Abstract
Several previous genome-wide and targeted association studies revealed that variants in the CHRNA5-CHRNA3-CHRNB4 (CHRNA5/A3/B4) gene cluster on chromosome 15 that encode the α5, α3 and β4 subunits of the nicotinic acetylcholine receptor (nAChRs) are associated with nicotine dependence (ND) in European Americans (EAs) or others of European origin. Considering the distinct linkage disequilibrium patterns in European and other ethnic populations such as African Americans (AAs), it would be interesting to determine whether such associations exist in other ethnic populations. We performed a comprehensive association and interaction analysis of the CHRNA5/A3/B4 cluster in two ethnic samples to investigate the role of variants in the risk for ND, which was assessed by Smoking Quantity, Heaviness Smoking Index, and Fagerström test for ND. Using a family-based association test, we found a nominal association of single nucleotide polymorphisms (SNPs) rs1317286 and rs8040868 in CHRNA3 with ND in the AA and combined AA and EA samples. Furthermore, we found that several haplotypes in CHRNA5 and CHRNA3 are nominally associated with ND in AA, EA, and pooled samples. However, none of these associations remained significant after correction for multiple testing. In addition, we performed interaction analysis of SNPs within the CHRNA5/A3/B4 cluster using the pedigree-based generalized multifactor dimensionality reduction method and found significant interactions within CHRNA3 and among the three subunit genes in the AA and pooled samples. Together, these results indicate that variants within CHRNA3 and among CHRNA5, CHRNA3, and CHRNB4 contribute significantly to the etiology of ND through gene-gene interactions, although the association of each subunit gene with ND is weak in both the AA and EA samples.
Keywords: Association analysis, CHRNA5, CHRNA3, CHRNB4, Interaction analysis, Nicotine dependence, Smoking
Introduction
Cigarette smoking is the most preventable cause of morbidity and death, resulting in more than 438,000 deaths in the United States in 2000 and approximately $193 billion in health-related economic losses (Mokdad and others 2004). Over the past four decades, cigarette smoking has caused an estimated 12 million deaths, including 4.1 million from cancers, 5.5 million from cardiovascular diseases, 2.1 million from respiratory diseases, and 94,000 infant deaths attributable to maternal smoking during pregnancy (www.cdc.gov/nccdphp/publications/aag/osh.htm.). Although cigarette smoke contains many noxious compounds, including carbon monoxide, acetaldehyde, and tobacco-specific nitrosamines, it is nicotine dependence (ND) that maintains the continued use of tobacco (Stolerman and Jarvis 1995; USDHHS 2000; WHO 2002). Twin and family studies reveal that genetics contributes significantly to ND, with an estimated mean heritability of 0.56 in adult smokers (Li and others 2003).
The psychopharmacologic effects of nicotine are mediated primarily by functionally diverse neuronal nicotinic acetylcholine receptors (nAChRs), a family of ligand-gated ion channels widely distributed in the brain (Gaimarri and others 2007; Picciotto and others 2000; Watkins and others 2000). These nAChRs are involved in numerous physiological functions both in the brain and in the periphery as well (Gotti and Clementi 2004). To date, 12 neuronal nAChR subunits have been identified, consisting of nine α (α2-α10) and three β (β2-β4) subunits. The human genes for all of these subunits except α8 have been cloned (Graham and others 2002). The 11 nAChR subunit genes are located on chromosomes 1, 4, 8, 11, 15, and 20. Of these, CHRNA5, CHRNA3, and CHRNB4 are grouped in a cluster on chromosome 15q24 (Raimondi and others 1992), in which CHRNA3 and CHRNA5 are located in a tail-to-tail configuration on opposite DNA strands and share some of their 3′-untranslated region (Duga and others 2001). Similarly, CHRNB3 and CHRNA6 are grouped in a cluster on chromosome 8p11. The clustered arrangement of CHRNA5/A3/B4 and CHRNB3/A6 could be related to control of the expression of these genes (Flora and others 2000; Xu and others 2006).
Several subunit genes have been investigated for association with ND and other smoking-related behaviors in human subjects [for reviews, see (Lessov-Schlaggar and others 2008; Li and Burmeister 2009)]. In a recent study, Saccone et al. (2007) reported associations of multiple single nucleotide polymorphisms (SNPs) in the CHRNA5/A3/B4 cluster with ND. However, the significance of these results did not survive correction for multiple testing. More recently, several genome-wide and candidate gene-based association studies provided further evidence for the association of variants of the CHRNA5/A3/B4 gene cluster with various nicotine-related behaviors (Berrettini and others 2008; Bierut and others 2008; Chen and others 2009; Greenbaum and others 2006; Saccone and others 2007; Schlaepfer and others 2008; Sherva and others 2008; Weiss and others 2008). In contrast, other studies have failed to reveal a significant association of this gene cluster with ND or other smoking-related phenotypes (Uhl and others 2008; Vink and others 2009).
Considering that the participants in these studies were primarily European Americans (EAs) or of European origin, and given the distinct differences in linkage disequilibrium (LD) patterns across ethnic populations, it remains unknown whether this gene cluster plays a role in the etiology of ND in other ethnic groups. The current study was designed to examine this possibility among a relatively large sample of African Americans (AAs) in comparison with EAs.
Materials and Methods
Participants and ND measures
Participants were recruited primarily from the Mid-South states of Tennessee, Mississippi, and Arkansas during 1999-2004. Extensive clinical and medical data were collected on each participant, including demographics (e.g., sex, age, race, biological relationships, weight, height, years of education, and marital status), medical history, smoking history and current smoking behavior, ND, and other relevant personal characteristics. The enrollment criteria for proband smokers were: 1) at least 21 years old, 2) smoker for at least five years, 3) consumer of an average of 20 cigarettes per day during the last 12 months, and 4) free of a self-reported history of diagnosis of any serious mental disorder; e.g., schizophrenia or other psychosis, bipolar disorder, or cognitive disorder (such as dementia or Alzheimer’s disease). Once a proband was recruited, siblings and parents were invited to join the study whenever possible. All participants provided informed consent; the study protocol and forms/procedures were approved by all participating Institutional Review Boards.
The ND of each smoker was ascertained by three of the measures used most commonly in tobacco research: Smoking Quantity (SQ; defined as the number of cigarettes smoked per day), the Heaviness of Smoking Index (HSI; 0-6 scale), and the Fagerström Test for ND score (FTND; 0-10 scale) (Heatherton and others 1991). The SQ provides a simple, quantified index of consumption (using a 0–3 point compressed format), whereas HSI includes one item addressing SQ and another assessing smoking urgency on awakening. The FTND score includes the HSI plus other indicators of behavioral propensity to smoke under various circumstances. Given the presence of overlap in the content of the three ND measures, there exist fairly robust correlations among them (r = 0.88 ~ 0.94) in both the AA and EA samples. The primary reasons for employing all three measures were that there is a lack of consensus regarding the best approach to assess ND as a phenotype and to permit maximum cross-referencing with previous studies of ND.
The participants were 1366 individuals from 402 AA families and 671 individuals from 200 EA families. The average age was 39.4 ± 14.4 (SD) years for the AA and 40.5 ± 15.5 years for the EA participants. The average nuclear family size was 3.14 ± 0.75 for AAs and 3.17 ± 0.69 for EAs. There were 1053 and 515 smokers in the AA and EA, samples, respectively, whereas all the rest had smoked fewer than 100 cigarettes in their lifetimes. The average HSI and FTND scores for smokers were 3.7 ± 1.4 and 6.26 ± 2.15, respectively for AAs and 3.9 ± 1.4 and 6.33 ± 2.22 for EAs. Age of smoking onset was 17.3 ± 4.7 (mean ± SD; years) and 15.5 ± 4.4, for the AA and EA samples, respectively. Years smoked were 20.4 ± 12.5 (mean ± SD) and 23.2 ± 13.5 for the AA and EA samples, respectively. The average number of cigarettes smoked per day was 19.4 ± 13.3 for AA smokers and 19.5 ± 13.4 for EA smokers. Detailed demographic and clinical characteristics of the two samples have been reported elsewhere (Li and others 2005; Li and others 2008b; Li and others 2006)
DNA samples, SNP selection, and SNP genotyping
DNA was isolated from the blood of each participant using a DNA Maxi kit from Qiagen, Inc. (Valencia, CA). A total of 22 SNPs, with 7 from CHRNA5, 11 from CHRNA3, and 4 from CHRNB4, were selected from either published papers (Berrettini and others 2008; Saccone and others 2007) or the NCBI SNP database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=snp) on the basis of the potential biological functions of the gene, high heterozygosity with a minor allele frequency (MAF) > 0.15, and association analysis results from other studies (Berrettini and others 2008; Saccone and others 2007), as well as an attempt to obtain uniform coverage of the gene/region. Detailed information about these SNPs (alleles, physical position, allele frequency, and primer/probe information from ABI) is summarized in Table 1.
Table 1.
Positions, nucleotide variation, allele frequency, and primer/probe sequences
| Gene | Marker number | dbSNP ID | Chrom. pos. | Alleles | SNP location | MAF (EA)* | MAF (AA)* | Primer and probe# |
|---|---|---|---|---|---|---|---|---|
| CHRNA5 | 1 | rs684513 | 76645455 | C/G | Intron1 | 0.217 | 0.158 | C_7051_10 |
| 2 | rs621849 | 76659916 | A/G | Intron1 | 0.375 | 0.425 | C_5864_10 | |
| 3 | rs637137 | 76661031 | A/T | Intron1 | 0.202 | 0.259 | C_5866_10 | |
| 4 | rs951266 | 76665596 | C/T | Intron2 | 0.417 | 0.117 | C_9510534_10 | |
| 5 | rs17408276 | 76668673 | C/T | Intron4 | 0.396 | 0.325 | C_34624626_10 | |
| 6 | rs16969968 | 76669980 | A/G | Exon5 | 0.424 | 0.205 | C_26000428_20 | |
| 7 | rs615470 | 76673043 | C/T | 3′ UTR | 0.325 | 0.342 | C_18757_10 | |
|
| ||||||||
| CHRNA3 | 8 | rs578776 | 76675455 | C/T | 3′ UTR | 0.242 | 0.342 | C_721253_10 |
| 9 | rs6495307 | 76677376 | C/T | Intron5 | 0.345 | 0.307 | C_1713433_10 | |
| 10 | rs1051730 | 76681394 | C/T | Exon5 | 0.4 | 0.11 | C_9510307_20 | |
| 11 | rs3743078 | 76681814 | C/G | Intron4 | 0.203 | 0.333 | C_27472090_10 | |
| 12 | rs1317286 | 76683184 | A/G | Intron4 | 0.41 | 0.3 | C_9510308_10 | |
| 13 | rs12914385 | 76685778 | C/T | Intron4 | 0.43 | 0.198 | C_12106059_10 | |
| 14 | rs2869546 | 76694400 | C/T | Intron4 | 0.333 | 0.322 | C_15836855_10 | |
| 15 | rs6495308 | 76694711 | C/T | Intron4 | 0.23 | 0.34 | C_222570_10 | |
| 16 | rs3743075 | 76696507 | A/G | Exon4 | 0.317 | 0.442 | C_27472089_10 | |
| 17 | rs8040868 | 76698236 | C/T | Exon2 | 0.457 | 0.364 | C_261698_10 | |
| 18 | rs6495309 | 76702300 | C/T | Promoter | 0.2 | 0.25 | C_30730895_10 | |
|
| ||||||||
| CHRNB4 | 19 | rs1948 | 76704454 | C/T | 3′ UTR | 0.267 | 0.117 | C_11941837_10 |
| 20 | rs12441088 | 76715319 | G/T | Intron1 | 0.25 | 0.3 | C_30730878_10 | |
| 21 | rs3813567 | 76721606 | C/T | Promoter | N/F | N/F | C_27488001_10 | |
| 22 | rs11637890 | 76722474 | C/G | Promoter | 0.347 | 0.067 | C_31121610_10 | |
Notes:
Based on the allele frequency from the NCBI SNP database
ID of primer/probe provided by Applied Biosystems Inc.
All SNPs were genotyped using the TaqMan assay in a 384-well microplate format (Applied Biosystems Inc., Foster City, CA). Briefly, 15 ng of DNA was amplified in a total volume of 7 μl containing an MGB probe and 2.5 μl of TaqMan universal PCR master mix. The amplification conditions were 2 min at 50°C and 10 min at 95°C followed by 40 cycles of 95°C for 25 sec and 60°C for 1 min. Allelic discrimination analysis was performed on the ABI Prism 7900HT Sequence Detection System. To ensure the quality of the genotyping, eight positive and negative control DNA samples were added to each 384-well reaction plate.
Association analysis
The PedCheck program (O’Connell and Weeks 1998) was used to detect any genotyping inconsistencies. To avoid potential bias, the 41 inconsistencies in the AA sample and 24 in the EA sample, detected in approximately 46,200 assays (i.e., 0.14% genotyping error) for 22 SNPs across all DNA samples, were excluded from statistical analysis. Pair-wise linkage LD between all possible SNP pairs was assessed using the Haploview (v. 4.0) program (Barrett and others 2005) with the option of determining haplotype blocks according to the definitions proposed by Gabriel et al. (2002). Associations between individual SNPs and the three ND measures were determined with the PBAT (v. 3.5) program using generalized estimating equations (Lange and others 2003). Associations between each ND measure and haplotypes from various SNP combinations were calculated using the FBAT (v. 1.7.3) program with the option of computing the P value of the Z statistic using Monte Carlo sampling under the null distribution of no linkage and no association (Horvath and others 2004). Three genetic models (additive, dominant, and recessive) were tested for both individual and multi-locus SNPs (i.e., haplotypes). For all PBAT and FBAT association tests, sex and age were used as covariates in the EA and AA samples and sex, age, and ethnicity as covariates in the combined sample. All significant associations were corrected for multiple testing according to the SNP spectral decomposition approach (SNPSpD)(Nyholt 2004) for individual SNP analysis and using Bonferroni correction by dividing the significance level by the number of major haplotypes (> 5.0% in frequency) for haplotype-based association analysis.
Interaction analysis of variants in CHRNA5/A3/B4 cluster
Gene–gene interactions of SNPs in the CHRNA5/A3/B4 cluster were analyzed using a newly developed pedigree-based generalized multifactor dimensionality reduction (PGMDR) method (Lou and others 2008). As with the previous association analyses, age and sex were included as covariates for the EA and AA samples and age, sex, and ethnicity as covariates in the pooled sample. Gene–gene interactions were examined for all two- to seven-locus models. The top-ranked interaction model was chosen for a given order interaction, and its P value of prediction accuracy (PA) was evaluated by a permutation test based on 10,000 shuffles of the adjusted phenotypic values. Because all P values reported here were based on permutation tests of each given interaction model, no correction was needed. On the other hand, we considered a P value of 0.0083 (i.e., 0.05/6) as an experiment-wide corrected one at the 0.05 significance level given that our gene–gene interaction analyses considered six interactive models for each sample. For detailed information on the PGMDR approach, please refer to the paper by Lou et al. (2008).
Power analysis
Power analysis for single-locus main effects as well as two-locus interactive effects was performed using the Quanto program (version 1.2.4; http://hydra.usc.edu/gxe/).
Results
Individual SNP-based association analysis
We evaluated 22 SNPs in the CHRNA5/A3/B4 cluster in two independent ethnic samples. Association of individual SNPs with the three ND measures was determined with the PBAT-GEE program (Lange and others 2003), and the results are shown in Table 2. For CHRNA3, we found marginal associations for rs1317286 with SQ and HSI (P = 0.02 and 0.05, respectively) and of rs8040868 with SQ (P = 0.019) in the combined sample. These findings were not significant after correction for multiple testing. As for CHRNA5 and CHRNB4, no significant association was detected for any individual SNP.
Table 2.
Allele frequencies and P values for associations of individual SNPs with three ND measures in the EA, AA, and combined samples
| European-American sample | African-American sample | Combined sample | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP ID | Alleles | Freq. | SQ | HSI | FTND | Freq. | SQ | HSI | FTND | Freq. | SQ | HSI | FTND |
| CHRNA5 | rs684513 | C/G | 0.81/0.19 | 0.16r | 0.16d | 0.10d | 0.80/0.20 | 0.23d | 0.37d | 0.47a | 0.81/0.19 | 0.29a | 0.39a | 0.36a |
| rs621849 | A/G | 0.59/0.41 | 0.18a | 0.28r | 0.38r | 0.56/0.44 | 0.20d | 0.094d | 0.061d | 0.57/0.43 | 0.38a | 0.13d | 0.063d | |
| rs637137 | A/T | 0.22/0.78 | 0.57d | 0.12a | 0.20a | 0.29/0.71 | 0.22d | 0.24d | 0.20d | 0.27/0.73 | 0.19d | 0.19d | 0.19d | |
| rs951266 | C/T | 0.34/0.63 | 0.17a | 0.14a | 0.44a | 0.10/0.90 | 0.76d | 0.89d | 0.81a | 0.18/0.82 | 0.35a | 0.29a | 0.48a | |
| rs17408276 | C/T | 0.35/0.65 | 0.16a | 0.40a | 0.71d | 0.13/0.87 | 0.61r | 0.51d | 0.58d | 0.19/0.81 | 0.23a | 0.65d | 0.69r | |
| rs16969968 | A/G | 0.37/0.63 | 0.17a | 0.088a | 0.26a | 0.04/0.96 | 0.62a | 0.51d | 0.61a | 0.14/0.86 | 0.15a | 0.11a | 0.22a | |
| rs615470 | C/T | 0.65/0.35 | 0.085a | 0.21a | 0.54a | 0.58/0.42 | 0.77d | 0.60d | 0.47d | 0.61/0.39 | 0.45a | 0.76d | 0.65d | |
|
| ||||||||||||||
| CHRNA3 | rs578776 | C/T | 0.27/0.73 | 0.78d | 0.38a | 0.64a | 0.54/0.46 | 0.61d | 0.51a | 0.39a | 0.46/0.54 | 0.61d | 0.37a | 0.36a |
| rs6495307 | C/T | 0.59/0.41 | 0.23d | 0.48d | 0.69d | 0.58/0.42 | 0.79d | 0.62a | 0.61a | 0.58/0.42 | 0.46d | 0.88d | 0.87r | |
| rs1051730 | C/T | 0.37/0.63 | 0.18d | 0.11d | 0.29d | 0.11/0.89 | 0.38d | 0.58d | 0.43d | 0.19/0.81 | 0.14d | 0.25a | 0.33d | |
| rs3743078 | C/G | 0.23/0.77 | 0.46d | 0.29a | 0.34a | 0.59/0.41 | 0.59d | 0.37r | 0.33d | 0.48/0.52 | 0.59a | 0.25d | 0.22d | |
| rs1317286 | A/G | 0.62/0.38 | 0.23d | 0.21d | 0.54d | 0.75/0.25 | 0.07d | 0.15d | 0.10d | 0.71/0.29 | 0.02 d | 0.05 d | 0.07d | |
| rs12914385 | C/T | 0.58/0.42 | 0.23d | 0.21d | 0.44d | 0.84/0.16 | 0.12d | 0.14d | 0.14d | 0.76/0.24 | 0.55d | 0.79r | 0.52d | |
| rs2869546 | C/T | 0.37/0.63 | 0.17d | 0.49d | 0.70d | 0.39/0.61 | 0.58r | 0.56d | 0.36r | 0.38/0.62 | 0.28d | 0.62d | 0.48d | |
| rs6495308 | C/T | 0.84/0.16 | 0.36d | 0.15d | 0.14d | 0.80/0.20 | 0.43a | 0.39d | 0.41d | 0.81/0.19 | 0.68d | 0.77d | 0.66d | |
| rs3743075 | A/G | 0.64/0.36 | 0.12a | 0.24a | 0.51a | 0.56/0.44 | 0.37d | 0.52d | 0.36r | 0.59/0.41 | 0.18a | 0.32a | 0.32d | |
| rs8040868 | C/T | 0.43/0.57 | 0.30a | 0.27d | 0.49d | 0.35/0.65 | 0.02 d | 0.039 d | 0.017 d | 0.37/0.63 | 0.019 d | 0.07a | 0.08a | |
| rs6495309 | C/T | 0.16/0.84 | 0.79d | 0.29d | 0.34a | 0.18/0.82 | 0.27d | 0.25d | 0.28d | 0.17/0.83 | 0.40d | 0.39d | 0.38d | |
|
| ||||||||||||||
| CHRNB4 | rs1948 | C/T | 0.31/0.69 | 0.15d | 0.25d | 0.42d | 0.22/0.78 | 0.79a | 0.43d | 0.34a | 0.25/0.75 | 0.29d | 0.42d | 0.31d |
| rs12441088 | G/T | 0.23/0.77 | 0.23a | 0.23a | 0.51a | 0.32/0.68 | 0.57d | 0.23d | 0.15d | 0.29/0.71 | 0.58d | 0.19d | 0.13d | |
| rs3813567 | C/T | 0.82/0.18 | 0.38d | 0.11a | 0.22a | 0.81/0.19 | 0.40d | 0.66d | 0.86d | 0.81/0.19 | 0.52d | 0.56r | 0.52a | |
| rs11637890 | C/G | 0.63/0.37 | 0.35d | 0.36d | 0.43d | 0.83/0.17 | 0.76d | 0.47d | 0.47a | 0.77/0.23 | 0.47d | 0.49d | 0.55d | |
Notes:
1) Significant associations at the 0.05 level before correction for multiple testing are given in bold.
2) Superscripts indicate genetic model used for analysis:
= additive
= dominant
= recessive.
3) For each ethnic-specific sample, age and sex were used as covariates; for the Combined sample, age, sex, and ethnicity were used as covariates.
We found substantial differences between the EA and AA samples in the allele frequencies for several SNPs (Table 2). The presence of these allele frequency discrepancies suggests genetic differences underlying ND. To eliminate any potential effect of ethnic differences on our association results, we performed independent association analyses within the individual samples. For the AA sample, rs8040868 within CHRNA3 was significantly associated with all three adjusted ND measures (P = 0.017 to 0.039); however, no association remained significant after correction for multiple testing. No significant association with ND was detected for any individual SNP in the three genes.
Haplotype block structure and LD analysis
The pair-wise D’ values of 22 SNPs for the three genes in a total of 77 kb of nucleotide sequence were determined using the Haploview program (Barrett and others 2005). On the basis of the block definition proposed by Gabriel et al. (2002), we found four blocks within the gene cluster in the AA, EA, and pooled samples (Figure 1). One block of 4.0 kb containing rs12441088 and rs3813567 in CHRNB4 was observed for the three samples; three other blocks differed across the AA and EA samples, indicating the presence of different genetic architectures.
Figure 1.
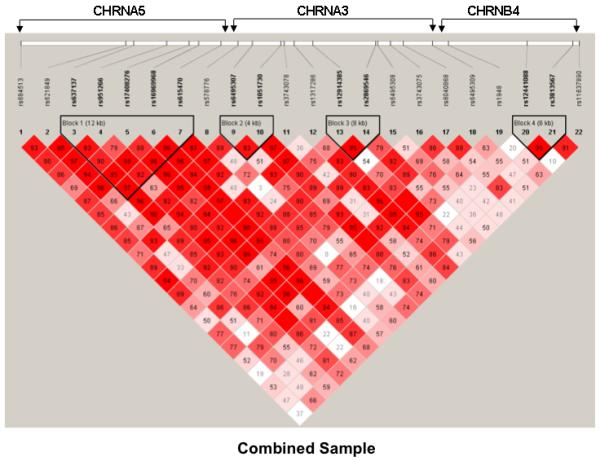
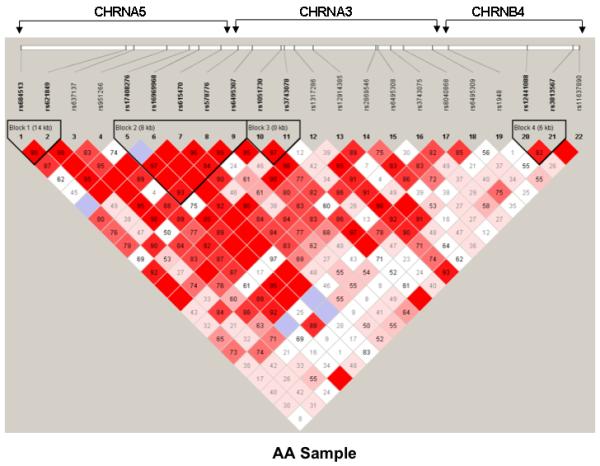
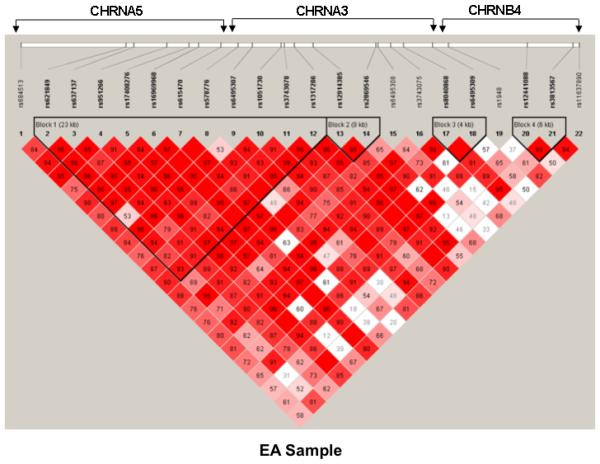
Haploview-generated LD patterns for 22 SNPs within the CHRNA5/A3/B4 cluster in the pooled (A), AA (B), and EA (C) samples. Pair-wise LD between all SNPs was evaluated using the Haploview program (Barrett and others 2005) with the option of determining haplotype blocks according to the criteria defined by Gabriel et al. (2002). The number in each box represents the D’ value for each SNP pair.
Haplotype-based association analysis
Using the FBAT program, we performed haplotype-based association analysis for all possible haplotypes in consecutive three-SNP combinations for the three ND measures. In the combined sample, we found a major haplotype, T-A-C, formed by SNPs rs17408276, rs16969968, and rs615470 in CHRNA5 at a frequency of 14%, that was significantly associated with SQ (P = 0.018) and HSI (P = 0.019) under the additive model (Table 3). We also found three major haplotypes in CHRNA3, G-C-T (11%; formed by rs1317286, rs12914385, and rs2869546), C-G-C (34%; formed by rs6495308, rs3743075, and rs8040868), and A-T-T (36%; formed by rs3743075, rs8040868, and rs6495309), which showed significant association with at least two of the three ND measures (P = 0.016 ~ 0.047; Table 3). However, none of these associations remained significant after correction for multiple testing of major haplotypes.
Table 3.
Haplotypes within CHRNA5 and CHRNA3 associated with three ND measures in the combined sample
| CHRNA5 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SQ |
HSI |
FTND |
|||||||||||||||||
| 5 | 6 | 7 | Freq | P- haplotype |
Z- score |
P- global |
No. of families |
P- haplotype |
Z- score |
P- global |
No. of families |
P- haplotype |
Z- score |
P- global |
No. of families |
||||
| T | A | C | 0.14 | 0.019 a | 2.33 | 0.073 | 121 | 0.018 a | 2.357 | 0.06 4 |
121 | 0.065 | 1.849 | 0.15 | 126 | ||||
| T | G | C | 0.47 | 0.33a | −0 .968 | 285 | 0.11a | −1.58 | 286 | 0.14 | −1.481 | 298 | |||||||
| CHRNA3 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | |||||||||||||
| G | C | T | 0.11 | 0.016 a | 2.4 | 0.064 | 125 | 0.041 a | 2.047 | 0.09 | 123 | 0.035 a | 2.108 | 0.24 | 132 | ||||
| A | C | C | 0.38 | 0.81a | −0.236 | 321 | 0.82a | 0.233 | 320 | 0.86a | 0.18 | 334 | |||||||
| C | G | C | 0.34 | 0.016 d | 2.402 | 0.16 | 200 | 0.038 d | 2.074 | 0.34 | 205 | 0.029 d | 2.171 | 0.31 | 218 | ||||
| C | C | T | 0.35 | 0.092a | −1.684 | 278 | 0.12a | −1.56 | 276 | 0.16a | −1.422 | 290 | |||||||
| A | T | T | 0.36 | 0.027 a | −2.2 | 0.17 | 278 | 0.047 a | −1.984 | 0.21 | 276 | 0.057r | −1.902 | 0.21 | 101 | ||||
| G | C | T | 0.34 | 0.083d | 1.729 | 196 | 0.18d | 1.353 | 208 | 0.19d | 1.314 | 217 | |||||||
Notes:
1) Only major haplotypes are shown; the one with a P value <0.05 is given in bold.
2) For the 0.05 significance level, significant P value after Bonferroni correction for four major haplotypes within both CHRNA5 and CHRNA3 is 0.0125.
3) Superscripts indicate the genetic model used in the analysis
= additive
= dominant
= recessive.
4) The ND measures were corrected for age, sex, and ethnicity.
In the AA sample, we found two major haplotypes in CHRNA3, with the first, G-C-T, formed by rs1317286-rs12914385-rs2869546, at a frequency of 14% showing significant associations with all the three ND measures (P = 0.016 ~ 0.043); and a second one, T-C-G, formed by rs2869546-rs6495308-rs3743075, at a frequency of 41%, revealing a significant association with SQ (P = 0.041; Table 4). Again, none of these associations remained significant after correction for multiple testing.
Table 4.
Haplotypes within CHRNA3 associated with three ND measures in the AA sample
| SQ | HSI | FTND | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 13 | 14 | 15 | 16 | Freq | P- haplotype |
Z- score |
P- global |
No. of families |
P- haplotype |
Z- score |
P- global |
No. of families |
P- haplotype |
Z- score |
P- global |
No. of families |
| G | C | T | 0.14 | 0.016 a | 2.397 | 0.12 | 126 | 0.043 a | 2.023 | 0.29 | 126 | 0.036 a | 2.09 | 0.35 | 128 | ||
| A | C | C | 0.37 | 0.91a | −0.114 | 242 | 0.83a | 0.217 | 243 | 0.91a | −0.114 | 245 | |||||
| T | C | G | 0.41 | 0.041 r | −2.046 | 0.06 | 100 | 0.10r | −1.63 | 0.18 | 100 | 0.18r | −1.355 | 0.19 | 100 | ||
| C | C | C | 0.33 | 0.34 | −0.963 | 70 | 0.45 | −0.76 | 70 | 0.29 | −1.069 | 70 | |||||
Notes:
1) Only major haplotypes are shown; the one with a P value <0.05 is given in bold.
2) For the 0.05 significance level, significant P value after Bonferroni correction for four major haplotypes is 0.0125.
3) Superscripts indicate the genetic model used in the analysis
= additive
= dominant.
4) The ND measures were corrected for age and sex.
Regarding the EA sample, we also found two haplotypes each in CHRNA5 and CHRNA3 that showed significant associations with HSI (P = 0.031~0.038) or FTND (P = 0.035~0.041) or both. None of these associations remained significant after correction for multiple testing.
Interaction analysis of CHRNA5/A3/B4 gene cluster
With the PGMDR method, we analyzed interactions for all possible combinations of two to seven SNPs within the CHRNA5/A3/B4 cluster. Table 6 shows the best interactive model for three ND measures in the AA and pooled samples. In the AA sample, we found the best interaction model for three loci (i.e., rs578776, rs3743078, and rs1317286 in CHRNA3) with a P value of 0.045 for HSI and 0.003 for FTND. For the four-locus model (i.e., rs684513 and rs615470 in CHRNA5; rs1317286 in CHRNA3; rs12441088 in CHRNB4), P values of 0.002 for SQ, 0.016 for HSI, and 0.041 for FTND were identified. Finally, for the six-locus model (i.e., rs684513 and rs621849 in CHRNA5; rs578776, rs1317286, and rs12914385 in CHRNA3; rs12441088 in CHRNB4), P values of 0.005 for SQ, 0.003 for HSI, and 0.044 for FTND were noted. All three models remained significant at the experiment-wide level for at least one ND measure.
Table 6.
Best interactive models for CHRNA5/A3/B4 cluster with ND in the AA and pooled sample on the basis of prediction accuracy and empirical P value from 10,000 permutations
| Sample | No. of loci |
Best Model | ND Measure |
Prediction Accuracy |
P value |
|---|---|---|---|---|---|
| AA | 3 | CHRNA3: rs578776; rs3743078; rs1317286 | HSI | 0.532 | 0.045 |
| FTND | 0.5482 | 0.003 | |||
| 4 |
CHRNA5: rs684513; rs615470 CHRNA3: rs1317286 CHRNB4: rs12441088 |
SQ | 0.553 | 0.002 | |
| HSI | 0.543 | 0.016 | |||
| FTND | 0.531 | 0.041 | |||
| 6 |
CHRNA5: rs684513, rs621849 CHRNA3: rs578776; rs1317286; rs12914385 CHRNB4: r12441088 |
SQ | 0.546 | 0.005 | |
| HSI | 0.549 | 0.003 | |||
| FTND | 0.529 | 0.044 | |||
| Pooled sample |
2 | CHRNA3: rs3743078; rs1317286 | SQ | 0.511 | 0.165 |
| HSI | 0.512 | 0.141 | |||
| FTND | 0.525 | 0.031 | |||
| 3 |
CHRNA5: rs621849 CHRNA3: rs3743078 CHRNB4: rs11637890 |
SQ | 0.530 | 0.011 | |
| HSI | 0.520 | 0.061 | |||
| FTND | 0.516 | 0.106 |
For the pooled sample, we identified two interactive models. The first consisted of two SNPs (rs3743078 and rs1317286) in CHRNA3, and the second consisted of SNPs rs621849 in CHRNA5, rs3743078 in CHRNA3, and rs11637890 in CHRNB4. However, neither model remained significant at the experiment-wide level after correction. We performed an identical interaction analysis on the EA sample, but no significant interactive model emerged.
Discussion
In the current study, we examined the genetic associations and interactions between variations in the CHRNA5/A3/B4 gene cluster and ND in two independent samples. Individual SNP-based association analyses revealed that rs8040868 in CHRNA3 was significantly associated with all the ND measures in the AA sample and with SQ in the combined sample, whereas rs1317286 revealed relationships with SQ and HSI in the combined sample. We also performed haplotype-based association analyses on all SNPs using a sliding window approach. In the CHRNA3 gene, one common haplotype (G-C-T) for SNPs rs1317286-rs12914385-rs2869546 in the combined and AA samples showed a significant positive association with ND. Moreover, we found haplotypes C-G-C/T, formed by rs6495308, rs3743075, and rs8040868, and A/G-T-T, formed by rs3743075, rs8040868, and rs6495309 in CHRNA5, to have a marginal association with ND in the combined and EA samples. Haplotype T-A-C, formed by rs17408276-rs16969968-rs615470 in CHRNA5, was significantly associated with SQ and HSI in the combined sample and only HSI in the EA sample, whereas the allelic T-G-C of this haplotype showed an inverse association with HSI in the EA sample. Although all the aforementioned haplotypes contain one or two SNPs of rs16969968, rs1317286, rs6495308, and rs8040868 that were found to have significant associations with ND in previous reports (Berrettini and others 2008; Saccone and others 2007), none of these associations remained significant in the present study after Bonferroni correction. Considering the fact that these subunit genes must assemble together in order to form functional nAChRs, we performed gene–gene interaction analysis on all SNPs in the gene cluster and found the three best interactive models in the AA sample and two in the pooled samples; only those for the AA sample remained significant at the experiment-wide level.
Recently, three independent genome-wide association studies (GWAS) provided strong evidence for associations between the variants of the CHRNA5/A3/B4 cluster and lung cancer (Amos and others 2008; Hung and others 2008; Liu and others 2008; McKay and others 2008; Thorgeirsson and others 2008; Wang and others 2008). Regarding the association of this gene cluster with ND, the results are less clear. One study (Thorgeirsson and others 2008) revealed a significant association of rs1051730 within CHRNA3 with smoking quantity (P = 5.0 × 10−16) in approximately 11,000 Icelandic smokers, whereas another study (Amos and others 2008) reported weak evidence of an association of SNPs rs1051730 and rs8034191 with smoking behavior in former but not current smokers. The third study (Hung and others 2008) found no association of SNPs 8034191 and rs16969968 with smoking dependence. Several other studies provide additional independent evidence pertaining to the association of variants of this gene cluster with ND. For example, Saccone et al. (2007) first reported significant associations of CHRNA5/A3/B4 cluster variants with ND in a large-scale candidate gene-based case-control study of 1050 cases and 879 controls of European ancestry where cases were defined as individuals with an FTND score of ≥ 4 and controls were individuals who smoked at least 100 cigarettes over a lifetime but had an FTND score of 0. Their results indicated rs16699968 (P = 0.0006), a nonsynonymous coding SNP in exon 5 of CHRNA5, and rs578776 (P = 0.0003) in the 3′-UTR of CHRNA3 showed the strongest associations. In another independent GWAS study, Berrettini et al. (2008) identified two other SNPs in CHRNA3 (rs1317286 and rs6495308) associated with cigarettes per day (CPD; P = 0.000003 and 0.00007, respectively) in three independent European samples, although none of the individual SNPs remained statistically significant after correction for multiple testing at the genome-wide level. Most recently, a candidate gene-based study (Weiss and others 2008) revealed a significant association of the variants in this cluster with degree of ND in participants of European origin who began daily smoking at or before the age of 16, but not among those who began daily smoking later, suggesting the association between the CHRNA5/A3/B4 region and ND is dependent on developmental stage.
There is no question that significant progress has been made in identifying the CHRNA5/A3/B4 cluster as a susceptibility locus for smoking and lung cancer. However, several important issues remain to be addressed. First, it is important to determine which variant(s) is responsible for the associations with ND and lung cancer. Within this region, rs1051730 has been associated with both ND (Saccone and others 2007; Thorgeirsson and others 2008; Weiss and others 2008) and lung cancer (Amos and others 2008; Hung and others 2008; Thorgeirsson and others 2008) whereas rs8034191 is associated primarily with lung cancer (Amos and others 2008; Hung and others 2008). In addition, several other SNPs, such as rs16969968, rs1317286, and rs6495308, are associated with ND. Of these, rs16969968 is most noteworthy because it represents a nonsynonymous α5 coding variant (e.g., aspartic acid to asparagine at position 398 of the polypeptide chain), resulting in a substitution of a negatively charged residue within the M3–M4 intracellular loop, a region thought to be involved in receptor trafficking. A recent function study (Bierut and others 2008) showed that the variant forms of the α5 subunit alter receptor function without affecting expression. In consideration of the extensive LD within this region, a resequencing effort is needed to address this issue. Second, it is necessary to determine if the CHRNA5/A3/B4 cluster also plays a significant role in the etiology of ND and lung cancer in other ethnic populations, such as those with Asian, African, and Hispanic origins, given that most participants to date have been of European descent, the current study being a notable exception. From information from International HapMap project and from the current study, we know LD structures differ greatly across ethnicities. Thus, it is important to determine if the association between this region and both lung cancer and ND exists, and whether the same or different functional variants are responsible (Briollais and others 2007).
More research is needed to determine which smoking-related characteristics are associated with the cluster. For example, ND was measured by SQ or SQ levels in the studies reported by Berrettini et al. (2008) and Thorgeirssone et al. (2008), whereas the study reported by Sacconne et al. (2007) focused on searching for susceptibility genes responsible for the transition from non-dependent to dependent smoking. On the other hand, Weiss et al. (2008) found a significant association of the same cluster with ND only in subjects who began daily smoking at or before the age of 16. Considering these findings in light of several published and unpublished studies that reveal no or weak evidence for the association of this cluster with ND (Vink and others 2009), additional research is needed to investigate the role of the cluster in a variety of smoking phenotypes. For example, it was recently reported that no association exists between this cluster and either smoking initiation or smoking cessation (Hung and others 2008; Thorgeirsson and others 2008; Uhl and others 2008).
Although the association of the CHRNA5/A3/B4 cluster with ND has received much attention recently, other nAChR subunit genes may play significant roles in the etiology of ND as well. For example, significant associations with various SNPs in CHRNA4 have been detected in at least three independent studies (Feng and others 2004; Hutchison and others 2007; Li and others 2005). Although no significant association of ND with CHRNB2 has been found in four independent studies (Feng and others 2004; Li and others 2005; Lueders and others 2002; Silverman and others 2000), recent research revealed that rs2072658 and rs2072661 in the 3′-UTR of CHRNB2 are associated with a reduced risk of smoking initiation, greater ability to quit smoking, and diminished early response to nicotine (Conti and others 2008; Ehringer and others 2007). Very recently, we showed a significant interaction between variants of CHRNA4 and CHRNB2 in affecting ND, concluding that CHRNB2 has a significant effect on ND when analyzed in interaction with CHRNA4. A smaller study in schizophrenic smokers suggested that CHRNA7 may be associated with smoking status (De Luca and others 2004), although the relevance of this finding to the general population of smokers is unknown. Variants in CHRNB1, CHRNB3, and CHRM1 (the second type of acetylcholine receptor found in both the brain and the periphery) also are associated with ND (Bierut and others 2007; Lou and others 2006). Greenbaum et al. (2006) reported a nominally significant association of ND and related behaviors with CHRNA7 and CHRNB2 in a relatively small sample.
Interactions among genetic loci are being appreciated increasingly in complex human diseases (Jung and others 2009). Recent examples of diseases or conditions associated with gene–gene interaction include coronary artery disease (Tsai and others 2007), type 2 diabetes (Qi and others 2007), Alzheimer’s disease (Fontalba and others 2009), schizophrenia (Gupta and others 2009), breast cancer (Briollais and others 2007), cervical cancer (Guzman and others 2008), autistic disorder (Ma and others 2009), and smoking addiction (Li and others 2008a; Tang and others 2009), to name a few. Interactive effects among genetic loci may exist without a significant main effect of any of them; in such cases, important genetic effects would have been missed if polymorphisms of such loci had not been modeled jointly (Jung and others 2009). Furthermore, in many cases, interactive effects of multiple genetic loci could be larger than the main effects at the individual loci (Robson and others 2004; Rodriguez and others 2006; Williams and others 2000).
Multifactor Dimensionality Reduction (MDR) is a method of detecting genetic interactions by exhaustively searching multi-locus combinations (Motsinger-Reif and others 2008; Ritchie and others 2001). In MDR, k (e.g., k=3) factors and their possible multifactor classes are represented in k-dimensional space. Each multifactor class in the space is then labeled ‘high risk’ if the cases-to-controls ratio meets or exceeds some threshold, or as ‘low risk’ if that threshold is not exceeded, thus reducing the k-dimensional space to one dimension with two levels (‘low risk’ and ‘high risk’) (Moore 2003). The best k-locus model is then selected, the model is evaluated against the testing group, and testing accuracy is calculated. The PA is then calculated for the testing set (Motsinger-Reif and others 2008). Pedigree-based generalized MDR (PGMDR), a new generalized MDR for pedigree data, is a non-parametric method based on the score of the generalized linear model, which permits adjustment for covariates and handling of both dichotomous and quantitative phenotypes (Lou and others 2008). A key advantage of PGMDR is that the method can handle different pedigree structures and sizes simultaneously in the presence of various patterns of missing data. In our study, by using PGMDR, three-locus, four-locus and six-locus interaction models were detected in the AA sample, and two-locus and three-locus interaction models were detected in the pooled sample (Table 6). It is intriguing that rs1317286, an SNP in the CHRNA3, which was associated with both SQ and HSI in the pooled sample, was included in four of these five best interaction models. Therefore, the interactive effects detected by PGMDR are potentially valuable, such that they not only lend support to the single-locus results, but also shed significant light on the joint effects among SNPs in the CHRNA5/A3/B4 gene cluster, which could be missed in single-locus analysis. Nevertheless, larger and more rigorous replication studies are necessary to establish such interactions. As pointed by Milne et al. (2008), replication in additional independent samples of observed gene–gene interaction is crucial. In particular, caution should be exercised during the replication because differences of LD between different study populations such as AA and EA may have important impacts on the detection of high-order gene–gene interactions. Even when significant interactive effects are observed in a replication study, caution is necessary in elucidating exactly what constitutes a replicated result and the biological meaning of such replication. Therefore, ideally, observed gene–gene interactions should not only be replicated from a statistical perspective but also be validated experimentally from a biological perspective.
For power calculations with Quanto for main effects, according to two recent studies (Saccone and others 2009; Schlaepfer and others 2008), the average genotypic relative risk (GRR) for the chromosome 15 CHRNA5/A3/B4 gene cluster SNPs was about 1.3. In the current study, rs1317286 in the CHRNA3 gene was associated only with SQ and HSI in pooled sample. Another SNP in the CHRNA3 gene, rs8040868, is associated only with SQ in the pooled sample and with all the three adjusted ND phenotypes in the AA sample. The MAFs for rs1317286 and rs8040868 were 0.300 and 0.364, respectively, in the AA sample and 0.410 and 0.457, respectively, in the EA sample. To achieve 80% power for detecting the main effect under the assumption of GRR = 1.3, we would need 1823, 1696 and 1136 case-sibling pairs for MAF = 0.2, 0.30, and 0.40, respectively. Thus, the sample sizes of the current study, ~ 930 sibpairs in the AA sample, and ~ 330 sibpairs in the EA sample, should be sufficient for detecting a GRR of 1.3 for an SNP with an MAF of 0.40 for the pooled sample (~ 1260 sibpairs). It should be remembered that the case-sibling design is used as a reference for above mentioned power analyses. Because among the 402 AA families, there were 187, 199, and 16 families with 0, 1, and 2 parents, respectively (Li and others 2006), and among the 200 EA families, there were 46, 131, and 23 families with 0, 1, and 2 parents (Li and others 2008b), respectively, our study provides a greater statistical power than a pure case-sibling design. Given that rs1317286, a SNP in the CHRNA3 gene with a main effect under the dominant model in the pooled sample was included in four of the five best interaction models detected by PGMDR, and another SNP, rs3743078 in the CHRNA3 gene, without a main effect, was detected in three of the five best interaction models, power calculation with Quanto for detecting interaction between two genetic loci, g and h, was performed for three sets of parameters: (1) in the absence of main effects for g and h and in the presence of an interactive effect between g and h (assuming Rg = 1, Rh = 1, and Rgh = 1.5, 2.0, 2.5, and 3.0); (2) in the absence of a main effect for g only, and in the presence of a main effect for h as well as an interactive effect between g and h (assuming Rg = 1, Rh = 1.25, and Rgh = 1.5, 2.0, 2.5, and 3.0); and (3) in the presence of main effects for g and h, as well as an interactive effect between g and h (assuming Rg = 1.25, Rh = 1.25, and Rgh = 1.5, 2.0, 2.5, and 3.0) under the dominant model for both loci and the MAF for gene h = 0.30. The required sample sizes were 717, 754, and 795 for Rgh = 2.0, 436, 464, and 496 for Rgh = 2.5 and 323, 347, and 375 for Rgh = 3.0 under these three respective sets of parameters when the MAF of gene g is 0.30. Therefore, our study appears to provide sufficient sample sizes to detect an Rgh ≥ 2.0 under all three sets of parameters for the AA sample (~ 930 sibpairs) and the pooled sample (~ 1260 sibpairs), using the case-sibling design as the reference.
In sum, this is the first genetic study aimed at investigating a potential association of the variants of the CHRNA5/A3/B4 cluster with ND in a sample of AA smokers. Although our results reveal a relatively weak association of the cluster with ND, we did find significant interaction among variants of the three subunit genes in affecting ND. As for the EA population, we did not identify significant interactions of variants in this cluster, although we found a nominal association of the cluster with ND. The small size of our EA sample may have contributed, in part, to the weak association.
Table 5.
Haplotypes within CHRNA5 and CHRNA3 associated with three ND measures in the EA sample
| CHRNA5 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SQ |
HSI |
FTND |
||||||||||||||
| 5 | 6 | 7 | Freq | P- haplotype |
Z- score |
P- global |
No. of families |
P- haplotype |
Z- score |
P- global |
No. of families |
P- haplotype |
Z- score |
P- global |
No. of families |
|
| T | A | C | 0.36 | 0.053a | 1.933 | 0.27 | 83 | 0.035 a | 2.111 | 0.12 | 83 | 0.16a | 1.394 | 0.32 | 83 | |
| T | G | C | 0.28 | 0.21a | −1.273 | 55 | 0.031 a | −2.157 | 55 | 0.073a | −1.795 | 55 | ||||
| CHRNA3 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 16 | 17 | 18 | |||||||||||||
| C | G | T | 0.083 | 0.092a | 1.661 | 0.19 | 12 | 0.038 a | 2.074 | 0.096 | 12 | 0.041 a | 2.038 | 0.17 | 12 | |
| C | G | C | 0.42 | 0.078a | 1.76 | 75 | 0.097a | 1.658 | 76 | 0.27a | 1.097 | 76 | ||||
| G | T | T | 0.077 | 0.24a | 1.175 | 0.45 | 13 | 0.033 a | 2.13 | 0.12 | 13 | 0.035 a | 2.105 | 0.19 | 13 | |
| A | C | T | 0.43 | 0.19a | 1.266 | 79 | 0.31a | 1.021 | 76 | 0.72a | 0.353 | 76 | ||||
Notes:
1) Only major haplotypes are shown; the one with a P value <0.05 is given in bold.
2) For the 0.05 significance level, significant P value after Bonferroni correction for three major haplotypes within CHRNA5 is 0.0167; value for four major haplotypes within CHRNA3 is 0.0125.
3) Superscripts indicate genetic model used in the analysis
= additive.
4) The ND measures were corrected for age and sex.
Acknowledgments
We are grateful for the invaluable contributions of clinical information and tissue samples by the participants in this study, as well as for the dedicated work of the research staff at different clinical sites. We also thank Dr. David L. Bronson for his excellent editing of this manuscript. This project was funded by National Institutes of Health grants DA-12844, DA-13783 and DA-025095.
References
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–22. doi: 10.1038/ng.109. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L. Variants in Nicotinic Receptors and Risk for Nicotine Dependence. Am J Psychiatry. 2008;165(9):1163–71. doi: 10.1176/appi.ajp.2008.07111711. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briollais L, Wang Y, Rajendram I, Onay V, Shi E, Knight J, Ozcelik H. Methodological issues in detecting gene-gene interactions in breast cancer susceptibility: a population-based study in Ontario. BMC Med. 2007;5:22. doi: 10.1186/1741-7015-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen J, Williamson VS, An SS, Hettema JM, Aggen SH, Neale MC, Kendler KS. Variants in nicotinic acetylcholine receptors alpha5 and alpha3 increase risks to nicotine dependence. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti DV, Lee W, Li D, Liu J, Van Den Berg D, Thomas PD, Bergen AW, Swan GE, Tyndale RF, Benowitz NL. Nicotinic acetylcholine receptor beta2 subunit gene implicated in a systems-based candidate gene study of smoking cessation. Hum Mol Genet. 2008;17(18):2834–48. doi: 10.1093/hmg/ddn181. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca V, Wang H, Squassina A, Wong GW, Yeomans J, Kennedy JL. Linkage of M5 muscarinic and alpha7-nicotinic receptor genes on 15q13 to schizophrenia. Neuropsychobiology. 2004;50(2):124–7. doi: 10.1159/000079102. [DOI] [PubMed] [Google Scholar]
- Duga S, Solda G, Asselta R, Bonati MT, Dalpra L, Malcovati M, Tenchini ML. Characterization of the genomic structure of the human neuronal nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster and identification of novel intragenic polymorphisms. J Hum Genet. 2001;46(11):640–8. doi: 10.1007/s100380170015. [DOI] [PubMed] [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(5):596–604. doi: 10.1002/ajmg.b.30464. others. [DOI] [PubMed] [Google Scholar]
- Feng Y, Niu T, Xing H, Xu X, Chen C, Peng S, Wang L, Laird N. A common haplotype of the nicotine acetylcholine receptor alpha 4 subunit gene is associated with vulnerability to nicotine addiction in men. Am J Hum Genet. 2004;75(1):112–21. doi: 10.1086/422194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora A, Schulz R, Benfante R, Battaglioli E, Terzano S, Clementi F, Fornasari D. Transcriptional regulation of the human alpha5 nicotinic receptor subunit gene in neuronal and non-neuronal tissues. Eur J Pharmacol. 2000;393(1-3):85–95. doi: 10.1016/s0014-2999(00)00040-6. [DOI] [PubMed] [Google Scholar]
- Fontalba A, Gutierrez O, Llorca J, Mateo I, Vazquez-Higuera JL, Berciano J, Fernandez-Luna JL, Combarros O. Gene-gene interaction between CARD8 and interleukin-6 reduces Alzheimer’s disease risk. J Neurol. 2009;256(7):1184–6. doi: 10.1007/s00415-009-5080-z. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. doi: 10.1126/science.1069424. others. [DOI] [PubMed] [Google Scholar]
- Gaimarri A, Moretti M, Riganti L, Zanardi A, Clementi F, Gotti C. Regulation of neuronal nicotinic receptor traffic and expression. Brain Res Rev. 2007;55(1):134–43. doi: 10.1016/j.brainresrev.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Gotti C, Clementi F. Neuronal nicotinic receptors: from structure to pathology. Prog Neurobiol. 2004;74(6):363–96. doi: 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Graham A, Court JA, Martin-Ruiz CM, Jaros E, Perry R, Volsen SG, Bose S, Evans N, Ince P, Kuryatov A. Immunohistochemical localisation of nicotinic acetylcholine receptor subunits in human cerebellum. Neuroscience. 2002;113(3):493–507. doi: 10.1016/s0306-4522(02)00223-3. others. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, Yakir A, Lancet D, Ben-Asher E, Lerer B. Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes. Mol Psychiatry. 2006;11(3):312–22. 223. doi: 10.1038/sj.mp.4001774. [DOI] [PubMed] [Google Scholar]
- Gupta M, Chauhan C, Bhatnagar P, Gupta S, Grover S, Singh PK, Purushottam M, Mukherjee O, Jain S, Brahmachari SK. Genetic susceptibility to schizophrenia: role of dopaminergic pathway gene polymorphisms. Pharmacogenomics. 2009;10(2):277–91. doi: 10.2217/14622416.10.2.277. others. [DOI] [PubMed] [Google Scholar]
- Guzman VB, Yambartsev A, Goncalves-Primo A, Silva ID, Carvalho CR, Ribalta JC, Goulart LR, Shulzhenko N, Gerbase-Delima M, Morgun A. New approach reveals CD28 and IFNG gene interaction in the susceptibility to cervical cancer. Hum Mol Genet. 2008;17(12):1838–44. doi: 10.1093/hmg/ddn077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86(9):1119–27. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26(1):61–9. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–7. doi: 10.1038/nature06885. others. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Allen DL, Filbey FM, Jepson C, Lerman C, Benowitz NL, Stitzel J, Bryan A, McGeary J, Haughey HM. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Arch Gen Psychiatry. 2007;64(9):1078–86. doi: 10.1001/archpsyc.64.9.1078. [DOI] [PubMed] [Google Scholar]
- Jung J, Sun B, Kwon D, Koller DL, Foroud TM. Allelic-based gene-gene interaction associated with quantitative traits. Genet Epidemiol. 2009;33(4):332–43. doi: 10.1002/gepi.20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics. 2003;4(2):195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- Lessov-Schlaggar CN, Pergadia ML, Khroyan TV, Swan GE. Genetics of nicotine dependence and pharmacotherapy. Biochem Pharmacol. 2008;75(1):178–95. doi: 10.1016/j.bcp.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, Duenes AS, Crews KM, Elston RC. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14(9):1211–9. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10(4):225–31. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98(1):23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Li MD, Lou XY, Chen G, Ma JZ, Elston RC. Gene-gene interactions among CHRNA4, CHRNB2, BDNF, and NTRK2 in nicotine dependence. Biol Psychiatry. 2008a;64(11):951–7. doi: 10.1016/j.biopsych.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, Elston RC. Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry. 2008b;13(4):407–16. doi: 10.1038/sj.mp.4002038. [DOI] [PubMed] [Google Scholar]
- Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC. A genomewide search finds major susceptibility Loci for nicotine dependence on chromosome 10 in african americans. Am J Hum Genet. 2006;79(4):745–51. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Vikis HG, Wang D, Lu Y, Wang Y, Schwartz AG, Pinney SM, Yang P, de Andrade M, Petersen GM. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100(18):1326–30. doi: 10.1093/jnci/djn268. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Mangold JE, Zhu J, Elston RC, Li MD. A combinatorial approach to detecting gene-gene and gene-environment interactions in family studies. Am J Hum Genet. 2008;83(4):457–67. doi: 10.1016/j.ajhg.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Ma JZ, Payne TJ, Beuten J, Crew KM, Li MD. Gene-based analysis suggests association of the nicotinic acetylcholine receptor beta1 subunit (CHRNB1) and M1 muscarinic acetylcholine receptor (CHRM1) with vulnerability for nicotine dependence. Hum Genet. 2006;120(3):381–9. doi: 10.1007/s00439-006-0229-7. [DOI] [PubMed] [Google Scholar]
- Lueders KK, Hu S, McHugh L, Myakishev MV, Sirota LA, Hamer DH. Genetic and functional analysis of single nucleotide polymorphisms in the beta2-neuronal nicotinic acetylcholine receptor gene (CHRNB2) Nicotine Tob Res. 2002;4(1):115–25. doi: 10.1080/14622200110098419. [DOI] [PubMed] [Google Scholar]
- Ma DQ, Rabionet R, Konidari I, Jaworski J, Cukier HN, Wright HH, Abramson RK, Gilbert JR, Cuccaro ML, Pericak-Vance MA. Association and gene-gene interaction of SLC6A4 and ITGB3 in autism. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.31003. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008 doi: 10.1038/ng.254. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RL, Fagerholm R, Nevanlinna H, Benitez J. The importance of replication in gene-gene interaction studies: multifactor dimensionality reduction applied to a two-stage breast cancer case-control study. Carcinogenesis. 2008;29(6):1215–8. doi: 10.1093/carcin/bgn120. [DOI] [PubMed] [Google Scholar]
- Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291(10):1238–45. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56(1-3):73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Motsinger-Reif AA, Reif DM, Fanelli TJ, Ritchie MD. A comparison of analytical methods for genetic association studies. Genet Epidemiol. 2008;32(8):767–78. doi: 10.1002/gepi.20345. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciotto MR, Caldarone BJ, King SL, Zachariou V. Nicotinic receptors in the brain. Links between molecular biology and behavior. Neuropsychopharmacology. 2000;22(5):451–65. doi: 10.1016/S0893-133X(99)00146-3. [DOI] [PubMed] [Google Scholar]
- Qi L, van Dam RM, Asselbergs FW, Hu FB. Gene-gene interactions between HNF4A and KCNJ11 in predicting Type 2 diabetes in women. Diabet Med. 2007;24(11):1187–91. doi: 10.1111/j.1464-5491.2007.02255.x. [DOI] [PubMed] [Google Scholar]
- Raimondi E, Rubboli F, Moralli D, Chini B, Fornasari D, Tarroni P, De Carli L, Clementi F. Chromosomal localization and physical linkage of the genes encoding the human alpha 3, alpha 5, and beta 4 neuronal nicotinic receptor subunits. Genomics. 1992;12(4):849–50. doi: 10.1016/0888-7543(92)90324-l. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson KJ, Lehmann DJ, Wimhurst VL, Livesey KJ, Combrinck M, Merryweather-Clarke AT, Warden DR, Smith AD. Synergy between the C2 allele of transferrin and the C282Y allele of the haemochromatosis gene (HFE) as risk factors for developing Alzheimer’s disease. J Med Genet. 2004;41(4):261–5. doi: 10.1136/jmg.2003.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez E, Mateo I, Llorca J, Sanchez-Quintana C, Infante J, Berciano J, Combarros O. Genetic interaction between two apolipoprotein E receptors increases Alzheimer’s disease risk. J Neurol. 2006;253(6):801–3. doi: 10.1007/s00415-005-0063-1. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, Agrawal A, Breslau N, Grucza RA, Hatsukami D. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(4):453–66. doi: 10.1002/ajmg.b.30828. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63(11):1039–46. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Wilhelmsen K, Pomerleau CS, Chasse SA, Rice JP, Snedecor SM, Bierut LJ, Neuman RJ, Pomerleau OF. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction. 2008;103(9):1544–52. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman MA, Neale MC, Sullivan PF, Harris-Kerr C, Wormley B, Sadek H, Ma Y, Kendler KS, Straub RE. Haplotypes of four novel single nucleotide polymorphisms in the nicotinic acetylcholine receptor beta2-subunit (CHRNB2) gene show no association with smoking initiation or nicotine dependence. Am J Med Genet. 2000;96(5):646–53. [PubMed] [Google Scholar]
- Stolerman IP, Jarvis MJ. The scientific case that nicotine is addictive. Psychopharmacology (Berl) 1995;117(1):2–10. doi: 10.1007/BF02245088. discussion 14-20. [DOI] [PubMed] [Google Scholar]
- Tang X, Guo S, Sun H, Song X, Jiang Z, Sheng L, Zhou D, Hu Y, Chen D. Gene-gene interactions of CYP2A6 and MAOA polymorphisms on smoking behavior in Chinese male population. Pharmacogenet Genomics. 2009;19(5):345–52. doi: 10.1097/fpc.0b013e328329893c. [DOI] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–42. doi: 10.1038/nature06846. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CT, Hwang JJ, Ritchie MD, Moore JH, Chiang FT, Lai LP, Hsu KL, Tseng CD, Lin JL, Tseng YZ. Renin-angiotensin system gene polymorphisms and coronary artery disease in a large angiographic cohort: detection of high order gene-gene interaction. Atherosclerosis. 2007;195(1):172–80. doi: 10.1016/j.atherosclerosis.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, David SP, Niaura R, Lerman C. Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Arch Gen Psychiatry. 2008;65(6):683–93. doi: 10.1001/archpsyc.65.6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDHHS . Reducing tobacco use: A report of the Surgeon General. US Department of Health & Human Services, Center for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promortion; Atlanta, Georgia: 2000. [Google Scholar]
- Vink JM, Smit AB, de Geus EJ, Sullivan P, Willemsen G, Hottenga JJ, Smit JH, Hoogendijk WJ, Zitman FG, Peltonen L. Genome-wide association study of smoking initiation and current smoking. Am J Hum Genet. 2009;84(3):367–79. doi: 10.1016/j.ajhg.2009.02.001. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008 doi: 10.1038/ng.273. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SS, Koob GF, Markou A. Neural mechanisms underlying nicotine addiction: acute positive reinforcement and withdrawal. Nicotine Tob Res. 2000;2(1):19–37. doi: 10.1080/14622200050011277. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . The World Health Report 2002. World Health Organization; 2002. [DOI] [PubMed] [Google Scholar]
- Williams SM, Addy JH, Phillips JA, 3rd, Dai M, Kpodonu J, Afful J, Jackson H, Joseph K, Eason F, Murray MM. Combinations of variations in multiple genes are associated with hypertension. Hypertension. 2000;36(1):2–6. doi: 10.1161/01.hyp.36.1.2. others. [DOI] [PubMed] [Google Scholar]
- Xu X, Scott MM, Deneris ES. Shared long-range regulatory elements coordinate expression of a gene cluster encoding nicotinic receptor heteromeric subtypes. Mol Cell Biol. 2006;26(15):5636–49. doi: 10.1128/MCB.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


