Abstract
We have started a space experiment using an experimental organism, the nematode Caenorhabditis elegans, in the Japanese Experiment Module, KIBO, of the International Space Station (ISS). The specimens were boarded by space shuttle Atlantis on mission STS-129 which launched from NASA Kennedy Space Center on November 16, 2009. The purpose of the experiment was several-fold: (i) to verify the efficacy of RNA interference (RNAi) in space, (ii) to monitor transcriptional and post-translational alterations in the entire genome in space, and (iii) to investigate mechanisms regulating and countermeasures for muscle alterations in response to the space environment. In particular, this will be the first study to utilize RNAi in space.
Introduction
Caenorhabditis elegans (C. elegans) is a free-living, non-parasitic soil nematode. It can be easily manipulated, observed and cultivated in the laboratory owing to its small size (an adult worm is approximately 1 mm in length), transparency and feeding on bacteria. A wealth of studies over the past few decades have resulted in C. elegans becoming a well-known model organism. For example, the complete cell-lineage, neuronal networks, muscle anatomy and genome sequence, make this an excellent in vivo model in which to conduct biological research both on Earth and in space. The first trials on the effects of the space environment, including cosmic radiations, upon C. elegans were performed using essentially standard culturing techniques (Johnson and Nelson, 1991; Nelson et al., 1994a,b; Hartman et al., 2001). Subsequently, a complete chemical liquid medium (CeMM) for use with C. elegans was prepared by Szewczyk et. al., (2003, 2006), which allowed study of the effects of surface tension in flight. During the Dutch Soyuz mission DELTA to the ISS in April 2004, an international collaboration of laboratories carried out the “FIRST International C. elegans Experiment in space” (ICE FIRST). One of the main goals of this experiment was to validate the biological response of C. elegans to 10 day spaceflight. Consistent with past experiments, animals displayed a normal rate of development in flight and returned in good apparent health. With the exception of a slight movement defect upon return to Earth, which appears to be due to altered muscle development in flight (Higashibata et al., 2006), no significant abnormalities were detected. Apoptosis also proceeded normally (Higashitani et al., 2005) and the rate of mutation associated with flight was below the experiments limits of detection (Zhao et al., 2006). These results appear similar to what is observed for humans and suggest that C. elegans can be used to study responses to spaceflight and may be developed as a biological sensor (Zhao et al., 2005; Custodia et al., 2001). At the molecular level, transcriptome and proteome analyses indicated decreased expression of muscle related genes and proteins, respectively, including altered expression of certain genes regulated by insulin and transforming growth factor-β (TGF-β) signalling in response to spaceflight (Higashibata et al., 2006, 2007, Selch et al., 2008).
C. elegans is the first animal in which RNA interference (RNAi) by double stranded RNA (dsRNA) was observed (Fire et al., 1998). RNAi is an evolutionarily conserved mechanism for silencing gene expression (Novina and Sharp 2004, Tomari and Zamore 2005). RNAi protects the genome from viruses and other insertable genetic elements and regulates gene expression during development. The antisense strand derived from the dsRNAs incorporates into an RNA-induced silencing complex that can either direct degradation of target mRNA or suppress the protein(s) it encodes from being expressed (Novina and Sharp 2004, Tomari and Zamore 2005). The discovery that RNAi works in mammalian cells has sparked intense investigation into its role in normal mammalian cell function, its use as a tool to understand or screen for genes functioning in cellular pathways in healthy and diseased cells and animals, and its potential for therapeutic gene silencing (Dykxhoorn and Lieberman 2005, Aigner 2006, Chakraborty 2007, DeVincenzo 2009). The ensuing results suggest RNAi may provide an important new therapeutic modality for treating infection, cancer, neurodegenerative disease, and other illnesses.
Here we introduce our current space experiment termed CERISE (C. elegans RNA interference Space Experiment) in the Japanese Experiment Module (JEM), called KIBO, on the International Space Station (ISS) beginning November 2009. A proposal for CERISE was reviewed and accepted by the International Space Life Sciences Working Group in 2004; the CERISE decal is shown in Fig. 1. The experimental aims are to verify the efficacy of RNAi in space and to analyze changes in the transcriptome and proteome in response to the space environment. We further aim to investigate mechanisms of and countermeasures for musclular alterations in response to spaceflight.
Fig. 1.

Decal for the CERISE experiment
Experimental Design
Nematode eggs were prepared using the alkaline bleach method with 0.5N KOH and 1.0% NaClO. After overnight incubation in M9 buffer containing 5mg/ L cholesterol at 20° C, the hatched L1 larvae were used for the space experiment. For most experiments, approximately 9,000 L1 were used for samples to be collected 4 days post-acitvation and 30 to 50 L1 were used for samples to be collected 8 days post-activation. For the muscle protein degradation studies, approximately 1,000 dauers, prepared according to the protocol of Hartman et al., (2001) with the exception that animals were cultured on 8X peptone NGM agar plates, were used with sample collection on day 4 post-activation. All larvae were maintained in the left compartment of the culturing bags in 2 ml S basal medium (Fig. 2). The right compartment of the culturing bag system, separated by a U-pin, contained bacterial feeds in 12 ml S basal medium at an OD600 of approximately 3.5. Bags were then stored in the Meas Exp. Unit A (15 bags per unit, Fig. 2). On 16th November 2009, Shuttle Atlantis (STS-129, ULF-3) launched from Kennedy Space Center, Florida. Upon reaching microgravity at KIBO on the ISS, flight crews activated the experiments by removing the U-pin (19th November), at which time four Meas Exp. Unit A were transferred into the Cell Biology Experiment Facility (CBEF) with or without 1 G rotation for either 4 or 8 days. After observation of the nematodes by microscopy, experiments were stopped by freezing and subsequent storage at −80° C in MELFI. For the 4 day culture experiments, the L1 or dauer larva grew to adulthood as the first generation, and for the 8 day experiments the L1 larvae’s second generations grew to adulthood under exposure to space environment from fertilization.
Fig. 2.
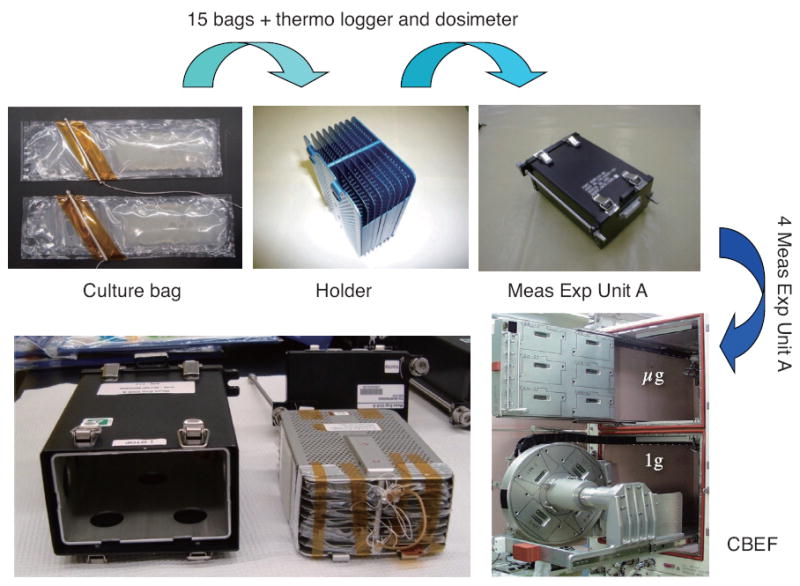
Experimental equipments and culture bags. The bags were made of polyethylene with heat shielding. Two compartments were separated with U-pin. Crew member removes the U-pin in space, and cultures are started in CBEF.
RNA interferences (RNAi)
To evaluate the efficacy of RNAi in the space environment, we used two target genes, Ce-rbx-1 (Sasagawa et al., 2003) and recombinant green fluorescent protein (GFP) genes. Strains AZ212 (ruls32; unc-119 (ed3), Praitis et al., 2001), whose integrated array is pAZ132 (pie-1∷GFP∷histone H2B fusion and unc-119 subclone), and PD4251 (ccls4251; dpy-20 (e1282), Fire et al., 1998), whose integrated array contains three plasmids: pSAK2 (myo-3 promoter driving a nuclear-targeted DFP-LacZ fusion), pSAK4 (myo-3 promoter driving mitochondrial-targeted GFP), and a dpy-20 subclone were used in this experiment. AZ212 GFP signals in the nuclei of oocytes and eggs, and PD4251 fluoresces in the nuclei and mitochondria of body wall muscles (Fig. 3). Double stranded RNA of Cerbx-1 and gfp genes were synthesized in Escherichia coli HT115 (DE3) with Litmus 28 plasmid vector in vivo system (Sasagawa et al., 2003). Following experiment activation and thus introduction of the nematodes to the bacteria, animals fed on the bacteria (termed feeding RNAi, Kamath et al., 2000) in the space environment. In addition, we have performed feeding RNAi against both asp-4 and asp-6, genes that encode aspartyl proteases, in order to study the effects of depletion of aspartyl protease on muscle protein degradation in response to microgravity.
Fig. 3.
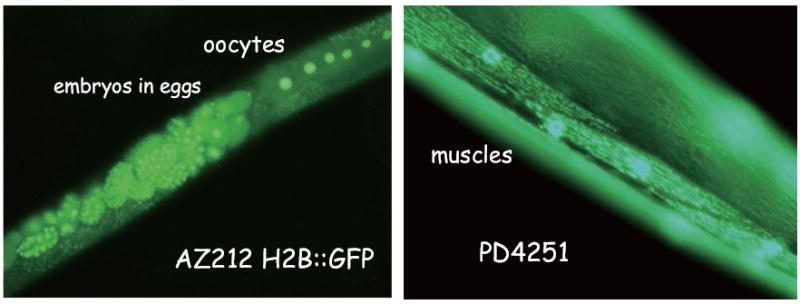
GFP recombinant strains used in this study to monitor RNAi activity and muscle structures. Nuclei of Oocytes and embryos are visualized in AZ212 (left panel). Nuclei and mitochondria of muscles are visualized in PD4251 (Right panel).
Transcriptome and Proteome analyses
To study the molecules and signals that are affected in response to the space environment, we have carried out transcriptome analyses using full genome DNA microarrays and proteome analyses, including post transcriptional modification. We have previously shown that both expression of the transcription factors for myosin heavy chains (MHC) and MHC genes themselves are down-regulated in response to spaceflight (Higashibata et al., 2006). Insulin and TGF-β regulated genes also displayed alterations (Selch et al., 2008). Additionally, proteomic analysis using 2-dimensional gel electrophoresis indicated that approximately 10-15% of spots (i.e. detectable proteins in the proteome) significantly increased or decreased in the flight samples compared with the ground control (Higashibata et al., 2007). While much information on the transcriptional and translational adaptations to spaceflight was gained from ICE FIRST, there were some experimental limitations such as mixed stage samples (non-synchronized culture) and late fixation (freezing after return to Earth). In the present experiments we, therefore, aim to confirm reproducibility of the above results by using a synchronized culturing system with on orbit fixation. We shall also perform comparative analysis of phosphorylated proteins between culturing at microgravity versus a 1 G control using defective mutants of stress activated MAPK p38 and JNK1(strains KU25: pmk-1 (km25) and VC8: jnk-1 (gk7)).
Muscle synthesis and degradation in space
It is well-known that the neuromuscular system is one of the physiologic systems most affected by spaceflight (Fitts et al., 2001). Muscles developed on Earth alter in morphology, contractile function and MHC gene expression during spaceflight, or unloading on Earth (Caiozzo et al., 1994; 1996; Criswell et al., 1996; Day et al., 1995; Harrison et al., 2003). In addition to depressed de novo synthesis, specific protein degradation systems, such as ubiquitin ligase(s)-mediated proteasomal degradation have been shown to be involved in skeletal muscle atrophy (Bodine et al., 2001; Gomes et al., 2001). For example, in denervated muscle of rats, mRNA levels of MHC I are decreased and mRNA levels of atrogin-1 (a ubiquitin ligase gene) are significantly increased (Horinouchi et al., 2005). However, the relative contributions of specific molecular changes within muscle to more global changes in muscle remains an area of active research. In C. elegans, we are studying the signal-transduction networks regulating muscle protein degradation (Szewczyk and Jacobson 2005, Szewczyk et al., 2007). At present we have demonstrated the existence of three distinct regulatory networks in C. elegans muscle. First, proteasome based degradation appears to be regulated by a molecular network tied to muscle cell depolarization. Second, autophagic based degradation appears to be regulated by a molecular network tied to growth factor signaling. Third, an unknown protease is regulated by a molecular network tied to muscle attachment to the extracellular matrix. In this flight experiment, investigations of both muscle development and atrophy in C. elegans are performed. We will utilize our standard methods to assess cytosolic muscle protein degradation in wild-type animals and in animals with blocked proteasomal degradation (MG132 inhibited) or blocked autophagic degradation (asp-4 and asp-6 RNAi). Additionally, as caspases are well known proteases and mutants for ced-3 are readily available, we will also examine cytosolic protein degradation in ced-3 mutant animals. Lastly, we have also included experiments that applied methyl-cellulose to the culture media to increase viscosity to understand whether increasing viscosity upregulates de novo muscle synthesis and / or represses muscle degradation in C. elegans.
Conclusion
We anticipate that the analyses of CERISE flight samples will provide us information on the physiological and molecular biological effects of microgravity / spaceflight on the experimental model C. elegans. In addition, it is essential to verify the efficacy of RNAi for its subsequent application in both future basic science and applied clinical use in the space environment. These samples have already been successfully launched, cultured and stored in MELFI Deep Freezer of ISS, and will be returned by Space Shuttle Endeavor (STS-130) in February, 2009.
Acknowledgments
The CERISE experiment has been conducted in the Japanese Experimental Module “Kibo” of the International Space Station. We have to thank crew members, lots of scientists and coordinators of JAXA and NASA for kindly and helpfully supporting our experiment. We also thank the Caenorhabditis elegans Genetic Center for the kind supply of mutant and recombinant strains. This work is supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and from the Japan Society for the Promotion of Science (JSPS). This study is also performed as part of the “Ground-Based Research Announcement for Space Utilization,” promoted by the Japan Space Forum. In addition, CM is supported by the International Advanced Research and Education Organization in Tohoku University. TE and NS’ participation are supported by the UK MRC (G0801271) and the US NIH NIAMS (AR054342).
References
- Aigner A. Gene silencing through RNA interference (RNAi) in vivo: Strategies based on the direct application of siRNAs. J Biotechnol. 2006;124:12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Caiozzo VJ, Baker MJ, Herrick RE, Tao M, Baldwin KM. Effect of spaceflight on skeletal muscle: mechanical properties and myosin isoform content of a slow muscle. J Appl Physiol. 1994;76:1764–1773. doi: 10.1152/jappl.1994.76.4.1764. [DOI] [PubMed] [Google Scholar]
- Caiozzo VJ, Haddad F, Baker MJ, Herrick RE, Prietto N, Baldwin KM. Microgravity-induced transformations of myosin isoforms and contractile properties of skeletal muscle. J Appl Physiol. 1996;81:123–132. doi: 10.1152/jappl.1996.81.1.123. [DOI] [PubMed] [Google Scholar]
- Chakraborty C. Potentiality of small interfering RNAs (siRNA) as recent therapeutic targets for gene-silencing. Curr Drug Tartget. 2007;8:469–482. doi: 10.2174/138945007780058988. [DOI] [PubMed] [Google Scholar]
- Criswell DS, Carson JA, Booth FW. Regulation of contractile protein gene expression in unloaded mouse skeletal muscle. J Gravit Physiol. 1996;3:58–60. [PubMed] [Google Scholar]
- Custodia N, Won SJ, Novillo A, Wieland M, Li C, Callard IP. Caenorhabditis elegans as an environmental monitor using DNA microarray analysis. Ann N Y Acad Sci. 2001;948:32–42. doi: 10.1111/j.1749-6632.2001.tb03984.x. [DOI] [PubMed] [Google Scholar]
- Day MK, Allen DL, Mohajerani L, Greenisen MC, Roy RR, Edgerton VR. Adaptations of human skeletal muscle fibers to spaceflight. J Gravit Physiol. 1995;2:47–50. [PubMed] [Google Scholar]
- DeVincenzo JP. Harnessing RNA interference to develop neonatal therapies: From Nobel prize winning discovery to proof of concept clinical trials. Early Human Dev. 2009;85:S31–S35. doi: 10.1016/j.earlhumdev.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Ann Rev Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fitts RH, Riley DR, Widrick JJ. Functional and structural adaptations of skeletal muscle to microgravity. J Exp Biol. 2001;204:3201–3208. doi: 10.1242/jeb.204.18.3201. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison BC, Allen DL, Girten B, Stodieck LS, Kostenuik PJ, Bateman TA, Morony S, Lacey D, Leinwand LA. Skeletal muscle adaptations to microgravity exposure in the mouse. J Appl Physiol. 2003;95:2462–2470. doi: 10.1152/japplphysiol.00603.2003. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Hlavacek A, Wilde H, Lewicki D, Schubert W, Kern RG, Kazarians GA, Benton EV, Benton ER, Nelson GA. A comparison of mutations induced by accelerated iron particles versus those induced by low earth orbit space radiation in the FEM-3 gene of Caenorhabditis elegans. Mutat Res. 2001;474:47–55. doi: 10.1016/s0027-5107(00)00154-8. [DOI] [PubMed] [Google Scholar]
- Higashibata A, Szewczyk NJ, Conley CA, Imamizo-Sato M, Higashitani A, Ishioka N. Decreased expression of myogenic transcription factors and myosin heavy chains in Caenorhabditis elegans muscles developed during spaceflight. J Exp Biol. 2006;209:3209–3218. doi: 10.1242/jeb.02365. [DOI] [PubMed] [Google Scholar]
- Higashibata A, Higashitani A, Adachi R, Kagawa H, Honda S, Honda Y, Higashitani N, Sasagawa Y, Miyazawa Y, Szewczyk NJ, Conley CA, Fujimoto N, Fukui K, Shimazu T, Kuriyama K, Ishioka N. Biochemical and Molecular Biological Analyses of space-flown nematodes in Japan, the First International Caenorhabditis elegans Experiment (ICE-First) Microgravity Sci Technol. 2007;19:159–163. doi: 10.1007/BF02919473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashitani A, Higashibata A, Sasagawa Y, Sugimoto T, Miyazawa Y, Szewczyk NJ, Viso M, Gasset G, Eche B, Fukui K, Shimazu T, Fujimoto N, Kuriyama K, Ishioka N. Checkpoint and physiological apoptosis in germ cells proceeds normally in spaceflown Caenorhabditis elegans. Apoptosis. 2005;10:949–954. doi: 10.1007/s10495-005-1323-3. [DOI] [PubMed] [Google Scholar]
- Horinouchi H, Kumamoto T, Kimura N, Ueyama H, Tsuda T. Myosin loss in denervated rat soleus muscle after dexamethasone treatment. Pathobiology. 2005;72:108–116. doi: 10.1159/000084113. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Nelson GA. Caenorhabditis elegans: a model system for space biology studies. Exp Gerontol. 1991;26:299–309. doi: 10.1016/0531-5565(91)90024-g. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2000;2:research 0002.1–0002.10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GA, Schubert WW, Kazarians GA, Richards GF. Development and chromosome mechanics in nematodes: results from IML-1. Adv Space Res. 1994a;14:209–214. doi: 10.1016/0273-1177(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Nelson GA, Schubert WW, Kazarians GA, Richards GF, Benton EV, Benton ER, Henke R. Radiation effects in nematodes: results from IML-1 experiments. Adv Space Res. 1994b;14:87–91. doi: 10.1016/0273-1177(94)90455-3. [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa Y, Uranno T, Kohara Y, Takahashi H, Higashitani A. Caenorhabdits elegans RBX1 is essential for meiosis, mitotic chromosomal condensation and segregation, and cytokinesis. Genes Cells. 2003;8:857–872. doi: 10.1046/j.1365-2443.2003.00682.x. [DOI] [PubMed] [Google Scholar]
- Selch F, Higashibata A, Imamizo-Sato M, Higashitani A, Ishioka N, Szewczyk NJ, Conley CA. Genomic response of the nematode Caenorhabditis elegans to spaceflight. Adv Space Res. 2008;41:807–815. doi: 10.1016/j.asr.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk NJ, Kozak E, Conley CA. Chemically defined medium and Caenorhabditis elegans. BMC Biotechnol. 2003;3:19, 1–7. doi: 10.1186/1472-6750-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk NJ, Peterson BK, Barmada SJ, Parkinson LP, Jacobson LA. Opposed growth factor signals control protein degradation in muscles of Caenorhabditis elegans. EMBO J. 2007;26:935–943. doi: 10.1038/sj.emboj.7601540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Johnsen R, Baillie D, Rose A. Worms in space? A model biological dosimeter. Gravit Space Biol Bull. 2005;18:11–16. [PubMed] [Google Scholar]
- Zhao Y, Lai K, Cheung I, Youds J, Tarailo M, Tarailo S, Rose A. A mutational analysis of Caenorhabditis elegans in space. Mutat Res. 2006;601:19–29. doi: 10.1016/j.mrfmmm.2006.05.001. [DOI] [PubMed] [Google Scholar]


