Abstract
In addition to their well-documented roles in the promotion of nonsense-mediated mRNA decay (NMD), yeast Upf proteins (Upf1, Upf2/Nmd2, and Upf3) also manifest translational regulatory functions, at least in vitro, including roles in premature translation termination and subsequent reinitiation. Here, we find that all upfΔ strains also fail to reinitiate translation after encountering a premature termination codon (PTC) in vivo, a result that led us to seek a unifying mechanism for all of these translation phenomena. Comparisons of the in vitro translational activities of wild-type (WT) and upf1Δ extracts were utilized to test for a Upf1 role in post-termination ribosome reutilization. Relative to WT extracts, non-nucleased extracts lacking Upf1 had approximately twofold decreased activity for the translation of synthetic CAN1/LUC mRNA, a defect paralleled by fewer ribosomes per mRNA and reduced efficiency of the 60S joining step at initiation. These deficiencies could be complemented by purified FLAG-Upf1, or 60S subunits, and appeared to reflect diminished cycling of ribosomes from endogenous PTC-containing mRNAs to exogenously added synthetic mRNA in the same extracts. This hypothesis was tested, and supported, by experiments in which nucleased WT or upf1Δ extracts were first challenged with high concentrations of synthetic mRNAs that were templates for either normal or premature translation termination and then assayed for their capacity to translate a normal mRNA. Our results indicate that Upf1 plays a key role in a mechanism coupling termination and ribosome release at a PTC to subsequent ribosome reutilization for another round of translation initiation.
Keywords: yeast, NMD, translation initiation, translation termination, ribosome reutilization
INTRODUCTION
The fidelity of protein synthesis is dependent on a functional translational apparatus and on the integrity of mRNA coding sequences. A premature termination codon (PTC) can disrupt mRNA integrity, generating an abbreviated open reading frame (ORF) that can give rise to a potentially toxic polypeptide fragment (Pulak and Anderson 1993). To obviate the accumulation of such polypeptide fragments, the offending mRNAs are recognized and degraded by the nonsense-mediated mRNA decay (NMD) pathway (Jacobson and Izaurralde 2007; Isken and Maquat 2008). Quality control by NMD requires the activities of the highly conserved UPF1, UPF2/NMD2, and UPF3 gene products in all eukaryotic cells, as well as those of the SMG1-9 genes in metazoans (Jacobson and Izaurralde 2007; Isken and Maquat 2008; Yamashita et al. 2009). Of the three Upf proteins, Upf1 appears to serve as the central regulator of the NMD pathway, with Upf2/Nmd2 and Upf3 regulating Upf1 function (Page et al. 1999; Maderazo et al. 2000).
While a requirement for the Upf proteins in promoting the rapid decay of nonsense mRNAs is well established, other functions for these proteins, and the possible connections between these functions and the promotion of mRNA decay, are only beginning to be understood. Notably, several experimental approaches have indicated that the Upfs have additional activities in translation. A possible role for these proteins in translation reinitiation emerged from analyses of premature termination in yeast cell-free extracts (Amrani et al. 2004). These studies demonstrated that a significant fraction of ribosomes terminating translation at PTCs in vitro reinitiated at proximal AUG codons, and that reinitiation was Upf1-dependent even though NMD was inactive in the cell-free lysates. Although these data suggest a positive role for the Upfs in regulating translation reinitiation, other data imply a translational repression function. Studies in yeast using CUP1 nonsense reporter constructs (Muhlrad and Parker 1994, 1999; Sheth and Parker 2006) and in mammalian cells using dual luciferase reporters (Isken et al. 2008) suggest that Upf1 association with a nonsense-containing mRNA represses its translation and that this event may be an obligatory precursor to subsequent mRNA decay. Additional experiments in yeast indicate that the Upf factors may also affect termination fidelity since nonsense codon read-through is enhanced when any of the three factors is inactive or absent (Weng et al. 1996a,b, 1998; Maderazo et al. 2000; Wang et al. 2001). Although the latter observation has been made with numerous nonsense alleles and with numerous Upf-deficient strains (Weng et al. 1996a,b, 1998; Maderazo et al. 2000; Wang et al. 2001), recent experiments demonstrate that the apparent loss of termination fidelity is an indirect consequence of the stabilization of the NMD-sensitive ALR1 mRNA and the resulting increase of intracellular Mg2+ levels (Johansson and Jacobson 2010). Finally, there is also evidence that Upf regulation of translation may not be limited to nonsense-containing mRNAs. The same study that implicated a Upf1 role in translation reinitiation in vitro also showed that the absence of Upf1 leads to decreased translation of a wild-type (WT) CAN1/LUC fusion mRNA (Amrani et al. 2004). Moreover, Upf1 appears to be required for optimal translation of some normal yeast transcripts in vivo (Muhlrad and Parker 1999), and a hyperphosphorylated mutant of Upf1 that is impaired for NMD leads to decreased translation of a wild-type LUC reporter mRNA in rabbit reticulocyte lysates (Isken et al. 2008).
Collectively, the effects of Upf deficiency on the translation of normal and PTC-containing mRNAs are indicative of an important but poorly understood function of the Upf proteins. Accordingly, we have explored further the role of the Upf factors in translation, both in vivo and in vitro. To do so, we first used chimeric PGK1/LUC nonsense alleles to test whether the Upf proteins played a role in post-termination reinitiation activity in intact cells. Having obtained experimental support for such a role, we then exploited the yeast cell-free translation system to elucidate the translational defects arising from NMD inactivation. These analyses revealed that extracts prepared from upf1Δ cells manifested a Upf1-complementable deficiency in apparent initiation activity. Additional experimentation strongly suggested that Upf1 is required in vitro for efficient termination and release of ribosomes at a PTC and their subsequent reutilization for additional rounds of translation.
RESULTS
Mutations in a PGK1/LUC reporter attenuate its reinitiation activity
We have previously shown that, in vitro, yeast ribosomes will reinitiate translation subsequent to premature termination, provided that Upf1 is present in the translation reaction (Amrani et al. 2004). This observation indicated that premature termination was inefficient and suggested that Upf1 might influence the extent to which prematurely terminating ribosomes remain associated with an mRNA (Amrani et al. 2004). Here, we used three approaches to understand the significance of Upf1-dependent translational reinitiation in vitro. First, we tested for reinitiation in vivo and evaluated its dependence on NMD factors. We then used a cell-free translation system to identify the steps in translation initiation that are affected in the absence of Upf1 activity. Finally, to understand the link between a factor involved in translation termination and its resulting initiation phenotype, we devised an in vitro assay to assess the abilities of ribosomes undergoing premature versus normal termination events to be reutilized in subsequent rounds of translation.
To determine whether Upf1 and other NMD factors affect post-termination reinitiation at a PTC in vivo, we constructed a reinitiation reporter and evaluated its expression in wild-type yeast strains and in strains devoid of NMD function. The reporter, a chimeric PGK1/LUC gene (Fig. 1A, construct 1) that encodes an mRNA subject to NMD (Fig. 1B), consists of the following: (1) the PGK1 5′-untranslated region (UTR); (2) the first 5% of the PGK1 open reading frame (ORF) followed by a UAG stop codon; (3) an 82-nucleotide (nt) PGK1 coding region sequence element whose first AUG codon, designated +19 AUG (relative to the U of the UAG), is out-of-frame with the upstream PGK1 ORF; (4) the LUC ORF lacking its normal start codon, fused in-frame with the +19 AUG; and (5) the 3′-UTR of PGK1. Luciferase assays, performed on extracts of wild-type yeast strains bearing this construct, yielded activity that was assigned a value of 100% for purposes of comparison with subsequent experiments (Fig. 1C). In this, and all other assays, luciferase activity was normalized to both total protein content of the sample, as well as to the level of the construct-specific mRNA. The derived values were thus expressed as relative light units (RLUs)/μg of protein per unit of mRNA.
FIGURE 1.
Reinitiation activity is reduced in strains bearing mutations in the NMD pathway. (A) Schematic representation of PGK1/LUC chimeric constructs 1–4. Start and stop codons are indicated, as are their reading frames and that of the LUC coding region, relative to the PGK1 start codon. (X) Indicates deletion or mutation of a particular feature; (+19 AUG) denotes the first of two in-frame PGK1 AUGs downstream from the PTC. Details of the constructs are given in Table 2. (B) Northern analysis of the PGK1/LUC mRNA expressed from either a low-copy (YCp) or high-copy (YEp) vector in wild-type (WT) cells or in isogenic cells harboring deletions of UPF1, UPF2/NMD2, or UPF3. Blots were stripped and reprobed for CYH2 transcripts as an internal control for NMD function (CYH2 pre-mRNA) and loading (CYH2 mRNA). (C) Relative luciferase activities of constructs 1–4 normalized for protein content and mRNA levels in the WT strain HFY114. The luciferase activity of construct 1 was assigned a value of 100%. (D) Reinitiation activity. Construct 1 was inserted into either a low-copy (YCp) or a high-copy (YEp) vector and transformed into the indicated strains. Relative luciferase activities, normalized for protein content and mRNA levels, are shown for YCp-bearing strains (black boxes) and for YEp-bearing strains (white boxes). The luciferase activity of construct 1 on a YCp plasmid in a WT strain was assigned a value of 100%. (Δ1) upf1Δ; (Δ2) nmd2Δ; (Δ3) upf3Δ; and combinations thereof. Vertical bars represent standard error of the mean. (E) Polysomal distribution of the PGK1/LUC reporter mRNA in WT and upf1Δ cells. (Top panel) WT cells; (middle panel) WT cells, long exposure; (bottom panel) upf1Δ cells. Note that the top and bottom panels were exposed for the same length of time. Polysomal and non-polysomal fractions are indicated, as is the position of the 80S peak (vertical arrow).
Our analyses indicated that the luciferase activity derived from the reporter represented reinitiation from the +19 AUG codon downstream from the partial PGK1 ORF or from one additional in-frame AUG located five codons downstream from the +19 AUG. First, we measured luciferase activity from cells containing construct 2 (Fig. 1A), in which the +19 AUG and the additional downstream AUG codon were deleted, and observed a fivefold decrease in luciferase expression relative to construct 1 (Fig. 1C). (The residual luciferase activity appears to derive from an in-frame AUG codon 82 nt into the LUC sequence [Fig. 2, construct 6].) Second, in a separate construct, the LUC ORF was placed out-of-frame with respect to the +19 AUG, as well as out-of-frame with the PGK1 ORF (Fig. 1A, construct 3). This construct yielded almost no detectable luciferase activity (Fig. 1C), indicating that the LUC ORF has to be in-frame with the +19 AUG to produce functional luciferase protein. Finally, deletion of the PGK1 stop codon (Fig. 1A, construct 4), which allows translation until a termination codon 75 nt into the LUC coding region, also resulted in a substantial decrease in luciferase activity (Fig. 1C). These results strongly implicate the +19 AUG codon in the expression of luciferase from a translational reinitiation event in the PGK1/LUC mRNA derived from construct 1.
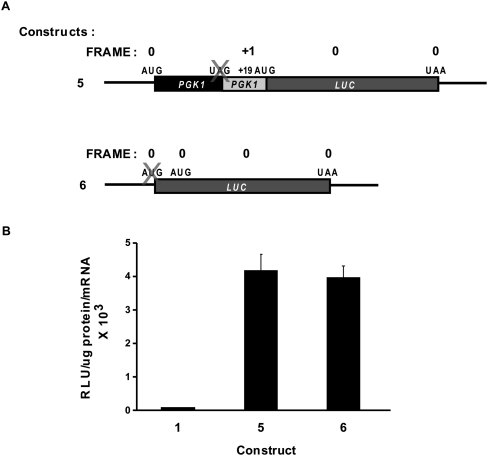
FIGURE 2.
Determination of ribosomal initiation efficiency and proportion of ribosomes that reinitiate downstream from the PGK1 stop codon. (A) Schematic representations of chimeric PGK1/LUC constructs 5 and 6. Labeling of start and stop codons and reading frame is as depicted in Figure 1, with “X” indicating deletion or mutation of a particular feature. (B) Luciferase activities of constructs 5 and 6 in the wild-type strain HFY114. Activity is expressed as RLU/microgram of protein per unit of mRNA. The activity of construct 1 (Fig. 1) is shown for purposes of comparison. Vertical bars represent standard error of the mean.
Reinitiation on nonsense-containing PGK1/LUC mRNA is sensitive to mutations in the NMD pathway
To assess whether NMD factors influence reinitiation events in vivo, PGK1/LUC construct 1, on a YCp plasmid, was transformed into isogenic yeast strains bearing deletions of UPF1, UPF2/NMD2, or UPF3, and combinations thereof. Although the mRNA derived from this construct is sensitive to NMD, as indicated by the observation that it is stabilized in yeast strains defective in this pathway (Fig. 1B), the expression of low-copy PGK1/LUC construct 1 in upf/nmd strains yielded a dramatic decrease in luciferase activity when compared to that in wild-type cells (Fig. 1D). It was important to determine whether this apparent decrease in luciferase activity was simply due to the increased stability of the PGK1/LUC mRNA and consequent increase in its steady-state level in these strains without a parallel increase in luciferase protein. To clarify this question, PGK1/LUC construct 1 was inserted into a high-copy YEp vector and transformed into the wild-type and upf/nmd mutant strains. Normalized values for luciferase activity in strains harboring the YEp construct were very similar to those in cells harboring the construct on a YCp plasmid (Fig. 1D). Notably, the increased levels of PGK1/LUC mRNA in wild-type cells (Fig. 1B) did not lead to any apparent decrease in normalized values of reinitiation activity, showing that mRNA levels do not affect overall reinitiation in this reporter. Additionally, the upf/nmd mutant strains showed a similar reduction in luciferase activity regardless of whether there was an increase in steady-state levels of the mRNA (Fig. 1B,D). As an additional control, we used sucrose gradient analysis to evaluate the polysomal distribution of the PGK1/LUC mRNA in wild-type and upf1Δ cells. These experiments manifested indistinguishable polysomal profiles for the PGK1/LUC mRNA in wild-type and upf1Δ cells, with >90% of the mRNA on polysomes in both cases (Fig. 1E). Collectively, these results indicate that the mutation-dependent variations in reinitiation activity evident in Fig. 1D are not simply a function of increases in the abundance of PGK1/LUC mRNA or of decreases in its overall association with ribosomes, and that, as was seen previously in cell-free translation reactions (Amrani et al. 2004), reinitiation in vivo is decreased in the absence of the key NMD factors.
A small percentage of ribosomes reinitiate downstream from the premature stop codon
To determine the proportion of ribosomes that initiate at the normal PGK1 AUG and are subsequently able to reinitiate, post-termination, on the PGK1/LUC mRNA, it was necessary to first assess the efficiency with which ribosomes initiated at the PGK1 AUG. We made a construct in which the premature PGK1 stop codon plus one nucleotide were deleted, resulting in a fusion ORF which begins at the PGK1 start codon and ends at the normal LUC terminator (Fig. 2A, construct 5). The luciferase activity of construct 1 was found to be ∼2.5% that of construct 5 (Fig. 2B), a comparison indicating that approximately one in 40 ribosomes reinitiated translation after encountering the premature PGK1 nonsense codon.
NMD is independent of translation reinitiation
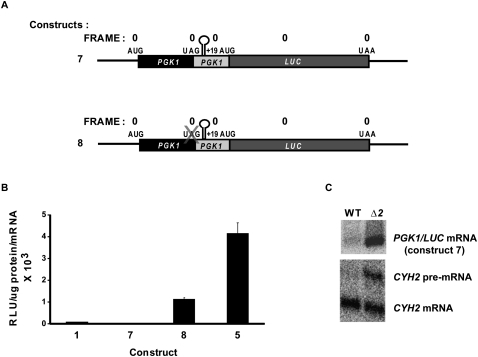
Since our data indicated that reinitiation efficiency is directly affected by the NMD factors and other experiments have shown that reinitiation can affect NMD (Zhang and Maquat 1997), it was of interest to determine whether rapid mRNA decay still occurred in the absence of reinitiation. To address this question, we introduced a strong stem–loop structure (ΔG = 58.3 kcal/mol) between the PGK1 stop codon and the PGK1 downstream +19 AUG (Fig. 3A, construct 7). This stem–loop structure bears a 25-nt-pair stem and 4-nt loop and is preceded by a 30-nt spacer between the stop codon and the stem–loop to ensure that the structure does not interfere with termination. mRNA derived from construct 7 displayed almost no luciferase activity (Fig. 3B) but continued to undergo NMD since it could be stabilized in an nmd2Δ strain (Fig. 3C). Further insight into the potency of the stem–loop structure used in these experiments was obtained from an additional construct that contains the stem–loop but lacks the premature PGK1 stop codon, generating an ORF that extends from the PGK1 initiator AUG, through the stem–loop and PGK1 downstream sequence, to the LUC terminator (Fig. 3A, construct 8). Luciferase activity derived from construct 8 was only 25% that of construct 5, which is identical but lacks the stem–loop (Fig. 3B). Collectively, the results of Figure 3 suggest that the strong stem–loop disrupts reinitiation (and presumably ribosome scanning) and substantially inhibits elongation, but is not sufficient to prevent NMD. Thus, we conclude that neither reinitiation nor any form of ribosome scanning beyond the termination codon is required for the promotion of NMD.
FIGURE 3.
A strong stem–loop structure does not eliminate nonsense-mediated mRNA decay. (A) Schematic representations of chimeric PGK1/LUC constructs 7 and 8. Labeling of start and stop codons and the reading frame is as depicted in Figure 1, with “X” indicating deletion or mutation of a particular feature. (B) Luciferase activities of constructs 7 and 8 in the WT strain HFY114. Activity is expressed as RLU/microgram of protein per unit of mRNA. Activities of constructs 1 and 5 are shown for purposes of comparison. Vertical bars represent standard error of the mean. (C) Northern analysis of PGK1/LUC construct 7 mRNA expressed from a low-copy (YCp) vector in WT cells or in isogenic cells harboring a deletion of UPF2/NMD2. Blots were stripped and reprobed for CYH2 transcripts as an internal control for NMD function (CYH2 pre-mRNA) and loading (CYH2 mRNA).
NMD-deficient extracts manifest a translation defect in vitro
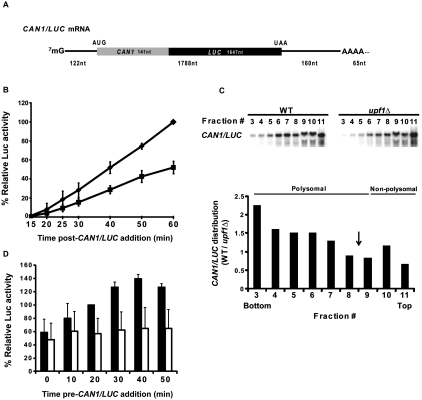
The in vivo results of Figures 1–3 are consistent with our earlier in vitro data (Amrani et al. 2004) and suggest that factors regulating NMD have an as-yet-undefined function that affects translation reinitiation. To characterize this role further, we returned to the analysis of yeast cell-free translation and evaluated the ability of non-nucleased extracts prepared from WT or upf/nmd mutant strains to translate a synthetic CAN1/LUC fusion mRNA (Fig. 4A) that harbors a full-length open reading frame (ORF) and remains stable during the course of in vitro translation (Amrani et al. 2004). Figure 4B shows that, relative to WT extracts, extracts from strains lacking Upf1 activity manifested reduced luciferase expression from the CAN1/LUC mRNA. Similar results were observed with extracts lacking Upf2/Nmd2 or Upf3 (data not shown). Northern analyses of WT and upf1Δ in vitro translation reactions fractionated on sucrose gradients demonstrated that the WT extract had higher relative amounts of CAN1/LUC mRNA in the heavier polysome fractions, thus confirming the relatively poor translation activity of the mutant extracts and suggesting that they initiated translation inefficiently (Fig. 4C).
FIGURE 4.
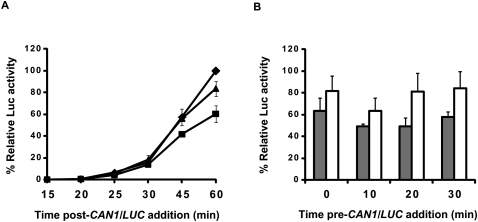
Translation of CAN1/LUC mRNA is decreased in upf1Δ extracts. (A) Schematic representation of the CAN1/LUC fusion mRNA. (B) Luciferase activities of WT (diamonds) and upf1Δ (squares) extracts prepared without micrococcal nuclease (MN) treatment and incubated with 100 ng of CAN1/LUC mRNA for the indicated times at 18°C. Relative luciferase activity was calculated using the WT 60-min values as 100%. (C, upper panel) Northern analysis of polysomal distribution of CAN1/LUC mRNA translated in vitro in WT or upf1Δ extracts. (Lower panel) Ratio of the polysomal distributions of the CAN1/LUC mRNA during in vitro translation. WT and upf1Δ extracts, prepared without MN treatment, were incubated with synthetic CAN1/LUC mRNA for 30 min at 18°C and then analyzed by fractionation on 7%–47% sucrose gradients and subsequent Northern blotting using a probe specific for the CAN1/LUC mRNA. The amount of CAN1/LUC mRNA (in arbitrary units) in each gradient fraction was determined by PhosphorImaging, and the y-axis depicts the ratios of those amounts in WT versus upf1Δ extracts. Sedimentation of the gradients was from right to left; the bars at the top of the panel indicate the polysomal and non-polysomal fractions. “Bottom” and “Top” refer to the regions of the gradients with the heaviest and lightest fractions, respectively. The vertical arrow depicts the position of the 80S ribosome peak. (D) Relative luciferase activities of WT (black boxes) and upf1Δ (white boxes) extracts treated as in B and incubated for 25 min after addition of CAN1/LUC mRNA at the indicated times. Relative luciferase activities were measured using WT 20-min values as 100%. Vertical bars in B and D depict standard deviation.
Further evidence for a translational deficiency in extracts from upf/nmd mutant strains followed from experiments that we dubbed “ribosome reutilization assays.” Here, translation of an extract's endogenous mRNA was allowed to proceed in the absence of any CAN1/LUC mRNA addition. At regular intervals (termed “time pre-CAN1/LUC addition” in Figs. 4–6), the translation reaction was supplemented with CAN1/LUC mRNA and incubated for 25 additional minutes to allow for synthesis of the luciferase fusion protein. Thus, luciferase activity obtained from the 25-min incubation with the reporter mRNA at each time point should reflect the relative functionality or availability of ribosomes and factors in the “preincubated” extracts to initiate and translate the exogenously added reporter mRNA. As shown in Figure 4D, WT extracts exhibit enhanced translatability of the reporter mRNA over time, whereas upf1Δ extracts (as well as nmd2Δ and upf3Δ extracts) (data not shown) fail to do so. These results suggest that, upon preincubation, WT extracts increase the availability or activity of a limiting component of the translation system, whereas the mutant extracts maintain over time a smaller and relatively unchanged pool of the functionally available component. One possible explanation for this observation, considered below, is that the increase in translation activity in WT extracts arises from translation termination and dissociation of ribosomal subunits from endogenous mRNAs since the extracts used in these assays have not undergone nuclease treatment. Thus, ribosomes in WT extracts might readily enter the pool of functional ribosomes over time, leading to an overall increase in CAN1/LUC expression, whereas the reutilization of ribosomes that were previously engaged in translation is somehow compromised in the mutant extracts.
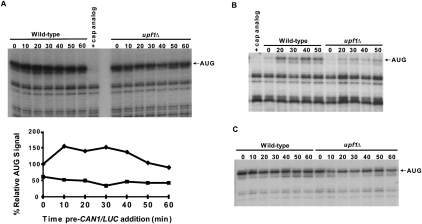
FIGURE 6.
Formation of 80S toeprints is diminished in upf1Δ extracts. (A) Cell-free translation extracts, prepared from WT and upf1Δ cells without MN treatment, were incubated at 18°C. CAN1/LUC mRNA (100 ng) was added at the indicated time points, and reactions were further incubated for 25 min, followed by addition of 2 mM cycloheximide (CHX) for 3 min. Reactions were frozen and subsequently analyzed by toeprinting. (Upper panel) The autoradiograph and (lower panel) the densitometry of the AUG band in WT (diamonds) and upf1Δ (squares) reactions normalized to the WT 0-min value. (B) Translation conditions were as in A, except that 2 mM CHX was added with 100 ng of CAN1/LUC mRNA at each time point, followed by incubation for 25 min. Lanes in A and B in which control reactions contained 2.7 mM cap analog are indicated. (C) Translation and incubation conditions were as in B, except that CHX was replaced with 2 mM GMP-PNP.
Upf1 affects translation in vitro
To test whether the decrease in translation of the CAN1/LUC reporter mRNA observed in upf1Δ extracts was due to the absence of Upf1 or resulted from an indirect effect of the lack of a functional NMD pathway, we assessed translation of the CAN1/LUC mRNA in upf1Δ extracts supplemented with either purified FLAG-Upf1 or BSA (as a control). As shown in Figure 5A, purified FLAG-Upf1 was sufficient to restore the translation activity of a upf1Δ extract to that approaching the activity of a WT extract. In addition, purified FLAG-Upf1 was also able to stimulate the translational capacity of a upf1Δ extract at different time points in a ribosome reutilization assay (Fig. 5B, cf. white boxes and gray boxes at each time point). These results indicate that Upf1 may play a direct role in enabling or enhancing the availability of functional ribosomes in vitro.
FIGURE 5.
Purified FLAG-Upf1 can complement translation defects in vitro. Cell-free translation extracts were prepared from WT or upf1Δ cells without MN treatment and preincubated for 10 min on ice with 270–360 ng of BSA or purified FLAG-Upf1, followed by incubation at 18°C in either of two modes: (A) addition of 100 ng of CAN1/LUC mRNA at t = 0 min and sampling for luciferase activity at the indicated time points or (B) addition of 100 ng of CAN1/LUC mRNA at the indicated time points and translation for 25 min at 18°C prior to measurement of luciferase activity. (A) Relative luciferase values were calculated as in Figure 4B for WT extracts supplemented with BSA (diamonds), upf1Δ extracts supplemented with BSA (squares), or upf1Δ extracts supplemented with FLAG-Upf1 (triangles). (B) Relative luciferase activities of upf1Δ extracts supplemented with BSA (gray boxes) or FLAG-Upf1 (white boxes) normalized to the corresponding WT+BSA values at each given time point. Vertical bars in A and B depict standard deviation.
Translation initiation is compromised in upf1Δ extracts
To understand the nature of the translation defect in mutant extracts, we evaluated their relative efficiencies of translation initiation on the CAN1/LUC mRNA using a primer extension inhibition (toeprinting) assay (Dmitriev et al. 2003; Amrani et al. 2004). WT and upf1Δ extracts were preincubated for different lengths of time and then supplemented with CAN1/LUC mRNA, as in Figure 4D. After incubating the extracts with the mRNA for 25 min, cycloheximide (CHX) was added to terminate the reactions and stall elongating 80S ribosomes. Toeprinting reactions were then performed using a radiolabeled oligonucleotide that allowed visualization of a gel band corresponding to a ribosome positioned at the initiator AUG codon (Fig. 6A, upper panel). Accumulation of this toeprint was sensitive to cap analog (Fig. 6A, upper panel), indicating that it arose from initiation activity, and WT extracts routinely showed a more intense toeprint than the upf1Δ extracts (Fig. 6A, upper and lower panels), reflecting better translation initiation.
Since the AUG toeprints could arise as a consequence of both de novo initiation and recycling of ribosomes on the same mRNA post-termination, we modified the assay such that CHX was added at the same time as the CAN1/LUC mRNA. Thus, only ribosomes undergoing de novo initiation (i.e., the first ribosome encountering the AUG codon) would contribute to the AUG toeprints. Figure 6B shows that, using this approach, only WT extracts exhibit a significant increase in the AUG toeprint within the first 20 min, whereas upf1Δ extracts exhibit only a minimal increase in the AUG toeprint band even at late time points, suggesting an absence of 80S ribosomes at the initiator AUG in these extracts. This result indicates further that the availability of functional ribosomes may be diminished in the absence of Upf1. To determine whether Upf1 affects an early or late step in initiation, we repeated the same assay, replacing CHX with the nonhydrolyzable GTP analog GMP-PNP, which allows 48S formation but inhibits joining of the 60S ribosomal subunit (Fig. 6C; Dmitriev et al. 2003). In this experiment, we observed that both WT and upf1Δ extracts exhibited strong AUG toeprint bands in the presence of GMP-PNP, indicative of the presence of 40S ribosomal subunits on the RNA, with the upf1Δ extracts showing a slightly diminished signal. Taken together, these results suggest that, although 40S loading is slightly impaired in upf1Δ extracts, Upf1 is likely to influence a late stage of translation initiation, namely, the joining of the 60S subunit to the 48S complex.
Initiation efficiency depends on prior termination events
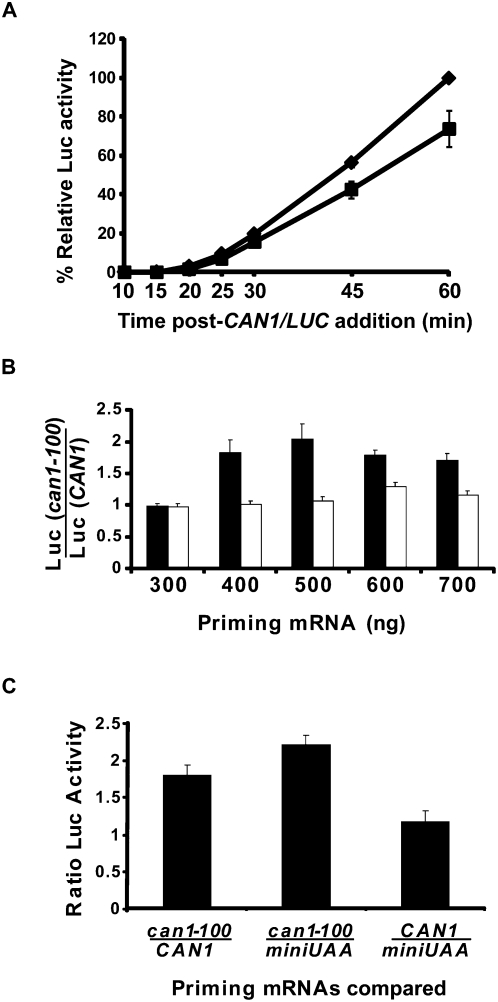
The results depicted in Figures 4–6 implicate Upf1 in the determination of translational initiation efficiency in vitro, but the mechanism underlying this activity and its relationship to the reinitiation defects seen in vivo (Figs. 1–3) and in vitro (Amrani et al. 2004) were unclear. The extracts used in our translation reactions are used without any prior nuclease treatment, and the ongoing translation of endogenous mRNAs includes that of a substantial number of mRNAs that are NMD substrates (He et al. 2003; Johansson et al. 2007). Since premature termination on the latter transcripts is altered in the absence of UPF1 (Amrani et al. 2004), we considered the possibility that these altered events might influence subsequent translation initiation activity in the mutant extracts. Accordingly, we devised an assay in which the translational capabilities of ribosomes that were allowed to undergo either normal or premature termination events were tested for their ability to subsequently translate the CAN1/LUC reporter mRNA. To achieve this, we first treated WT and upf1Δ extracts with micrococcal nuclease to eliminate the endogenous mRNAs that would act as competitors of an exogenously added translational template. With this treatment, the ability of upf1Δ extracts to translate the CAN1/LUC mRNA in vitro resembled that of WT extracts for the first 30 min of incubation (Fig. 7A) (i.e., for the time period analyzed in subsequent assays), confirming our earlier observation that MN-treated extracts showed similar AUG toeprints of the CAN1/LUC transcript (Amrani et al. 2004). We then primed these extracts with excess amounts of either synthetic WT CAN1 mRNA or its nonsense-containing counterpart (can1-100 mRNA). We rationalized that providing an excess of the priming mRNA templates would allow most of the functional ribosomes in a given extract to be involved in translation and thereby undergo either normal or premature translation termination events. Furthermore, these terminating ribosomes might be differentially predisposed for further rounds of translation in these extracts. We tested this difference by measuring the luciferase activity generated by the subsequent addition of the CAN1/LUC reporter mRNA to the reactions containing extracts translating the WT or PTC-containing mRNAs. Determining the ratio of the luciferase activities obtained from translation reactions primed with the can1-100 versus the CAN1 mRNA for a given extract provided a measure of the efficiency of ribosome release from the PTC and their subsequent reutilization. Figure 7B depicts the ratios of luciferase activity obtained in WT and upf1Δ extracts from reactions that were first primed with varying amounts of the can1-100 or CAN1 mRNAs. The results of this experiment indicate that WT extracts manifest as much as twofold more translation of the CAN1/LUC reporter mRNA when primed with high levels of the can1-100 mRNA versus the CAN1 mRNA, whereas the translation of the reporter mRNA in upf1Δ extracts is unaffected by the nature of the priming mRNA. This result suggests that in WT extracts, but not in upf1Δ extracts, the termination and release of ribosomes from premature stop codons generates a pool of ribosomes with greater recycling ability than ribosomes released from the normal terminator.
FIGURE 7.
Initiation efficiency depends on prior termination events. Translation extracts prepared from WT or upf1Δ cells were treated with MN for 5 min at 25°C. (A) Luciferase activities of WT (diamonds) and upf1Δ (squares) extracts incubated with 100 ng of CAN1/LUC mRNA for the indicated times at 18°C. Relative luciferase activity was calculated using the WT 60-min values as 100%. (B) Treated extracts were incubated with the designated concentrations of the priming CAN1 or can1-100 mRNA for 7 min at 18°C, followed by addition of 300 ng of CAN1/LUC mRNA and incubation for an additional 25 min. Ratios of luciferase activities calculated from reactions programmed with can1-100 and CAN1 mRNAs are shown for WT (black boxes) and upf1Δ (white boxes) extracts. (C) CAN1 or can1-100 mRNA (500 ng of either) or 50 ng of miniUAA mRNA was incubated with WT extracts for 7 min, followed by addition of 300 ng of CAN1/LUC mRNA and further incubation for 25 min at 18°C. Ratios of luciferase activity were calculated as designated on the x-axis. Vertical bars in all panels depict standard deviation.
Since the ORF in the PTC-containing mRNA is shorter than its WT counterpart, in principle, the smaller size of the ORF (and thus greater frequency of terminating ribosomes), rather than the nature of the termination event itself, could be responsible for the increased luciferase activity in translation reactions primed with the can1-100 mRNA in WT extracts. Hence, we repeated the assay in WT extracts using either the can1-100 mRNA or the miniUAA mRNA (Amrani et al. 2004). The latter is a transcript that contains the same ORF as the can1-100 mRNA, but which has a normal termination event because the short ORF is followed by a normal 3′-UTR (Amrani et al. 2004). A comparison of the ratios of luciferase activities obtained with the two RNAs showed that priming with the can1-100 mRNA yielded enhanced translation of the CAN1/LUC mRNA relative to priming with the miniUAA mRNA (Fig. 7C, middle box). The experiments in Figure 7C (right box) also demonstrate that the luciferase output obtained when reactions were primed with the miniUAA mRNA were comparable to those primed with the CAN1 mRNA. Taken together, these results indicate that premature termination events, not the length of the ORF, are responsible for the increased luciferase activity in this system and reflect the possibility that more ribosomes, or more active ribosomes, were released from the nonsense-containing transcript than from the WT or pseudo-WT mRNA.
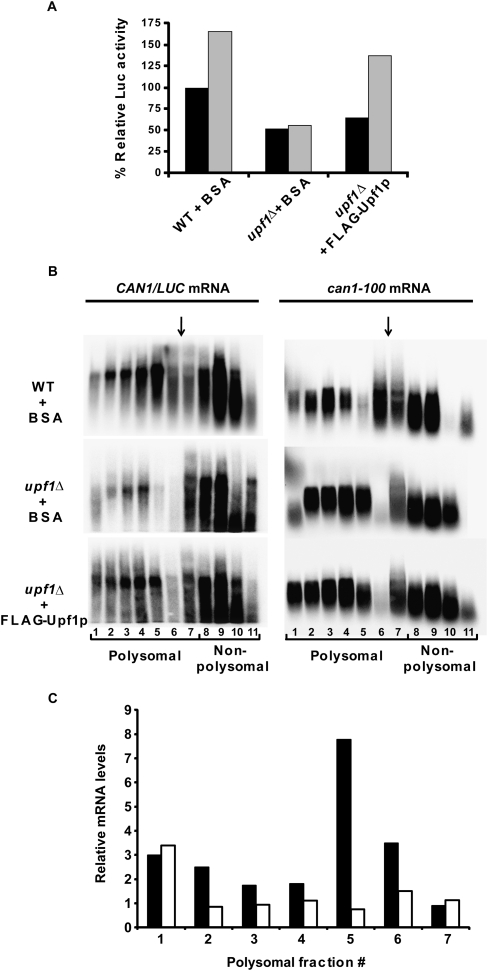
To test whether Upf1 is involved in the increase of translation activity subsequent to premature termination, we primed upf1Δ extracts with the CAN1 or can1-100 mRNAs in the presence of purified FLAG-Upf1 or BSA and measured the responses of the respective extracts to CAN1/LUC mRNA that was added subsequently. We found that addition of purified FLAG-Upf1 was sufficient to enhance luciferase activity in upf1Δ extracts primed with can1-100 mRNA, similar to that observed with WT extracts (Fig. 8A, cf. gray boxes in WT+BSA and upf1Δ +FLAG-Upf1). To further analyze the translation status of the CAN1/LUC mRNA in reactions primed with the can1-100 mRNA in the presence of FLAG-Upf1 or BSA, we fractionated the translation reactions on sucrose gradients and analyzed the fractions by Northern blotting for their levels of specific transcripts. These experiments showed that, in WT extracts (supplemented with BSA), the CAN1/LUC mRNA was distributed throughout the sucrose gradient, including the polysome and lighter fractions (Fig. 8B, left panel). In upf1Δ extracts (supplemented with BSA), there was significantly less CAN1/LUC mRNA in the polysome fractions, with most of the mRNA being confined to the sub-80S fractions. However, addition of FLAG-Upf1 to the upf1Δ extracts restored the polysome distribution of the CAN1/LUC mRNA to that seen in the WT control. Comparison of the distribution of the CAN1/LUC mRNA in upf1Δ extracts supplemented with or without FLAG-Upf1 showed up to threefold more CAN1/LUC mRNA in polysomal fractions of extracts that contained FLAG-Upf1 (Fig. 8C, black boxes). To determine whether the decreased translation of the secondary CAN1/LUC mRNA was due to overall decreased translational capacity of the nuclease-treated upf1Δ extracts, we also analyzed the distribution of the can1-100 mRNA (Fig. 8B, right panel; Fig. 8C, white boxes) used to prime the reactions. We observed a similar distribution pattern for the primary can1-100 mRNA in extracts containing or lacking Upf1, further indicating that decreased CAN1/LUC translation in upf1Δ extracts is most likely a consequence of translational termination at a PTC in the absence of Upf1.
FIGURE 8.
Purified FLAG-Upf1 restores efficient translational activity to upf1Δ extracts. Cell-free translation extracts were prepared from WT or upf1Δ cells and treated with MN for 5 min at 25°C. Extracts were incubated with 270 ng of BSA or FLAG-Upf1 for 10 min on ice and added to translation mixes containing 500 ng of CAN1 (A) or can1-100 (A–C) mRNA and incubated for 7 min at 18°C. CAN1/LUC mRNA (300 ng) was then added and reactions were further incubated for 25 min at 18°C. (A) Relative luciferase activities in the reactions detailed on the x-axis primed with the (black boxes) CAN1 or (gray boxes) can1-100 mRNAs. (B) can1-100–primed translation reactions were set up as in A, terminated by the addition of CHX (2 mM), and subsequently fractionated on 7%–47% sucrose gradients. The fractions were then analyzed by Northern blotting using a DNA probe specific for the CAN1/LUC (left panel) or the can1-100 (right panel) mRNA. Polysomal and non-polysomal fractions are indicated, as is the position of the 80S peak (vertical arrow) in each blot. (C) Relative mRNA levels in polysome fractions. The amount of CAN1/LUC (black boxes) or can1-100 mRNA (white boxes) in each polysomal fraction of B was determined by phosphorimaging. The y-axis depicts the ratios of those amounts in upf1Δ extracts supplemented with FLAG-Upf1 versus BSA.
Addition of WT ribosomal subunits enhances translation in upf1Δ extracts
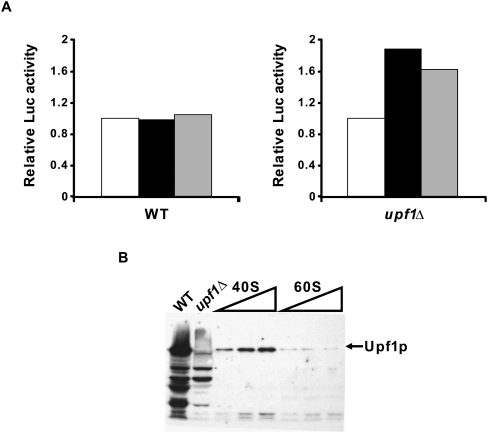
Although Upf1 has been shown to cosediment with polyribosomes (Peltz et al. 1993b; Atkin et al. 1995; Yamashita et al. 2009), relatively little is known about the nature of this association. Since the results of Figures 7 and 8 suggested that Upf1 might enhance the ability of ribosomes to recycle to another round of translation, we evaluated the effect of adding purified WT ribosomal subunits to upf1Δ extracts. We reasoned that the absence of Upf1 in these extracts might lead to the accumulation of defective 40S and/or 60S subunits and that the possible amelioration of the translation defects upon addition of the WT subunits would shed light on which subunit was affected in the absence of Upf1. Figure 9A (right panel) shows that addition of purified 40S or 60S subunits to non-nucleased upf1Δ extracts was sufficient to enhance translation of the CAN1/LUC mRNA. WT extracts (Fig. 9A, left panel), on the other hand, did not show any increases in translation upon addition of either ribosomal subunit. The stimulatory activity of 40S and 60S subunits in upf1Δ extracts suggested that either both subunits were affected by the absence of Upf1 or that only one subunit was affected but that the other contained bound Upf1. Indeed, Figure 9B shows that Upf1 preferentially copurified with 40S subunits, whereas it was markedly reduced in the 60S subunit fraction. Since the subunit purification protocol (Algire et al. 2002) involves high salt washes, this result indicates that Upf1 can form a stable association with 40S subunits, and probably accounts in part for the increase in luciferase activity observed upon their addition to upf1Δ extracts. In turn, this suggests that the stimulatory activity of the purified 60S subunits may reflect a relative inactivity of the endogenous 60S subunits in upf1Δ extracts.
FIGURE 9.
40S and 60S ribosomal subunits enhance translation in upf1Δ extracts. Cell-free translation extracts were prepared from WT or upf1Δ cells. (A) Extracts were supplemented with 100 ng of CAN1/LUC mRNA and then incubated with BSA (white boxes), purified 40S subunits (black boxes), or purified 60S subunits (gray boxes) for 60 min at 18°C. Luciferase activity is depicted relative to the value obtained with the BSA control for each extract. The graph depicts the average of two experiments. (B) Western blot of increasing amounts of purified 40S (5, 10, and 15 μg) and 60S (5, 10, and 15 μg) subunit preparations probed with polyclonal anti-Upf1 antibody. WT and upf1Δ extracts (5 μg each) were run as controls.
DISCUSSION
In addition to well-documented roles targeting nonsense-containing mRNAs for rapid decay (Jacobson and Izaurralde 2007; Franks and Lykke-Andersen 2008), the Upf factors of the NMD pathway also appear to have translational regulatory functions. Results from both yeast and mammalian cells have implicated possible roles for Upf1, Upf2/Nmd2, and Upf3 in the regulation of translational termination, repression, and reinitiation (Czaplinski et al. 1998; Muhlrad and Parker 1999; Maderazo et al. 2000; Wang et al. 2001; Isken et al. 2008; Ivanov et al. 2008). To consider the possible general translational involvement of these factors in yeast, we sought in this study to first examine whether in vitro results supporting a Upf role in translational reinitiation (Amrani et al. 2004) could be recapitulated in vivo. Using a series of PGK1/LUC reporter constructs, we defined a cell-based assay in which ∼2.5% of the ribosomes that prematurely terminate the PGK1 ORF subsequently reinitiate and translate the LUC ORF. Relative to wild-type cells, strains that lacked the function of any one of the three UPF genes manifested markedly reduced reinitiation activity in this assay (Fig. 1D), but reinitiation per se played no role in activating NMD (Fig. 3). These observations not only served to confirm and elaborate the earlier results we had obtained in cell-free extracts (Amrani et al. 2004), but also led us to investigate in greater detail the mechanism by which the Upf proteins influence translational activity.
Prior studies indicated that Upf1 regulation of translation might not be limited to nonsense-containing mRNAs. For example, yeast cells expressing WT-like CUP1 reporters showed reduced Cup1 levels when UPF1 was deleted (Muhlrad and Parker 1999). Furthermore, in vitro translation of the WT synthetic CAN1/LUC mRNA in yeast extracts was found to require Upf1 (Amrani et al. 2004), and mammalian cell-free extracts containing hyperphosphorylated Upf1 showed reduced translation of synthetic Renilla LUC mRNA (Isken et al. 2008). Consistent with these observations, we not only confirmed that optimal translation of the CAN1/LUC mRNA is Upf1-dependent, but that Upf2/Nmd2 and Upf3 are also required (Fig. 4; data not shown). Moreover, using a novel assay for ribosome reutilization, we found that elimination of any one of the Upf proteins led to reduced availability of ribosomes for translation initiation on CAN1/LUC mRNA (Fig. 4D; data not shown). The significance of these results was underscored by the observation that the translational defect(s) of upf1Δ extracts could be overcome by the addition of purified recombinant Upf1 (Figs. 5, 8). Collectively, these experiments suggested that Upf1 plays a direct role in regulating the initiation efficiency of ribosomes that enter a new cycle of translation.
A further understanding of the apparent initiation defect was obtained by attempting to identify the stage of initiation that was impaired. Toeprinting analyses of the CAN1/LUC initiator AUG codon indicated that although upf1Δ extracts had fewer 80S ribosomes at the AUG than did WT extracts, the two extracts exhibited similar 40S recruitment, suggesting that a loss of Upf1 affects subunit joining (Fig. 6). Indeed, the addition of purified WT 40S or 60S ribosomal subunits to upf1Δ extracts stimulated translation in these extracts, but not in WT extracts (Fig. 9A). Further analyses showed that Upf1 was preferentially associated with 40S subunits, suggesting that enhanced translation observed upon 40S addition could, in part, be explained by the ribosome-bound Upf1 (Fig. 9B). In combination with the toeprinting results, these data implied that (1) the increase in luciferase activity detected upon addition of 40S subunits to upf1Δ extracts was most likely Upf1-mediated and (2) the stimulation of translation occurring in upf1Δ extracts upon addition of 60S subunits was probably attributable to the accumulation of inactive 60S subunits in those extracts.
It is becoming increasingly apparent that factors involved in regulating translation may have multiple functions and that this multitasking may link the initiation pathway to those for termination and/or NMD (Amrani et al. 2008; Saini et al. 2009). Moreover, it has been shown that (1) eIF3, eIF1, and eIF1A, as well as the eIF3j subunit, not only play a role in initiation, but also stimulate the in vitro dissociation of post-termination 80S complexes (Pisarev et al. 2007); (2) at elevated temperatures, yeast strains harboring the temperature-sensitive prt1-1 mutation (in eIF3) lose their abilities to both initiate translation and promote NMD (Welch and Jacobson 1999); (3) yeast cells with mutations in both SUI1 (encoding eIF1) and UPF1 manifest synthetic lethality (Harger and Dinman 2004); and (4) phosphorylated mammalian Upf1 can interact with eIF3, and this interaction appears to inhibit cap-dependent in vitro translation (Isken et al. 2008). Consistent with these observations, we find that yeast upf1Δ extracts are unable to recycle ribosomes efficiently from a nonsense-containing mRNA (Figs. 7, 8), a result suggesting that events at termination can influence the subsequent activity of ribosomal subunits (Khoshnevis et al. 2010; Pisarev et al. 2010). Since our in vitro translation system does not support NMD (e.g., see Fig. 8B; Amrani et al. 2004), we hypothesize that Upf1's role in promoting ribosome dissociation and/or recycling from nonsense-containing mRNAs represents an independent function that occurs concurrently or prior to its role in promoting decapping. How these events may be linked remains to be elucidated. Since premature termination in yeast is considerably less efficient than normal termination (Amrani et al. 2004), one possibility is that the post-peptide hydrolysis recruitment of eIF3, and perhaps other initiation factors, to a prematurely terminating ribosome is Upf1-dependent. Inclusion of Upf1 would allow termination complex remodeling and efficient 80S dissociation, yielding a 40S subunit capable of reinitiating translation and a 60S subunit capable of being primed for subsequent translation cycles (Amrani et al. 2006).
It remains to be seen whether the association between Upf1 and 40S subunits reflects a cause or consequence of ribosome dissociation, whether other NMD factors show a similar association with ribosomal subunits, and what other factors may be present in a Upf-dependent manner. Upf1 has intrinsic ATPase and helicase activities (Czaplinski et al. 1995), and it will be of interest to test whether these activities are principally required for ribosome remodeling at a premature termination codon or for the stimulation of mRNA decay.
MATERIALS AND METHODS
Yeast strains, vectors, and transformations
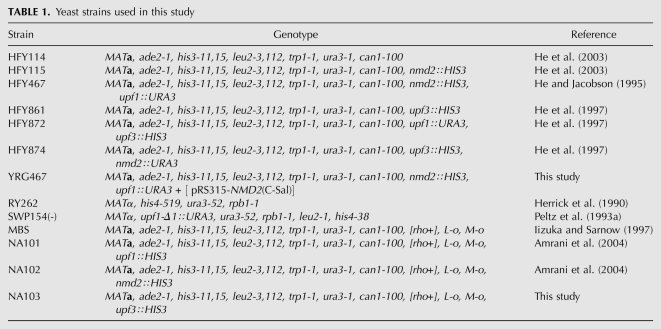
The yeast strains used in this study, and their genotypes, are listed in Table 1. Strain YRG467 was constructed by transforming HFY467 with plasmid pRS315-NMD2(C-Sal) (He et al. 1996). For in vivo studies, strains were grown in selective media (Amberg et al. 2005) at 30°C, or at 24°C for temperature-sensitive strains. Yeast transformations were performed by the rapid protocol (Soni et al. 1993), and the yeast vectors utilized included pRIP1 (Parker and Jacobson 1990), pRS314 (Sikorski and Hieter 1989), YEplac22 (Gietz and Sugino 1988), and pRS316 (Sikorski and Hieter 1989). For in vitro translation experiments, MBS and strains derived from it (NA101, NA102, and NA103) were grown in YEPD at 25°C for extract preparation (Amrani et al. 2004).
TABLE 1.
Yeast strains used in this study
Plasmid-borne expression constructs
The constructs used for in vivo expression analyses are described in Table 2. The oligonucleotides used in this study are listed in Table 3. Construct 1, a chimeric PGK1/LUC gene (Fig. 1A), consists of the PGK1 5′-untranslated region (UTR); the first 5% of the PGK1 open reading frame (ORF) followed by a UAG stop codon (contextually a premature termination codon); an 82-nt PGK1 sequence element (derived from a 106-nt segment previously designated as a DSE) (see Table 2; Peltz et al. 1993a) that contains two AUG codons (out-of-frame with the upstream PGK1 ORF) fused in-frame to the LUC ORF lacking its normal start codon; and the 3′-UTR of PGK1. All constructs for in vivo luciferase expression experiments contain a 5′-BamHI site and a 3′-HindIII site and were subcloned into vectors by excising with BamHI/HindIII. PCR fragments 1 and 2 were generated using the mini-PGK1 allele as a template. This allele contains a 58-nt tag in its 3′-UTR to allow later detection of its mRNA on a Northern blot (Peltz et al. 1993a). Fragment 1 was generated using the M13/pUC Reverse Amplification primer (BRL) and primer 3-106-82 and digested with BamHI and PstI restriction enzymes. Fragment 2 was generated using primer 5-PPGKH and the M13/pUC Forward Amplification primer (BRL) and digested with PstI and HindIII. Fragment 3 was generated by PCR using pXP1 as a template (a kind gift from Paul Dobner, University of Massachusetts Medical School) and primers 5-PSLUCP and 3-PLUCSP, and digested with PstI. Fragments 1 and 2 were ligated into pGEM-3Z(f)+ (Promega) and cut with BamHI/HindIII. The resulting construct was cut with PstI, incubated with calf intestinal alkaline phosphatase (CIAP), and ligated with fragment 3 to generate construct 3. Construct 1 was generated by digesting construct 3 with SalI to remove a 10-base-pair (bp) fragment and religating to close. BamHI/HindIII fragments containing construct 3 and construct 1 were then subcloned into the centromeric yeast vectors pRIP1 (cut with BamHI/HindIII) and pRS314 (cut with SmaI), and the high-copy plasmid, YEplac112 (cut with BamHI/HindIII). The resulting constructs were transformed into strains HFY114, YRG467, HFY115, HFY861, HFY467, HFY872, and HFY874.
TABLE 2.
Pertinent features of constructs used in this study
TABLE 3.
Oligonucleotide primers used in this study
Construct 2 was generated with the Chameleon Site-Directed Mutagenesis Kit (Stratagene) using the following strategy: plasmid pRIP1 containing construct 1 was digested with BamHI/SalI to yield an 800-bp fragment containing the 5′-end of construct 1 from the PGK1 start to the junction of the 82-nt PGK1 sequence element and LUC and an 8-kb fragment containing the remainder of the construct and pRIP1. The smaller fragment was subcloned into pGEM-3Z(f)+ to yield pGEM-3Z(f)+/PGKBS. The resulting plasmid was mutagenized with primers PGKDATG and pGEM-SEL. A BamHI/SalI fragment containing the mutagenized construct was then ligated with the 8-kb fragment described above to yield construct 2. Construct 5 was generated similarly, with primers PGKΔTAG and pGEM-SEL, followed by mutagenic primer FUSION. These constructs were also placed into pRS314 and YCplac22. Construct 4 was generated using primers PGKDELINIT and PGKΔTAG. Construct 6 was generated as follows: An ∼560-bp fragment was made by PCR using pGEM-3Z(f)+/PGKBS as a template and primers M13 reverse and PGKUAGSAL2, and cut with BamHI/SalI. This fragment was ligated into the 8-kb fragment described above to yield construct 6. All of the above constructs were transformed into HFY114, YRG467, HFY115, HFY861, HFY467, HFY872, and HFY874.
To generate construct 7, construct 1 was subcloned into pGEM-3Z(f)+ and cut with NheI. Linkers NHELINKERA and NHELINKERB were annealed together and ligated into the NheI site. These constructs were sequenced for orientation such that NHELINKERA reads 5′ → 3′. The resulting construct [pGEM-(Sal)lucNHE] was digested with StuI to cut within the NHE linker and treated with CIAP. Oligonucleotide primers HPR1A and HPR2A were annealed together, and HPL1A and HPL2A were annealed together. The resulting short double-stranded fragments (designated HairpinR and HairpinL, respectively) were isolated on a 15% acrylamide gel and purified by the crush-and-soak method (Maxam and Gilbert 1977). Ligations were performed such that HairpinR and HairpinL were ligated together for 0 and 5 min before addition of the StuI-cut pGEM-(Sal)lucNHE. Colonies from the above ligations were screened by colony PCR for 162-bp inserts containing HairpinR and HairpinL (as opposed to double HairpinR or double HairpinL inserts) and sequenced for orientation such that HairpinL was 5′ of HairpinR. The resulting BamHI/HindIII fragment–containing construct 7 was subcloned into YCplac22. This construct was transformed into HFY114, YRG467, HFY115, HFY861, HFY467, HFY872, and HFY874. Construct 8 was made by deleting the UAG stop codon of construct 7 using the Quick-Change Site-Directed Mutagenesis Kit (Stratagene). The mutagenic primers were DSTOP1 and DSTOP2. This construct was transformed into yeast strains HFY114, YRG467, HFY115, HFY861, HFY467, HFY872, and HFY874.
The CAN1/LUC (“Fusion”) construct used as a template for the preparation of synthetic mRNA for in vitro translation experiments has been described elsewhere (Amrani et al. 2004). pRS316pA-CAN1 and pRS316pA-can1-100 templates were derived by ligating an EcoRI/HindIII fragment of plasmid CFE86 (containing the complete CAN1 gene; a kind gift from Chunfang Li) or plasmid CFE87 (which contains the complete can1-100 gene; a kind gift from Chunfang Li) into pRS316pA linearized by cleavage with EcoRI and HindIII. To generate pRS316pA, oligonucleotide primers Xho-pA-Asp (5′-[phos]TCGAG(A)75G-3′) and Asp-pT-Xho (5′-[phos]GTACC(T)75C-3′) were annealed, alkaline phosphatase-treated (Roche), and ligated into XhoI/Asp718-linearized pRS316.
Measurement of luciferase activity in cells, Bradford assays, and Northern analyses
Yeast cultures (30 mL) were grown in selective medium overnight at 24°C to an OD600 of 0.7–1.2. Four milliliters of each culture was removed, and the cells were pelleted and frozen at −70°C for later RNA extraction. An additional 20 mL of each culture was pelleted and washed twice with 1× PBS (0.01 M NaH2PO4, 0.15 M NaCl). The cells were resuspended in 2 mL of 1× PBS and split between two microcentrifuge tubes. These were then pelleted and stored at −70°C to await glass bead extraction. The frozen cell pellets were resuspended in 200 μL of luciferase extraction buffer (LEB) (0.1 M KPO4 at pH 7.8, 1 mM DTT, 1 mM PMSF). Acid-washed glass beads (200 mg; Sigma 425–600 μm) were added, and the tubes were then vortexed for 30 sec and placed on ice for 30 sec, alternately, for 5 min. The tubes were then spun in a microcentifuge at 14,000 rpm for 5 min at 4°C, and the supernatant (extract) was placed into a fresh tube on ice. The following components were then added to a luminometer cuvette: 100 μL of LEB, 20 μL of extract, and 350 μL of luciferase assay buffer (LAB) at pH 7.8 (25 mM glycylglycine at pH 7.8, 15 mM KPO4at pH 7.8, 15 mM MgSO4, 4 mM EGTA, 2 mM ATP, 1 mM DTT). Assays were performed in a Monolight 2010 luminometer by injecting 100 μL of 200 μM D-luciferin and potassium salt (Analytical Luminescence Labs) in 25 mM glycylglycine (pH 7.8) and measuring light output for 10 sec. Extracts were stored at −70°C following luciferase assays until Bradford assays were performed. Bradford assays were performed on a Beckman DU7400 Spectrophotometer with the Bio-Rad Protein Assay Kit according to the microassay protocol using 10 μL of extract per assay. RNA was prepared from frozen cell pellets as described previously (Herrick et al. 1990) and quantitated on a Beckman DU7400 Spectrophotometer. Aliquots (10 μg) of total RNA were loaded onto a 1% agarose/formaldehyde/MOPS gel and electrophoresed, blotted, and hybridized as described previously (Herrick et al. 1990). Transcripts derived from the different constructs were detected with an oligonucleotide (Peltz et al. 1993a) that hybridizes to the 3′-UTR of the respective construct mRNAs but does not hybridize to the endogenous PGK1 mRNA. mRNA signals were quantitated using a Betascope (Betagen) blot analyzer. mRNA levels were normalized for loading by stripping the blots and rehybridizing with a random-primed probe targeted to the CYH2 mRNA. Final luciferase activity was expressed as relative light units (RLUs) of 20 μL of extract, per microgram of protein in 20 μL of extract, per unit of construct mRNA in 10 μg of total RNA, normalized for loading differences with the CYH2 mRNA signal. Each set of assays performed on a given day included three cultures of construct 1 in strain HFY114 on a YCp plasmid, and each Northern blot from each set of assays contained the RNAs from these three cultures. The average luciferase activity (expressed in RLU/microgram of protein per unit of mRNA) of these three cultures was assigned a value of 100% to which all other luciferase activities in a set of assays were compared.
Extract preparation, synthetic mRNA, in vitro translation, and toeprinting assays
Saccharomyces cerevisiae strains MBS, NA101, NA102, and NA103 were used to make extracts for in vitro translations as described (Iizuka and Sarnow 1997; Wu et al. 2007; He et al. 2008). Capped and polyadenylated CAN1/LUC mRNA was synthesized with the SP6 mMessage mMachine Kit (Ambion) as per the manufacturer's protocol, from HindIII-linearized plasmids. Capped and polyadenylated CAN1 and can1-100 mRNAs were synthesized in vitro from KpnI-linearized pRS316pA-CAN1 and pRS316pA-can1-100 plasmids using the T7 mMessage mMachine Kit (Ambion) as per the manufacturer's protocol. Translation assays were as described (Amrani et al. 2004; Wu et al. 2007), except that each assay was performed at 18°C without prior treatment of the extract with micrococcal nuclease (MN), unless otherwise specified, and used 100 μg of extract and 100 ng of the CAN1/LUC mRNA unless specified otherwise. When used, MN pre-treatment of extracts was performed for 5 min at 25°C (Amrani et al. 2004). Protein concentrations of translation extracts were measured using the BCA Protein Assay Kit (Pierce). Translation reactions were incubated as specified in the figure legends and frozen in a dry ice/ethanol bath until luciferase assays were to be performed. The latter were done with a TD20/20 luminometer (Promega), as described previously (Amrani et al. 2004). For translation reactions involving FLAG-Upf1 purified from yeast (a kind gift from Stephanie Kervestin), 270–360 ng of FLAG-Upf1 or BSA was preincubated with extracts on ice for 10 min prior to adding the rest of the components of the translation reactions. For add-back of purified 40S and 60S ribosomal subunits, 5 μg of purified 40S subunits, 10 μg of purified 60S subunits, or BSA was added to the reaction without prior incubation with extracts. For toeprinting reactions, translations were terminated by incubation for 3 min with 2 mM cycloheximide or GMP-PNP (Sigma), aliquotted, frozen in a dry ice/ethanol bath, and stored at −70°C until reverse transcription reactions were to be performed (Amrani et al. 2008) with oligomer #105 (5′-CGTCTGTGGTCTGTTTGTGAAGCC-3′). All in vitro translation experiments, except add-back experiments with ribosomal subunits, were performed at least three times with independently prepared extracts. Add-back experiments with ribosomal subunits were done at least twice with independently prepared extracts.
Sucrose gradient analyses
For in vitro mRNA:ribosome association assays, the equivalent of six translation reactions were incubated for 30 min at 18°C, stopped by the addition of 2 mM CHX, and fractionated on an 11 mL of a 7%–47% sucrose gradient. The gradients were scanned at A254, and the resulting absorbance profiles were used to determine the position of the polysomal and non-polysomal fractions (Mangus and Jacobson 1999; Amrani et al. 2008). RNA was extracted from each fraction and analyzed by Northern blotting as described previously (Mangus and Jacobson 1999), using random-primed probes directed against the LUC ORF or the 3′ half of the can1-100 ORF (Amrani et al. 2008). All experiments were done at least twice with independently prepared extracts. Analyses of polysome distributions in vivo were performed as described previously (Mangus and Jacobson 1999).
Ribosome subunit preparation and Western blot analysis
We used a procedure for purification of 40S and 60S ribosomal subunits that had been described previously (Algire et al. 2002), except that we used 9L of strain MBS grown in YPD at 25°C to an OD600 of 1.0. Fractions enriched for 40S and 60S subunits were concentrated in Amicon Ultra-15 100K NMWL filter devices (Millipore). Protein concentrations were measured using the BCA Protein Assay Kit (Pierce). The 40S fraction contained 18S rRNA but no detectable 25S rRNA, and the 60S fraction contained 25S rRNA but no detectable 18S rRNA, as visualized on a 1% formaldehyde-agarose gel stained with ethidium bromide. Upf1 association with 40S and 60S ribosomal subunits was assessed by Western blot analysis using the methods and the anti-Upf1 antibody described previously (Mangus and Jacobson 1999).
ACKNOWLEDGMENTS
We thank Meng-Jiao Shi, Paul Dobner, Marcus Johansson, and Feng He for technical advice and reagents; members of the Jacobson laboratoy for comments on the manuscript; and Stephanie Kervestin for a gift of FLAG-Upf1 protein. This work was supported by grants to A.J. from the National Institutes of Health and the Human Frontier Science Program.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1987710.
REFERENCES
- Algire MA, Maag D, Savio P, Acker MG, Tarun SZ Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, et al. 2002. Development and characterization of a reconstituted yeast translation initiation system. RNA 8: 382–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke DJ, Strathern JN 2005. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, Jacobson A 2006. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7: 415–425 [DOI] [PubMed] [Google Scholar]
- Amrani N, Ghosh S, Mangus DA, Jacobson A 2008. Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453: 1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin AL, Altamura N, Leeds P, Culbertson MR 1995. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell 6: 611–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Weng Y, Hagan KW, Peltz SW 1995. Purification and characterization of the Upf1 protein: A factor involved in translation and mRNA degradation. RNA 1: 610–623 [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett 533: 99–104 [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A 1988. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Harger JW, Dinman JD 2004. Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA 10: 1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A 1995. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 9: 437–454 [DOI] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A 1996. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA 2: 153–170 [PMC free article] [PubMed] [Google Scholar]
- He F, Brown AH, Jacobson A 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol 17: 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- He F, Amrani N, Johansson MJ, Jacobson A 2008. Qualitative and quantitative assessment of the activity of the yeast nonsense-mediated mRNA decay pathway. Methods Enzymol 449: 127–147 [DOI] [PubMed] [Google Scholar]
- Herrick D, Parker R, Jacobson A 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 10: 2269–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Sarnow P 1997. Translation-competent extracts from Saccharomyces cerevisiae: Effects of L-A RNA, 5′ cap, and 3′ poly(A) tail on translational efficiency of mRNAs. Methods 11: 353–360 [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2008. The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat Rev Genet 9: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE 2008. Interactions between UPF1, eRFs, PABP, and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 27: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Izaurralde E 2007. Nonsense-mediated mRNA decay: From yeast to metazoans. In Translational control in biology and medicine(ed. Mathews MB et al. ), pp. 655–687 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Johansson MJO, Jacobson A 2010. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev 24: 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MJ, He F, Spatrick P, Li C, Jacobson A 2007. Association of yeast Upf1p with direct substrates of the NMD pathway. Proc Natl Acad Sci 104: 20872–20877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H 2010. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep 11: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo AB, He F, Mangus DA, Jacobson A 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol 20: 4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Jacobson A 1999. Linking mRNA turnover and translation: Assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods 17: 28–37 [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W 1977. A new method for sequencing DNA. Proc Natl Acad Sci 74: 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R 1999. Recognition of yeast mRNAs as ‘nonsense containing‘ leads to both inhibition of mRNA translation and mRNA degradation: Implications for the control of mRNA decapping. Mol Biol Cell 10: 3971–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P 1999. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol 19: 5943–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Jacobson A 1990. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MATa1 gene are involved in promoting rapid mRNA decay in yeast. Proc Natl Acad Sci 87: 2780–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz SW, Brown AH, Jacobson A 1993a. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev 7: 1737–1754 [DOI] [PubMed] [Google Scholar]
- Peltz SW, Trotta C, He F, Brown A, Donahue JL, Welch E, Jacobson A 1993b. Identification of the cis-acting sequences and trans-acting factors involved in nonsense-mediated mRNA decay. In Protein synthesis and targeting in yeast(ed. Tuite M et al. ), pp 1–10 Springer, Berlin [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, Anderson P 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 7: 1885–1897 [DOI] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Murray JA 1993. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr Genet 24: 455–459 [DOI] [PubMed] [Google Scholar]
- Wang W, Czaplinski K, Rao Y, Peltz SW 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 20: 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Jacobson A 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J 18: 6134–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW 1996a. Genetic and biochemical characterization of mutations in the ATPase and helicase regions of the Upf1 protein. Mol Cell Biol 16: 5477–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW 1996b. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol 16: 5491–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Czaplinski K, Peltz SW 1998. ATP is a cofactor of the Upf1 protein that modulates its translation termination and RNA binding activities. RNA 4: 205–214 [PMC free article] [PubMed] [Google Scholar]
- Wu C, Amrani N, Jacobson A, Sachs MS 2007. The use of fungal in vitro systems for studying translational regulation. Methods Enzymol 429: 203–225 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. 2009. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 23: 1091–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Maquat LE 1997. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J 16: 826–833 [DOI] [PMC free article] [PubMed] [Google Scholar]