aguilera C, Aguilera A 2008. Biogenesis of mRNPs: Integrating different processes in the eukaryotic nucleus. Chromosoma 117: 319–331 [DOI] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A 2008. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci 13: 483–491 [DOI] [PubMed] [Google Scholar]
- Moore MJ 2005. From birth to death: The complex lives of eukaryotic mRNAs. Science 309: 1514–1518 [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H 2002. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L 2000. The TT8 gene encodes a basic helix–loop–helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palusa SG, Ali GS, Reddy ASN 2007. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J 49: 1091–1107 [DOI] [PubMed] [Google Scholar]
- Papp I, Mur LA, Dalmadi Á, Dulai S, Koncz C 2004. A mutation in the Cap Binding Protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol Biol 55: 679–686 [DOI] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS 2005. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA 2010. Nuclear networking fashions pre-messenger RNA and primary microRNA transcripts for function. Trends Cell Biol 20: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW 2009. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136: 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Wagner DR 2001. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13: 1263–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska KD, Simpson CG, Ciesiolka A, Szewc L, Lewandowska D, McNicol J, Szweykowska-Kulinska Z, Brown JWS, Jarmolowski A 2010. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res 38: 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN 2007. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol 58: 267–294 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Köcher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 6: 558–566 [DOI] [PubMed] [Google Scholar]
- Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S 2009. Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138: 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga S, Furumizu C, Yokoyama R, Kurata T, Sato S, Kato T, Tabata S, Suzuki M, Komeda Y 2008. The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development 135: 1751–1759 [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU 2004. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131: 1491–1501 [DOI] [PubMed] [Google Scholar]
- Smith MR, Willmann MR, Wu G, Berardini TZ, Möller B, Weijers D, Poethig RS 2009. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci 106: 5424–5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y 1996. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Snoek LB, Proveniers MCG, Peeters AJM 2009. The many functions of ERECTA. Trends Plant Sci 14: 214–218 [DOI] [PubMed] [Google Scholar]
- Vaucheret H 2008. Plant ARGONAUTES. Trends Plant Sci 13: 350–358 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crété P, Vaucheret H 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Voinnet O 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136: 669–687 [DOI] [PubMed] [Google Scholar]
- Wang X, Chang Y, Li Y, Zhang X, Goodrich DW 2006. Thoc1/Hpr1/p84 is essential for early embryonic development in the mouse. Mol Cell Biol 26: 4362–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Y, Zhang X, Goodrich DW 2007. An allelic series for studying the mouse Thoc1 gene. Genesis 44: 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC 2005. Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, de Paiva GR, Laux T, Koltunow AM, Apuya N, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB 1994. Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell 6: 1713–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu Z, Lu F, Dong A, Huang H 2006. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yelina NE, Smith LM, Jones AM, Patel K, Kelly KA, Baulcombe DC 2010. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci (in press). doi: 10.1073/pnas.0911341107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Kato A, Torii KU, Komeda Y 1998. The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J 15: 301–310 [DOI] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X 2005. Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]