Abstract
MSL3 resides in the MSL (male-specific-lethal) complex that upregulates transcription by spreading the H4K16 acetyl-mark. We discovered a DNA-dependent interaction of MSL3 chromodomain with the histone H4K20 monomethyl-mark. Structure of a ternary complex shows DNA minor groove accommodates the histone H4 tail, and monomethyllysine inserts in a four-residue aromatic cage in MSL3. Histone H4K16 acetyl-mark antagonizes MSL3 binding, suggesting MSL function is regulated by a combination of post-translational modifications.
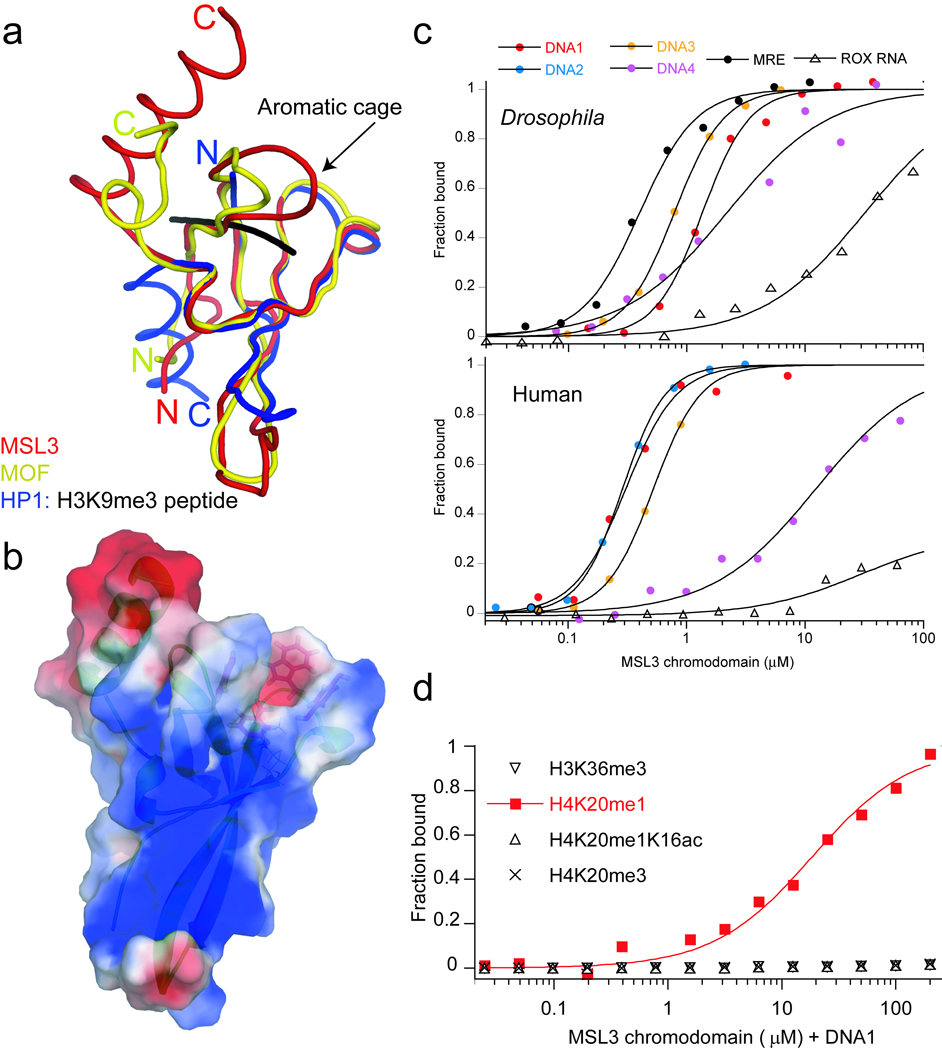
The MSL complex is necessary for dosage compensation on the male X chromosome in fruit flies 1,2. MOF and MSL3 are MSL subunits conserved from flies to humans3–5, each contains a chromodomain implicated in chromatin targeting 6–9. The deletion of MSL3 chromodomain leads to disruption of MSL targeting and spreading in vivo 8. To gain functional insights, we characterized the atomic structure of the Drosophila MSL3 chromodomain. Residues 10–91 constitute a contiguous module that is larger than HP1 or MOF chromodomains. It contains a surface unusually rich in positive charge, indicating potential nucleic acid binding (Fig. 1a,b and supplementary Table 1) 7,10. We observed two distinct conformations of the Phe56 and Trp59 side chains, rendering a four-residue aromatic cage accessible or occluded (Supplementary Fig. 1). Despite presence of aromatic cage, we and others could not verify an MSL3 chromodomain interaction with any histone methyl-marks 11.
Figure 1. MSL3 chromodomain structure and specificity.
(a) Superposition of the Drosophila MSL3 with MOF (PDB: 2BUD) and HP1 bound to a H3K9me3 peptide (PDB:1KNE). (b) MSL3 surface electrostatic potential. (c) Specificity for nucleic acids. (d) DNA-dependent specificity for the H4K20 monomethyl-mark.
To verify interactions with nucleic acids, we used fluoresceinated DNA and RNA in polarization assays (Supplementary Table 2). The MSL3 chromodomain preferentially binds DNA with affinity in the sub-micromolar range (Fig. 1c and Supplementary Table 2). It binds to MRE DNA (the GA-rich CES11D1) 1 with an affinity of 0.4 µM which is 80-fold tighter than binding to RNA, and markedly better than binding to GC-rich DNA. DNA containing a tandem repeat of the AGGTCA element, which is associated with site-specific transcription factors such as PPARγ-RXRα nuclear receptors 12, binds similar to MRE DNA. The binding isotherms for DNA1-DNA3 fit with a Hill coefficient of 2 (Fig. 1c), suggesting two non-interacting DNA binding sites on each of the MSL3 chromodomains. By contrast, Drosophila HP1 and Polycomb or the human CHD1 chromodomains did not bind DNA. Consistent with the fluorescence results, we found that NMR resonances of the human MSL3 chromodomain (Supplementary Fig. 2) completely disappear upon addition of DNA1, indicative of a substantially larger complex.
We then asked whether the pre-assembled complex of MSL3 with DNA would more effectively interact with a methylated histone peptide. We found both Drosophila and human chromodomain-DNA complexes preferentially bound the H4K20me1 peptide with a KD ~ 10–18 µM (Fig. 1d and Supplementary Table 2). Interestingly, neither the H4K20me3 nor the unmodified H4 peptide bound, indicating the interaction is sensitive to the level of methylation. Unlike H4K20me3 which marks heterochromatin, H4K20me1 is widely distributed in the body of genes (reviewed in 13), a region also associated with MSL complex localization 1,8. The H4K16 acetyl-mark and H4K20 methyl-mark are thought to be mutually exclusive on Drosophila chromosomes 14. We found the MSL3 chromodomain does not bind to a peptide containing the H4K20 monomethylation and H4K16 acetylation (Fig. 1d). This suggests either the chromodomain or its DNA effector is sensitive to neutralization of the Lys16 side chain.
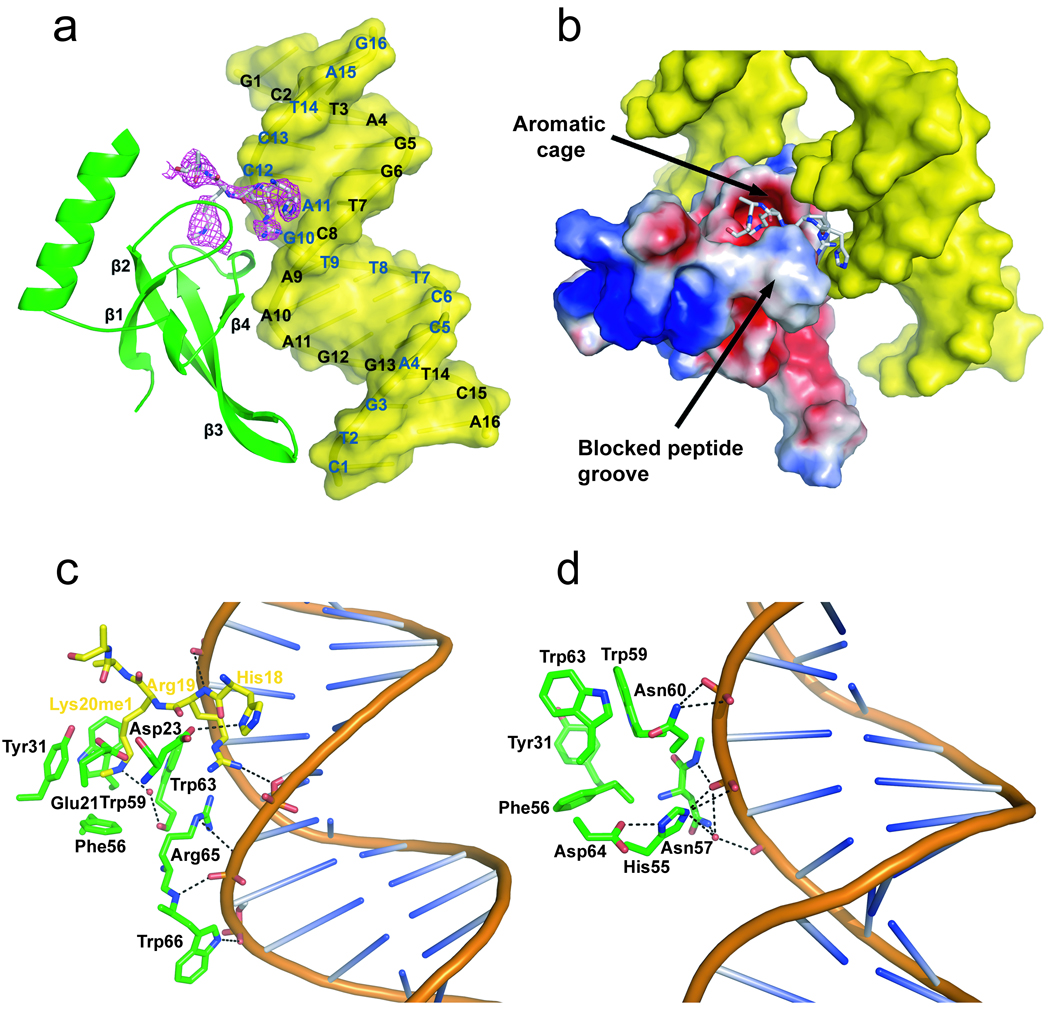
To delineate the mechanism of DNA-dependent peptide recognition, we solved the ternary structure of the human chromodomain with DNA2 and histone peptide at 2.3 Å resolution (Fig 2a and supplementary Table S1). MSL3 interacts exclusively with the minor groove and DNA backbone leading to 540 Å2 buried surface area. Within each asymmetric unit we identified two DNA interaction surfaces in MSL3. The interaction sites for DNA are adjacent to the aromatic cage (Fig. 2, and supplementary Fig. 3). These constitute Arg65 and Trp66 at one face, and His55, Asn57, and Asn60 at the other face (Fig. 2c, d). Arg65 mediates polar interactions with both the DNA and histone H4 tail (Fig. 2c). The R65A mutant results in a two-fold drop in affinity for DNA (Supplementary Table 2). Mutations R65A in conjunction with H55A decrease affinity for DNA by more than 10-fold (Supplementary Table 2). Superposition of MSL3 chromodomain and the sequence non-specific DNA-binding protein Sac7d from archaea 15 shows Arg65 in MSL3 and Arg42 in Sac7d contact the DNA minor groove. Although MSL3 chromodomain and Sac7d bind DNA with similar affinity, they exploit different DNA geometry (Supplementary Fig . 4a).
Figure 2. Structure of the MSL3 chromodomain in complex with DNA and the H4K20me1 peptide.
(a) Ribbon diagram of chromodomain (green) bound to DNA2 (yellow) and peptide (yellow); electron density from the refined |2Fo−Fc| map at 0.7 σ is in pink. The sphere within the aromatic cage is a water molecule. (b) Surface electrostatic potential of the chromodomain and its interactions with one peptide and two DNA duplexes. Residues (green) that constitute the primary (c) and secondary (d) DNA recognition surfaces. Residues Tyr31, Phe56, Trp59 and Tyr63 constitute an aromatic cage.
Residues Tyr31, Trp59 and Trp63 reinforce the walls, and Phe56 forms the floor of the MSL3 aromatic cage, which is 4 Å-deep and has a volume of 340 Å3. The lack of binding to the trimethylated H4 tail suggests access to the aromatic cage is regulated. His18 and Arg19 of the peptide position within the DNA minor groove, and the monomethyllysine inserts in the centroid of the aromatic cage (Fig. 2b). We found one ordered water molecule that hydrogen-bonded to the methylammonium moiety of Lys20 as well as to the amide group of Trp63. Although we did not observe electron density for Lys16 or Arg17 in the H4 peptide, the antagonizing effect of H4K16 acetylation can be explained by the destabilization of the interaction between the H4 peptide and DNA. The observation that residues N-terminal to the methyllysine are critical for binding was previously shown for the HP1 chromodomain 10 and human CHD1 tandem chromodomains 16. Here, we found the MSL3 chromodomain requires DNA for discriminatory interactions with the histone H4 tail, and a favorable interaction is restricted to the monomethylated peptide.
As compared to the deletion of the MSL3 chromodomain which rendered transgenic flies 50% viable, the W59G mutation led to 80% viability and a near-intact chromatin binding 8. We measured a two-fold decrease in affinity for DNA and no change in H4K20me1 binding in the W59G mutant (supplementary Table S2 and Fig. S2). Another study found that F56A mutation led to 64% viability of flies 9. Although we measured a subtle decrease in the affinity of F56A mutant for DNA, the binding to the H4K20me1 peptide was compromised (Supplementary Table 2). By relying on NMR analysis, we found neither W59G nor F56A mutation disturb the global structure of the MSL3 chromodomain (Supplementary Fig. 2), suggesting their phenotypes correspond to defects in chromatin targeting.
Chromatin undergoes compaction and decompaction for the appropriate control of transcription levels 17. ChIP-Seq analysis of the human genome has reported the presence of the H3K36me3 and H4K20me1 marks downstream of transcription start sites 18. As the MBT motif in the L3MBTL1 protein binds specifically to the H4K20me1 mark in recombinant nucleosome arrays 19, we modeled this potential interaction for the MSL3 chromodomain. Using the minor groove base pairs (A4–A10 in Fig. 2a,c) we superimposed the ternary complex on a minor groove adjacent to a H4 tail in the tetranucleosome structure 20. This superposition revealed the MSL3 chromodomain, the histone peptide and the other minor groove (Fig. 2d) register closely with the histone H4 tail and the DNA minor groove from an adjacent nucleosome (Supplementary Fig. 4c). Therefore, the corecognition of a methylated histone H4 tail and two adjacent nucleosomes may contribute to targeting of the MSL complex in vivo.
Supplementary Material
Acknowledgements
We thank Katrina Clines and Maks Chruszcz for technical support, and Thomas Conrad and Asifa Akhtar for initiating biological investigations. This work was supported by grants from NIH (GM070558) and AHA (0740058N) to S.K..
Footnotes
Author contributions. D.K., B.J.B, and S.K. designed, performed and analyzed experiments; V.C. assisted with DNA purification and mutagenesis. F. R. and P. H. assisted with crystallography. B.J.B and S.K. wrote the paper.
Accession codes. Protein Data Bank: Coordinates for the free and ternary complex of the MSL3 chromodomain have been deposited with accession codes 3M9Q and 3M9P, respectively.
References
- 1.Alekseyenko AA, et al. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straub T, Grimaud C, Gilfillan GD, Mitterweger A, Becker PB. PLoS Genet. 2008;4:e1000302. doi: 10.1371/journal.pgen.1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith ER, et al. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Mol Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucchesi JC. Curr Opin Genet Dev. 2009;19:550–556. doi: 10.1016/j.gde.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akhtar A, Zink D, Becker PB. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen PR, et al. J Biol Chem. 2005;280:32326–32331. doi: 10.1074/jbc.M501347200. [DOI] [PubMed] [Google Scholar]
- 8.Sural TH, et al. Nat Struct Mol Biol. 2008;15:1318–1325. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buscaino A, Legube G, Akhtar A. EMBO Rep. 2006;7:531–538. doi: 10.1038/sj.embor.7400658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs SA, Khorasanizadeh S. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, et al. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra V, et al. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Mizzen CA. Biochem Cell Biol. 2009;87:151–161. doi: 10.1139/O08-131. [DOI] [PubMed] [Google Scholar]
- 14.Nishioka K, et al. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 15.Robinson H, et al. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- 16.Flanagan JF, et al. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 17.Li B, Carey M, Workman JL. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Barski A, et al. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Trojer P, Reinberg D. Cell Cycle. 2008;7:578–585. doi: 10.4161/cc.7.5.5544. [DOI] [PubMed] [Google Scholar]
- 20.Schalch T, Duda S, Sargent DF, Richmond TJ. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.