Abstract
Recent progress in an emerging area of designing aptamer and nanomaterial conjugates as molecular diagnostic and drug delivery agents in biomedical applications is summarized. Aptamers specific for a wide range of targets are first introduced and compared to antibodies. Methods of integrating these aptamers with a variety of nanomaterials, such as gold nanoparticles, quantum dots, carbon nanotubes, and superparamagnetic iron oxide nanoparticles, each with unique optical, magnetic, and electrochemical properties, are reviewed. Applications of these systems as fluorescent, colorimetric, magnetic resonance imaging, and electrochemical sensors in medical diagnostics are given, along with new applications as smart drug delivery agents.
Keywords: DNA, aptamer, nanomaterials, diagnostics, biosensor, drug delivery, biomedical, targeted therapeutics
1. Introduction
Recent developments in nanostructured material synthesis and engineering have made a huge impact on a number of fields including nanoelectronics, photonics, biology, and medicine [1, 2]. One of the main reasons for such an impact is that every nanomaterial, including metallic nanoparticles (NPs), quantum dots (QDs), carbon nanotubes (CNTs), and magnetic nanoparticles has unique physical and chemical properties that can be used for various applications. Of particular relevance to this review are optical, magnetic, electronic, thermal, catalytic, and mechanical signals generated by these nanomaterials, making them ideal candidates for signal transductions in medical diagnostics.
In order for nanomaterials to be used as clinical probes, it is necessary to provide them with target recognition capability; this task is generally accomplished by functionalizing the nanomaterials with biomolecules. Proteins such as antibodies are well known targeting molecules to be conjugated onto the nanomaterials. Recently functional nucleic acids are an emerging class of molecules to be used together with nanomaterials for biomedical applications [3–6].
Functional nucleic acids are DNA or RNA molecules that can interact with or bind to a specific analyte, resulting in conformation change or catalytic reaction [7, 8]. Since DNA is more stable and cheaper to produce than RNA, this review is mainly focused on functional DNA conjugated nanomaterials. Functional DNA molecules include DNAzymes, aptamers, and aptazymes. DNAzymes (also called catalytic DNA or deoxyribozymes elsewhere) are DNA molecules that can catalyze many chemical and biological reactions in the presence of specific molecules, mostly metal ions, as cofactors. Aptamers, on the other hand, are nucleic acid molecules that can specifically bind to chemical or biological molecules. Aptazymes are a combination of DNAzyme and aptamer. Once functional DNAs are combined with nanostructured materials, it can endow the nanomaterials with target recognition capability allowing the hybrid system to be used as a sensor.
The target recognition ability of functional DNA has already been used extensively with nanomaterials to develop biosensors [9–11] for various target molecules to detect DNA [12–14], RNA [15], and metal ions [3, 16–30]. Among them, aptamers modified with nanomaterials have great potential to be used for clinical diagnostics as they can detect diverse targets ranging from small molecules and proteins to intact viruses and cells.
In this review, various nanomaterial-aptamer systems for potential application in clinical diagnostics will be covered. First, aptamers will be introduced and their advantages and disadvantages will be discussed in comparison with antibodies. Following that, methods of combining aptamers with different nanomaterials to produce diagnostic agents, such as fluorescence, colorimetric, surface enhanced Raman scattering (SERS), magnetic resonance imaging (MRI), and electrochemical detection, will be summarized. Finally, recent progress in using the aptamers-nanomaterials conjugates for targeted drug delivery will be covered.
2. Overview of aptamers
2.1 Aptamer
Aptamers can be considered as nucleic acid analogue of antibodies; they can bind with high affinity and specificity to a broad range of targets, such as small molecules, proteins, viruses, or even specific types of cells. The concept of nucleic acid binding to target molecules or proteins was first introduced when a RNA with high affinity and selectivity to viral or cellular proteins was observed during the investigation on HIV and adenovirus in 1980’s [31]. Aptamers can possess strong affinities to target molecules, with dissociation constants down to nanomolar or picomolar ranges. Currently, nucleic acid aptamers have been selected for more than 150 targets [32], including small molecules such as cocaine [33] and aspartame [34], growth factors [35–38], peptides [39, 40], toxins [41, 42], viral proteins [43, 44], and cells and bacteria [45–47]. Among them some aptamers received approval from the US Food and Drug administration (FDA) for vascular endothelial growth factor (VEGF)-165 isoform [48] and ocular vascular disease [49]. Recently, targeted drug delivery using polymeric nanoparticle (NP) and aptamer conjugates on prostate cancer cells has been demonstrated in vivo [50], showing that aptamers have great potential in therapeutics and diagnostics [31, 48, 49, 51–57].
2.2 In vitro selection of aptamer
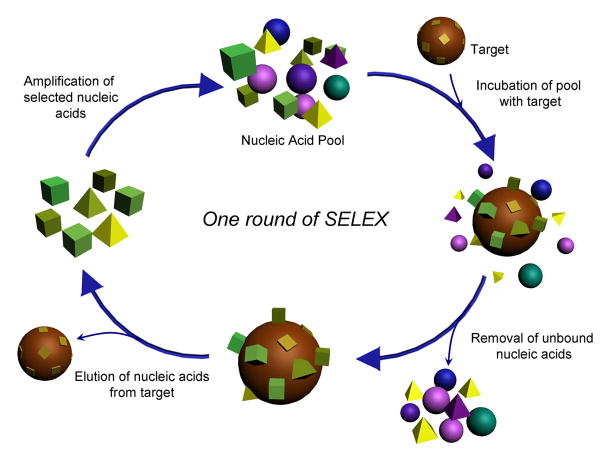
Aptamers can be obtained via isolation from synthetic combinatorial libraries of nucleic acids through an in vitro selection process called SELEX (systematic evolution of ligands by exponential enrichment) [51, 58, 59]. The selection process starts with generating random sequences of 1014–1015 oligonucleotide strands which are chemically synthesized and amplified with polymerase chain reaction (PCR) (Figure 1). The nucleic acid library is incubated with a target molecule, often immobilized onto a solid state matrix such as a gel or a column so that the DNA or RNA having affinity to the target molecule can be captured. The target-bound nucleic acid molecules are separated from the unbound strands in the pool, following which the bound DNA or RNA strands are eluted from the target molecule and amplified via PCR to seed a new pool of nucleic acids, which is then enriched with sequences having higher affinity to the target. The next round of selection process is usually performed under more stringent conditions (such as lower target concentration and shorter time for binding). After ~ 10–20 rounds of the selection processes, the oligonucleotide with the highest affinity to the target molecule can be obtained. Furthermore, cell-based SELEX has been recently reported which generates aptamers with high binding coefficients from living cells [32, 60]. This method allows selection of aptamers specific for the molecules present in the targeted cell. Since this work can be performed without the prior knowledge of the molecules on the cells, it not only allows early diagnostics but also opens opportunities for identification of new cancer cell markers.
Fig. 1.
Schematic depiction of SELEX. Nucleic acid pool containing 1014–1015 nucleic acids with random sequence is incubated with a target molecule. After removing unbound nucleic acids, nucleic acids are eluted from the target molecule. The pool for the next round is prepared after amplification and mutation of the selected nucleic acids. Selection can be completed after about 15 rounds of selection process.
2.3 Advantages of aptamers
Antibodies are being used in the majority of biomedical applications. Aptamers are an emerging class of molecules with several important advantages [51–55]. First, aptamers are efficient at binding to both large molecules such as proteins [61], cells [62], and small molecules such as nucleotides [63, 64], organic dyes [65], amino acids [66], and metal ions [67, 68], while antibodies are generally competent in binding to mostly larger molecules. Therefore there is a potential niche market for aptamers in diagnostics and drug delivery when small molecules are the targets. Second, aptamers are selected in vitro, and therefore can be used to select for a wide range of targets, including toxic or non-immunogenic molecules. Furthermore, the selection process can be performed under non-physiological condition including extremely high or low temperatures or pHs. These properties are difficult to be obtained with antibodies since they are produced in vivo. Third, once an aptamer is selected, it can be obtained in a large amount through chemical synthesis which is cost-effective and has minimal batch to batch difference in activity. It is easy to chemically modify aptamers with a variety of fluorophores, electrochemical or Raman reporters, or functional groups on either 5′ or 3′ end, on the bases, or even on the backbone of DNA[69]. Moreover, aptamers can be incorporated into one, two or three dimensional DNA-based nanostructures [70–75] which also have strong potential to be used for clinical and diagnostic applications [76–78]. In designing microarrays, aptamers can provide additional advantages. For example, as the size of aptamers is much smaller (~1–2 nm, <10 kDa) than those of antibodies (~10 nm and ~155 kDa), aptamers have higher surface density [57] and less steric hindrance [51], which helps to increase the binding yield. In addition, non-specific adsorption happens much less for nucleic acid-immobilized surfaces compared to protein immobilized surfaces [79], which facilitates the engineering process. Finally, aptamers are much more stable to heat [80], pH [50], and organic solvents [81] than antibodies and, unlike antibodies, can be denatured and renatured multiples times without significant loss of activity [82]. These properties make aptamers an excellent choice for biomedical applications.
2.4 Challenges facing aptamer-based diagnostics
Although aptamers are excellent candidates as diagnostic and drug delivery agents, there are still a few issues that have yet to be addressed for their practical applications. One issue is that nucleic acid aptamers could be vulnerable to nuclease degradation in cells or in blood [56]. This problem can be more significant for RNA aptamers because RNA molecules are much more vulnerable to hydrolysis in biological fluid. To overcome this problem, several strategies have been proposed to increase the stability of aptamers, such as modification of aptamers with 2′-aminopyrimidine [83], 2′-fluoropyrimidine [83], or 2′-O-methyl nucleotides [84]. Another limitation aptamers might have is the necessity to limit its administration to local regions, since there is a possibility that proteins in untargeted organs can also be affected. However, some preliminary results showed the possibility of systematic delivery [51,52]. Finally, one of the major challenges is the development of general methods to convert the highly specific molecular recognition between aptamers and their targets into detectable signals. Conjugation of aptamers with nanomaterials can be an ideal way to overcome this hurdle.
3. Conjugation of nanomaterials and aptamers for biosensing and diagnostics
3.1. Fluorescence based sensors
Aptamers can be easily modified with a variety of organic dyes and thus fluorophore-based detection method has been the most widely demonstrated [3, 5, 6, 85]. Since this review mainly focuses on the nanomaterial-based detection, fluorophore-based detection will not be discussed here.
Quantum dots (QDs) or semiconductor NPs are one type of fluorescent nanomaterials with several unique optical properties [86–88]. As compared to organic fluorescent dyes, quantum dots are more photostable and the wavelength of the emitted light can be controlled by changing their size or materials. Furthermore, QDs have very broad excitation and sharp emission ranges, making it possible to excite different QDs with a single wavelength and yet result in a variety of emission wavelengths. Based on these attractive properties, QDs have been broadly used for bioimaging and diagnostics.
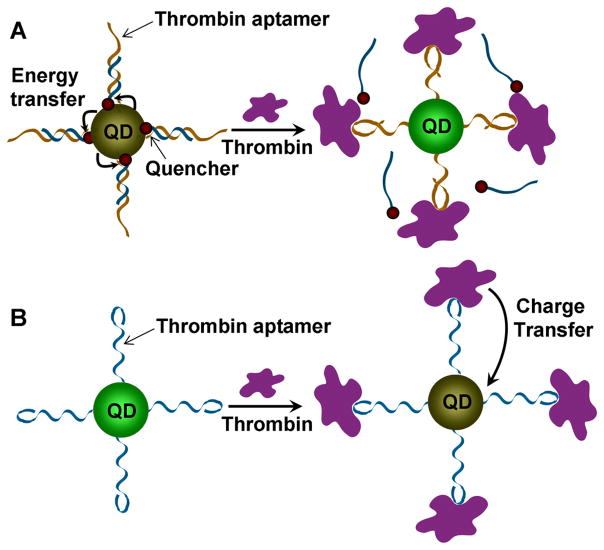
QDs functionalized with DNA [89–91] have been used for the detection of DNA [92] and real time monitoring of hybridization procedure [93]. The first QD conjugated with aptamer was reported by Ellington and co-workers for energy transfer based detection of thrombin (see Figure 2A) [94]. In this work, thrombin aptamers functionalized on QDs were hybridized with a complementary DNA strand labeled with a quencher at the end. In the absence of thrombin, the fluorescent signal of QDs was quenched because the QDs were placed in close proximity to the quenchers, resulting in the energy transfer from the quantum dot to the quencher. The presence of thrombin, however, recovered the signal as thrombin binding to the aptamer induced the release of the complementary strand containing the quencher.
Fig. 2.
Aptamer-QD based fluorescent sensors. (A) Thrombin aptamer is conjugated onto QD and hybridized to a complementary DNA with a quencher. Fluorescence of QD is quenched due to energy transfer from the QD to the quencher. The complementary DNA containing the quencher can be released after introduction of thrombin, inducing recovery of the fluorescence from QD. (B) Thrombin aptamers are conjugated to QDs. The fluorescence of QDs can be quenched as thrombin binds to aptamer due to the charge transfer from thrombin to QD.
A different type of QD-aptamer sensor was developed by Strano and co-workers (see Figure 2B) [95]. In this work, they conjugated QDs with thrombin-specific aptamers. When thrombin was added, the interaction of thrombin with aptamers brought them close to QDs. Selective quenching of fluorescence occurred due to the charge transfer occurring from thrombin to the QDs. The sensor was very sensitive with the detection limit of 1 nM and had high selectivity.
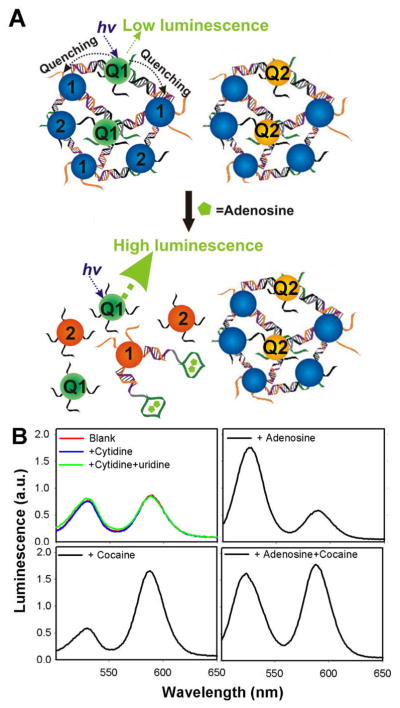
For medical applications, it is advantageous to perform multiplexed detection of more than one analyte using a single sensor in an “one-pot” process. Our group reported multiplexed detection of adenosine and cocaine using both QDs and gold nanoparticles (AuNPs) conjugated with aptamers (Figure 3) [96]. In this work, aggregates of QDs with emission peak at 525 nm and AuNPs were formed by DNA aptamer specific for adenosine. In parallel, aggregates of QDs with emission peak at 585 nm and AuNPs were formed by a cocaine specific DNA aptamer. While fluorescence from the sensor composed of the mixture of both assemblies was initially quenched, the signal was enhanced at 525 nm with the addition of adenosine or at 585 nm in the presence of cocaine. Fluorescence increase at both 525 nm and 585 nm was observed when both of the analytes were present.
Fig. 3.
Multiplexed detection of analytes using QDs. (A) Aptamer conjugated QD-AuNP aggregates are formed with two different QDs emitting at different wavelengths (QD1 and QD2). QDs in aggregated states are quenched due to the energy transfer from each QD to AuNPs. The fluorescence of QD1, however, is recovered after introduction of a target analyte (e.g. adenosine) owing to their binding with aptamers. (B) Introduction of adenosine and/or cocaine induces the fluorescence increase of QD1 (emission at 525 nm) and/or QD2 (emission at 585 nm), respectively. On the other hand, control analytes (cytidine and uridine) do not induce fluorescence signal increase from QDs. Reproduced with permission of ref. 90, copyright of American Chemical Society.
It was recently reported that carbon nanotubes (CNT) also have fluorescence in the near-IR range which is advantageous for cell imaging by avoiding the high background fluorescence from the organelles in the cells [97]. Therefore, aptamer conjugated CNTs also have potential to be used as a fluorescence tag for cancer cell imaging. First, a photodynamic therapy agent generating singlet oxygen (1O2) in the presence of α-thrombin was demonstrated using a photosensitizer labeled α-thrombin aptamer and single wall nanotube (SWNT) complex[98]. As the aptamer wrapped around the SWNT in the absence of target molecule, the labeled photosensitizer was placed close to SWNT quenching the generation of singlet oxygen. On the other hand, the interaction between thrombin and aptamer induced release of photosensitizer from SWNT, generating singlet oxygen in the presence of light source. The same group further extended the methodology for detection of biomolecular interactions based on fluorescence. [99]
Furthermore, fluorophore labeled DNA can also be immobilized on a solid surface. This provides several advantages such as low fluorescence background, regeneration, and long term storage compared to solution based fluorescence sensors [25]. In addition, the performance of fluorescence sensors can be engineered by the property of the material on which aptamer is immobilized. As the sensitivity and volume of the sample can be critical for some biomedical diagnostic applications, we investigated the influence of materials using Pb2+ specific DNAzyme labeled with fluorescein and Au surface as a model system. By immobilizing DNAzyme on a planar Au substrate, we could reach an order of magnitude improvement (1 nM) in the detection limit due to lower background, compared to solution based sensors (10 nM) [100]. Au coated nano capillary membrane (NCAM) further enhanced fluorescent signal twelve times due to the increased surface area provided by the nanopore walls of the membrane and increased surface roughness. This NCAM sensor also allowed regenerations by rehybridization of fresh DNA strand and storage up to 30 days without significant loss of activity [101]. Solid supported fluorescent sensors can also incorporate ratiometric fluorescence internal controls which allow to standardize the number of molecules immobilized on the surface and to perform real time detection of target molecules [102]. In addition, aptamers or DNAzymes can be incorporated into microfluidic devices which have advantages of generating low waste volume output, consuming less amount of detecting material, and having facile regeneration capabilities [103, 104]. The Pb2+ DNAzyme integrated NCAM based microfluidic device consumed 4.2 picoliter of DNAzyme for each detection with detection limit of 11 nM [105]. Finally, by changing the design of the microfluidic device, multiple detections could be made in a single cycle with minimal detection variation [106].
3.2. Colorimetric sensor
As fluorescence based sensors rely on the emission of the fluorophores or quantum dots after excitation at a certain wavelength, it is necessary to have analytical equipments such as fluorimeter or fluorescence microscope for detection. For real-time and on-site use, however, it is more convenient if detection can be carried out without any equipment. Colorimetric detection provides an advantage in this regard, since it allows detection to be made by naked eye [16, 107–111]. Novel metallic nanoparticles, such as gold or silver nanoparticles, are ideal materials that allow colorimetric detection [112]. AuNPs have very high extinction coefficient, making their color distinguishable without any instrument at only a few nanomolar concentration. Dispersed AuNPs smaller than 100 nm in solution originally have reddish color. When AuNPs aggregate, their color changes from red to blue due to their surface plasmon resonance shift to a higher wavelength [113].
There are two different methods for designing colorimetric sensors based on aptamer and AuNPs. The labeled method is based on directed assembly (or disassembly) of AuNPs due to analyte-specific cleavage or conformational change of the DNA molecules [8, 16, 113]. Two batches of AuNPs, each chemically functionalized with a different DNA strand via Au-thiol chemistry, are mixed together resulting in dispersed AuNPs due to the strong negative charge of DNA covering AuNP surface. In the presence of a bridging DNA strand, complementary to both DNA strands functionalized on AuNP surface, AuNPs aggregate with their color changing from red to blue.
The label-free method takes advantage of different adsorption properties of single stranded (ss) DNA and double stranded (ds) DNA onto citrate modified AuNPs [114]. Citrate modified AuNPs are naturally unstable in the presence of NaCl and can easily aggregate. Since ssDNA is flexible and can partially uncoil its strand, it can be readily adsorbed on AuNPs surface resulting in the enhancement of electrostatic repulsion between AuNPs, which in turn stabilizes AuNPs even in the presence of NaCl. On the other hand, as dsDNA is stiff and covered with negatively charged phosphate backbone, it will repel the negatively charged citrate modified AuNPs, resulting in aggregation of AuNPs.
Both labeled and label-free methods have been used for colorimetric detection of DNA [12–14], RNA [15], and various metal ions [16–19, 24, 26, 28, 29] when DNA or DNAzymes are incorporated into the systems. Recently, a systematic comparison of the two methods using the same DNAzyme has been reported, providing a practical guidance in the choice of methods [115].
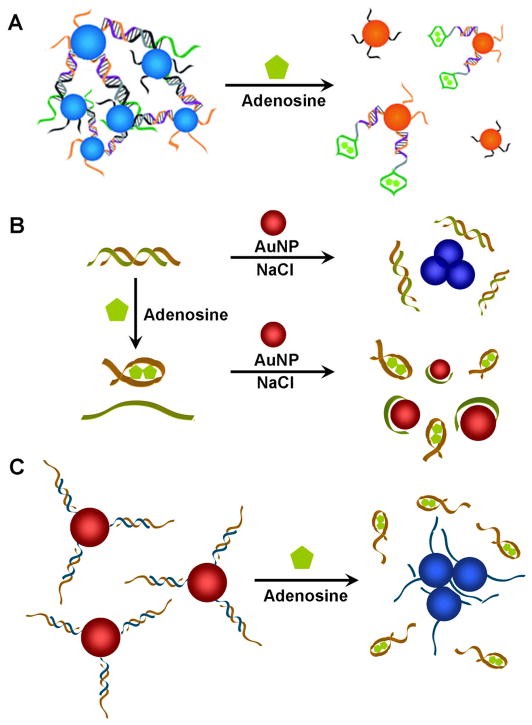
Aptamers can also be incorporated into both systems for colorimetric detection. In 2006, we reported colorimetric sensors specific for adenosine and cocaine using the labeled method [111]. In the absence of target, the aptamer-modified AuNPs remained aggregated, appearing blue; in the presence of the target, the AuNPs disassembled, resulting in red color (see Figure 4A). The color change occurred in less than 1 minute. The labeled method has been used for detection of other target molecules such as small molecules and proteins [108, 110]. Dong, Wang, and coworkers demonstrated that labeled aptamer-AuNP colorimetric system could be used for detection of thrombin with extremely high sensitivity with detection limit of 14 fM using dot-blot arrays [116]. Finally, we reported multiplex colorimetric sensing system based on smart AuNPs responsive to multiple stimuli with controlled cooperativity [117].
Fig. 4.
Aptamer-AuNP based colorimetric sensors. (A) AuNPs functionalized with oligonucleotides have been used for labeled sensors. AuNPs are initially aggregated with adenosine specific aptamer strands. The introduction of adenosine stimulates disassembly of AuNPs, changing the color of AuNPs from blue to red. (B) The interaction between unmodified AuNPs and unlabeled ssDNA strand has been used to make label-free sensors. In the presence of adenosine, dsDNA containing an adenosine aptamer strand (brown) releases ssDNA (green) which can be adsorbed onto unmodified AuNPs. AuNPs remain dispersed with red color even in the presence of NaCl due to enhanced stability of AuNPs provided by ssDNA. In the absence of adenosine, dsDNA with adenosine aptamer strand stays hybridized. The electrostatic repulsion between dsDNA and AuNP as well as the stiffness of dsDNA make them ineffective in preventing NaCl induced charge screening on AuNP surface, causing aggregation of AuNPs with color change from red to blue. (C) Both labeled and label-free method has been combined to make an adenosine sensor. AuNPs are chemically functionalized with DNA (blue) and hybridized with adenosine aptamer strand (brown). In the presence of adenosine, aptamer strands are released from AuNPs decreasing the stability of AuNPs. The resulting aggregation of AuNPs induces color change from red to blue.
Fan and co-workers reported colorimetric detection of ATP using unmodified AuNPs and ATP aptamers based on label-free method (see Figure 4B) [118]. AuNPs aggregated with blue color in the absence of ATP but remained dispersed displaying red color in its presence. The sensor had high selectivity and sensitivity with detection limit of 0.6 μM. This label-free aptamer based colorimetric detection method was also used for other analytes such as potassium [19], cocaine [118], thrombin [119].
Li and coworkers reported a detection method based on the combination of labeled and label-free methods (see Figure 4C). They first chemically functionalized short DNA strands on AuNPs and hybridized those strands with long partially complementary aptamer strands. The long aptamer DNA provides additional negative charge to enhance the stability of AuNPs at a certain concentration of NaCl. Introduction of target molecules induced interaction between target molecule and aptamer strand resulting in the release of long aptamer strands. The targeted release of aptamer strand decreased the number of negatively charged DNAs on AuNPs, lowering the stability of AuNPs. This turn-on colorimetric sensor, changing its color from red to blue in the presence of analytes, had high selectivity and a detection limit of 10 μM [120, 121].
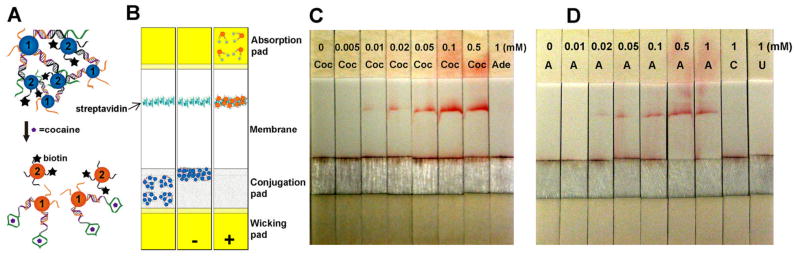
While AuNP based colorimetric sensors provide a way to carry out detection without the need for analytical equipment, it still requires careful handling of micro-liter scale solutions, which is not trivial for most users, especially those at home. Therefore it is preferred to develop more user-friendly diagnostic kits, such as dip-stick tests. Since the discovery of antibodies, there have been a number of reports converting antibody based detection onto the lateral flow devices [122] and many of the kits, such as home pregnancy test, are commercially available. In contrast, few reports demonstrating nucleic acid based lateral flow devices have appeared in the literature. For example, Ioannou and Christopoulos et al. used DNA functionalized AuNPs to analyze the hybridization of DNA on lateral flow devices [123]. We developed a lateral flow device based on labeled AuNP and aptamer system (see Figure 5) [124]. The lateral-flow device is composed of four components, a wicking pad, a glass fiber conjugation pad, a membrane, and an absorption pad. Biotin labeled AuNP aggregates containing cocaine aptamers were dropped onto the conjugation pad, and streptavidin was immobilized on a specific part of the membrane to capture biotinylated AuNPs. When the wicking pad of lateral flow device was dipped into a sample solution containing cocaine, the solution flowed along the conjugation pad, rehydrated and induced disassembly of AuNP aggregates. Since biotin was labeled on disassembled AuNPs, the AuNPs could be captured on the part of the membrane with streptavidin forming a red line. When the concentration of cocaine in the solution increased, the intensity of the red line formed on the membrane became higher and the estimated detection limit was ~10 μM. In addition to the simplicity in testing without the need of precise solution transfer, the method was also more sensitive than solution-based tests, due to the integration of binding, separation, and detection on a simple test paper-like platform. Red lines did not appear with control analytes, showing good selectivity of the lateral flow device. In the process, we discovered a new design of the lateral flow device taking advantage of the physical size difference of nanoparticles in various assembly states, which provides a critical control for assessing the performance of the device. The lateral flow device could also be applied to other target molecules, such as adenosine, by using aptamer specific for those molecules. Since aptamers for a broad range of molecules have been obtained, this method is general enough to be adapted to develop “dip-stick” tests for any analyte for which an aptamer can be obtained. In addition, the lateral flow device also performed well in human blood serum, showing its compatibility with biological samples. Additionally, Pelton, Li and coworkers reported that aptamers can be immobilized on cellulose surface as a potential platform for detection of medically relevant biomolecules [125], making it possible for medical diagnostics.
Fig. 5.
Simple dip stick tests based on labeled AuNP-aptamer lateral flow device. (A) Schematic illustration of cocaine aptamer induced AuNP disassembly and incorporation of such system onto the lateral flow device. Aggregates of AuNPs functionalized with both cocaine aptamer and biotin are disassembled in the presence of cocaine due to the binding between aptamer and cocaine. (B) The structure of lateral-flow device; the device is composed of a wicking pad, a glass fiber conjugation pad, a membrane, and an absorption pad. Biotin labeled AuNP aggregates containing cocaine aptamers are dropped onto the conjugation pad and the wicking pad is dipped into a sample solution with cocaine. The solution flows along the conjugation pad, rehydrates and induces disassembly of AuNP aggregates. (C) The dipstick test for cocaine. In the presence of cocaine, disassembled AuNPs with biotin are captured onto streptavidin immobilized on the membrane, producing a red line. (D) The lateral flow device can be generally applied with other aptamers, such as adenosine aptamers, for adenosine sensing. Reproduced with permission of ref. 109. Copyright Wiley-VCH Verlag GmbH & Co. KGaA.
3.3. Surface Enhanced Raman Scattering (SERS)
Raman scattering is an optical methodology utilizing the inelastic scattering of a photon. It has several advantages over fluorescence as it can provide structural information of a molecule, does not suffer from photobleaching, and has a much narrower spectra (<1 nm, half widths). As Raman cross section is inherently weak and inefficient (10−30 cm2 per molecule vs 10−16 cm2 per molecule for fluorescence), there has been limitation in utilizing Raman scattering as a tool for highly sensitive detection [126]. In 1974, however, it was reported that Raman scattering can be significantly amplified when molecules are absorbed on metal surface, due to the extremely high electromagnetic fields produced on hot spots generated on the surface of metals [127]; this phenomenon is called Surface Enhanced Raman Scattering (SERS). SERS becomes more efficient on rough metal surfaces or on metal nanoparticle aggregates due to localized surface plasmon resonance [128].
In 1997, Nie and Kneipp independently demonstrated the possibility of detecting single molecules based on SERS [126, 129]. Using Ag colloidal nanoparticles, Nie reported that the enhancement factor of 1014–1015 and the cross section of 10−16 cm2 per molecule can be obtained which are comparable to fluorescence [126]. Using AuNPs nanoparticles coated with Ag as surface enhancement promoter and Raman reporters, highly sensitive and selective multiplexed detection of DNA was reported by Mirkin and co-workers [130]. Halas et al. demonstrated that SERS spectra can be obtained from DNA without any Raman reporters although the spectra obtained are mostly identical for several model DNA strands, which are dominated by adenine [131].
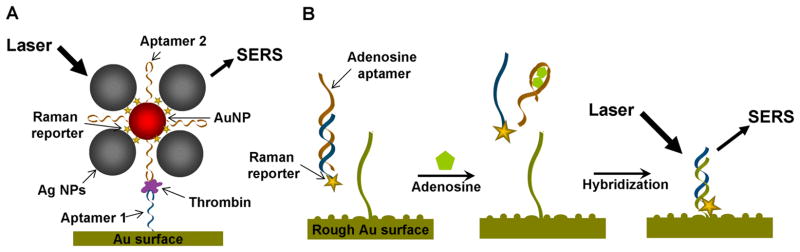
The detection of proteins or small molecules using SERS has also been reported using DNA aptamers. Wang, Dong, and co-workers used aptamers to detect α-thrombin by SERS (See Figure 6A) [132]. Thrombin has two binding sites specific for two different DNA aptamers with high affinity [61, 133]. A substrate functionalized with aptamer 1 was first treated with thrombin and then incubated in AuNPs functionalized with aptamer 2 and SERS reporter. As a result, AuNPs were attached to the surface through aptamer-thrombin interaction. After Ag NP deposition, the NPs became larger, resulting in highly enhanced Raman scattering and thus highly sensitive signal. The detection limit was reported as 0.5 nM and the system had high selectivity for β- and γ-thrombins. Jiang, Yu, and co-workers demonstrated a different methodology to detect adenosine using aptamers [134]. They used a dsDNA composed of both an adenosine aptamer strand and a partially complementary short DNA strand with a Raman reporter (Figure 6B). In the presence of adenosine, the aptamer strand underwent structure switching, resulting in denaturation of dsDNA. Therefore the Raman probe labeled oligonucleotide was released in solution. The released strand later hybridized to the complementary DNA which was previously immobilized on a rough gold surface. This chain of processes resulted in an enhanced Raman signal.
Fig. 6.
Detection of thrombin by SERS. (A) SERS based thrombin sensing. Thrombin is first conjugated on thrombin aptamer 1 (blue) immobilized on Au surface. AuNPs with both Raman reporters and thrombin aptamer 2 (orange) is then attached on the thrombin pre-conjugated on aptamer 1 via the interaction between thrombin and aptamer 2. After Ag NP deposition, amplified Raman signal can be obtained. (B) SERS based adenosine sensing. In the presence of adenosine, structure switching of adenosine aptamer strand (orange) of dsDNA causes the release of the short DNA strand with Raman reporter (blue). The DNA with Raman reporter is hybridized to the DNA immobilized on rough Au surface (green), inducing amplified Raman scattering.
SERS can not only be used in vitro, but it can also be used in vivo for tumor targeting. Nie and co-workers recently reported that polyethylene glycol (PEG) modified 80 nm AuNPs which incorporated both Raman reporter and antibody modification could be used as SERS nanoparticle for targeted imaging of tumors [135]. Gambhir and coworkers also demonstrated non-invasive whole body Raman imaging and in vivo tumor imaging using SERS nanoparticles and single wall CNTs [136].
3.4. Magnetic Resonance Imaging (MRI)
While both fluorescence and SERS have been used in imaging in vivo, the penetrating length is limited. MRI, on the other hand, does not have this limitation and allows 3D imaging of the whole human body with clinically available instruments all over the world. Currently, MRI is mostly effective only when the difference in tissues, such as between a solid tumor and a regular tissue, is large, with little molecular information. For future MRI imaging, it is preferred to use MRI to detect molecular markers of the tumor before the tumor develops. To achieve this goal, new smart MRI contrast agents responsive to small molecular markers are required.
Superparamagnetic iron oxide nanoparticles are a new class of MRI contrast agents, catching the attention of many researchers due to their advantageous properties over gadolinium-based contrast agents. It has been shown in literature that superparamagnetic iron oxide nanoparticles can be coupled to biological molecules including thiol modified oligonucleotides for active targeting. Weissleder and co-workers demonstrated that cross-linked dextran iron oxide (CLIO) nanoparticles can be functionalized with DNA which can be used to detect sequence specific oligonucleotide by MRI [137, 138]. CLIO nanoparticles change the spin-spin relaxation time (T2) of surrounding water protons. In addition, researchers observed that these nanoparticles become more efficient in decreasing T2 when assembled into bigger clusters. This observation has been confirmed by many different assembly and disassembly systems [137, 139–141]. Based on this principle it is possible to distinguish assembled and dissembled states of the nanoparticles and therefore detect the analytes which results in such transition.
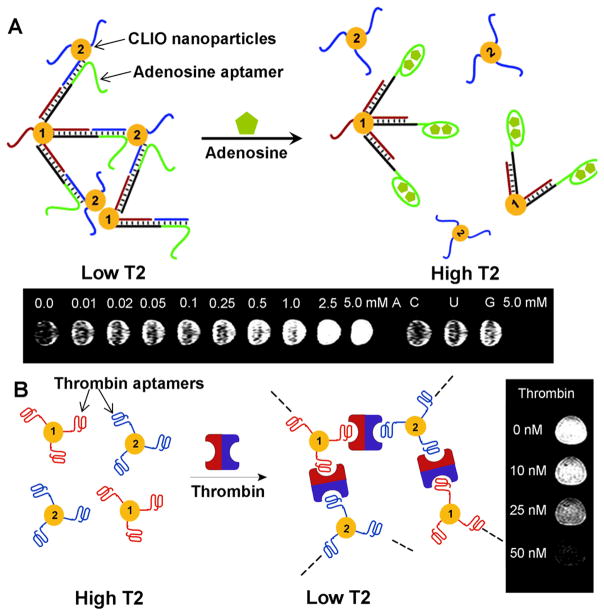
Recently we have developed a method for detecting chemical and biological molecules using aptamer functionalized CLIO nanoparticles. Adenosine aptamer could induce disassembly of CLIO nanoparticle aggregates in the presence of adenosine which was observed by MRI with an increase in the brightness of T2-weighted MR images (Figure 7A) [142]. The control analytes without any activity on adenosine aptamer did not show any change in MR images. The CLIO nanoparticles functionalized with mutated aptamer (which does not interact with adenosine) did not show any change in MR image, either. In order to demonstrate the response on proteins instead of small molecules and observe the change in contrast, we have tested the system for thrombin [143]. The CLIO nanoparticles functionalized with thrombin aptamers assembled in the presence of thrombin, which could be observed by the decrease of brightness in T2 weighted MR images (Figure 7B). Such an aptamer functionalized superparamagnetic iron oxide nanoparticles are suitable for non-invasive in vivo imaging of small molecular markers, making early diagnosis and treatment of diseases possible before the progression of the disease to late stages.
Fig. 7.
Smart MRI contrast agents based on aptamer functionalized CLIO nanoparticles. (A) MRI based small molecule sensing. Aggregates of CLIO nanoparticles were formed by hybridization of two strands of DNA (brown and blue) functionalized on CLIO nanoparticles with the complementary DNA containing adenosine aptamer (green). The CLIO nanoparticle aggregates disassembled in the presence of adenosine, due to the structure switching of aptamers after binding to adenosine, which could be observed by the increase of T2 value and brighter MR images. (B) MRI based protein sensing. The CLIO nanoparticles functionalized with two different thrombin aptamers (red and blue) assemble in the presence of thrombin, which could be observed by the decrease of T2 value and darker MR images. Figure 7A is reproduced with permission from ref. 127, copyright of Wiley-VCH Verlag GmbH & Co. KGaA. Figure 7B is reproduced with permission from ref 128, copyright of the American Chemical Society.
3.5. Electrochemical detection
Most of the optical detection methods require highly transparent sample solution as the color of the solution might interfere with the optical signal generated from sensors. For biomedical diagnostics on biological samples, such as blood, it is necessary to find an alternative method. Electrochemical detection provides another important route for non-transparent samples since the signal transduction can be accomplished by non-optical means [144].
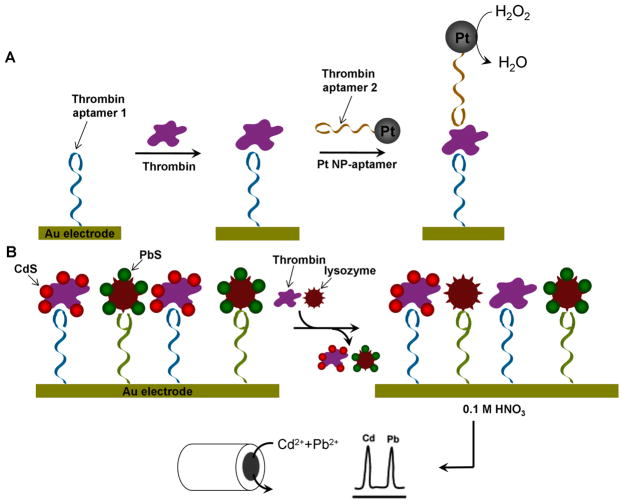
Electrochemical detection is carried out by measuring changes in redox states of redox active probes. As the labels can be easily attached to DNA, electrochemical method has been used intensively to detect DNA [145–147]. There are several different strategies reported for electrochemical detection of analytes using aptamers. One takes advantage of the redox state change induced by the enzyme immobilized on DNA aptamers. Since thrombin has two binding sites for two different aptamers [61, 133], it is possible to immobilize thrombin on Au electrode with one aptamer and bind enzyme which is tethered on the other aptamer. An example of using Pt NPs as catalytic labels is shown in Figure 8A [148]. The thrombin aptamer 1 was first immobilized on Au electrode and allowed to bind to thrombin. The Pt NP tethered with thrombin aptamer 2 was then introduced to form thrombin-aptamer-Pt NP sandwich complex. The Pt NPs catalyzed H2O2 reduction to H2O, resulting in cathodic currents which enabled thrombin detection with a detection limit as low as 1 nM. Similar methods using biocatalysts were also reported [149, 150] but Pt NPs showed higher sensitivity, probably due to the effectiveness of the Pt NP catalyst. It is also possible to carry out electrochemical sensing based on the conformation change of redox reporter labeled aptamers immobilized on Au electrode for detection of small molecules [151–153] and protein [154–157].
Fig. 8.
Electrochemistry based multiplexed detection of proteins. (A) Electrochemical sensor for the detection of thrombin, based on the redox change induced by the Pt NPs immobilized on DNA thrombin aptamers. The Pt NPs catalyze H2O2 reduction to H2O, resulting in cathodic currents, enabling thrombin detection. (B) Multiplexed detection of thrombin and lysozyme. Aptamers for both proteins are immobilized on Au surface and CdS labeled thrombin and PbS labeled lysozyme are then bound on the aptamers, respectively. In the presence of protein analyte, the protein replaces corresponding QD labeled protein on the surface. The amount of proteins in sample solution can be identified by monitoring the remaining QD by electrochemical stripping detection. Reproduced with permission of ref. 143, copyright of American Chemical Society.
The electrochemical sensors mentioned above are highly sensitive. To extend the methodology to multiplex detection, Wang and co-workers used protein labeled QDs with different chemical compositions [158]. In this work, they immobilized two different aptamers on a Au surface which were specific for thrombin and lysozyme, and then introduced both thrombin and lysozyme labeled with CdS and PbS QDs, respectively (see Figure 8B). The protein analytes (either thrombin or lysozyme) were added afterwards, replacing corresponding QD labeled proteins on the surface with sample analytes. By monitoring the remaining QDs via electrochemical stripping detection, it was possible to identify the proteins in the sample solution. Using this method, sub-picomolar detection limit was achieved with high selectivity.
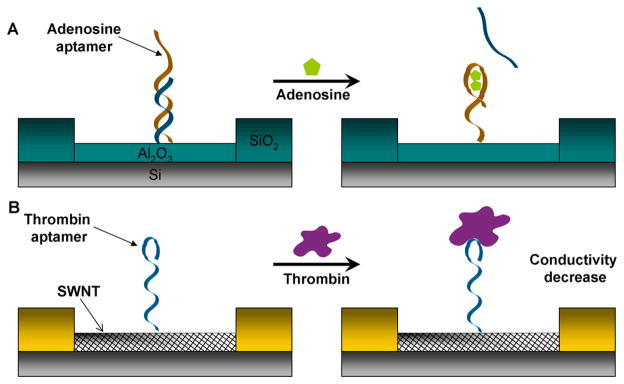
Willner and co-workers demonstrated label-free detection of adenosine mono-phosphate (AMP) using ion-selective field effect transistors (ISFET) [159]. In this work, they immobilized adenosine selective aptamers on Al2O3 gate surface and hybridized aptamers with short complementary DNA strands (see Figure 9A). The addition of AMP induced structure switching of aptamer strand, causing release of short partially complementary DNA strand. This released complementary strand altered the charge associated on the gate, changing the source to drain current. Gate-to-source potential (Vgs) of the device increased as the concentration of AMP became higher and the detection limit of the device was 50μM.
Fig. 9.
Detection of chemical and biological molecules based on aptamer conjugated electronic devices. (A) Label-free detection of AMP using ion-selective field effect transistors. Adenosine aptamer (brown) is immobilized onto Al2O3 gate surface and hybridized with a partially complementary DNA strand (blue). In the presence of adenosine, complementary strand is released, inducing change of the source-to-drain current. (B) CNT based field effect transistor sensor for the detection of thrombin. Thombin aptamers (blue) are immobilized on CNT field effect transistor. The introduction of thrombin decreases the conductance of field effect transistor.
Carbon nanotube (CNT) is an interesting nanomaterial with either metallic or semiconductive properties based on its structure, such as chirality or diameter [160–162]. As CNTs have the size (c.a. 1–2 nm diameter) comparable to most biomolecules as well as interesting physical and chemical properties, there has been intense investigation to conjugate CNTs with biomolecules to form a hybrid system. It has been reported that DNA can either noncovalently bind to the external part of CNT walls after electrical, thermal, or sonication treatment [163], or covalently bind to the carboxyl groups introduced on the CNT sidewalls after oxidization treatment [164–167]. Bifunctional reagents can also be used to introduce functional groups on DNA [168, 169].
CNTs can be used as a conducting channel in field effect transistors (FET). As the interaction between biomolecules and CNTs can modulate the conductivity of the FET, detection can be carried out based on the change of electrical signal. Detection of DNA has been demonstrated using CNT based FETs.
Lee and co-workers were the first to report the detection of adenosine using aptamer conjugated single wall carbon nanotube (SWNT) field effect transistor (FET) [170]. In their work, they modified the surface of CNT with CDI-Tween via hydrophobic interaction [169] and covalently attached 3′-amine modified thrombin aptamer DNA on it (Figure 9B). As soon as thrombin was dropped on the surface of the device, the conductance of the device decreased abruptly and soon saturated, which might be attributed to the screening of negative charges of DNA aptamers or positive charge induced by the introduction of thrombin. The detection limit was as low as 10 nM and the device had selectivity over the protein elastase, which is another serine protease with molecular weight and isoelectric point similar to thrombin.
It is reported that aptamers work much more efficiently than antibodies in CNT FET sensors. Tamiya and co-workers compared the performance of IgE specific aptamer and monoclonal antibody for the detection of immunoglobulin E (IgE) on CNT FET system [171]. Even though they could observe IgE induced conductance decrease for both aptamers and antibodies systems, the aptamer system worked much more effectively than antibody with the detection limit of 250 pM. The reason is mainly due to the small size of aptamers (1–2 nm) compared to antibodies (~10 nm) that allows target protein to bind to CNT within the electrical double layer in physiological conditions, making the detection possible. This method is versatile and its application for the detection of microorganism was also reported using Escheuchia-coli (E. coli) as an example [172]. By employing RNA based E. coli aptamers, the study demonstrated the detection of E. Coli in less than 20 minutes with selectivity over salmonella.
4. Targeted drug delivery in cells using aptamer-nanomaterial conjugates
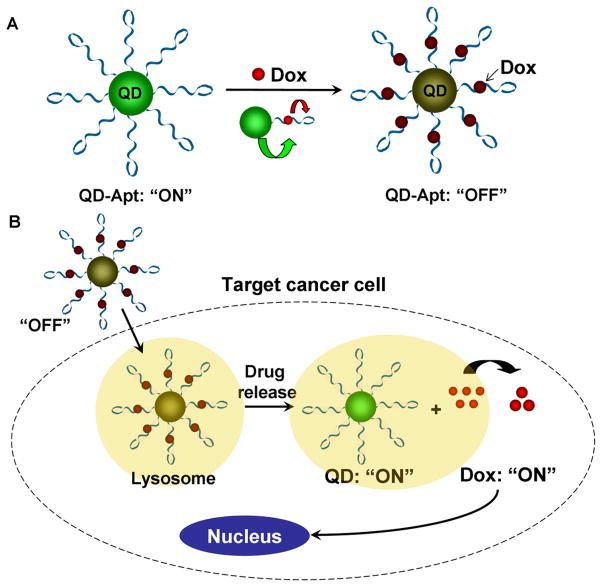
In addition to molecular diagnostics and imaging discussed above, the aptamer-nanomaterial conjugates have also been applied in targeted drug delivery, making it possible for therapeutic applications. Farokhzad, Langer, and co-workers reported a smart QD-aptamer conjugate which serves both as a fluorescence imager and a drug delivery vehicle (See Figure 10) [173]. The QD-aptamer conjugate was composed of 3 components; QD, prostate cancer (PCa) cell specific RNA aptamer, and doxorubicin (Dox) which is a commonly used anthracycline drug with fluorescence when intercalated into CG pairs of double stranded oligonucleotide. Aptamers were conjugated to QDs and Dox was intercalated to the aptamer strand [174], forming QD-aptamer (Dox) system. QD-aptamer (Dox) system was initially ‘off’ since the fluorescence of QD was transferred to Dox via energy transfer and the fluorescence of Dox also was quenched by the dsRNA aptamer due to energy transfer as well. Once injected into cancer cell, QD-aptamer (Dox) system gradually released Dox induced by the binding of target molecule onto RNA aptamer. This Dox release recovered the fluorescence of the QD. So the QD-aptamer (Dox) system not only allows targeted drug delivery, but also enables monitoring drug release and imaging target cells. A similar methodology has also been applied to superparamagnetic iron oxide nanoparticles for prostate cancer imaging and therapy [175].
Fig. 10.
QD-aptamer conjugate serving as both a fluorescence imaging agent and a drug delivery vehicle. (A) The conjugate is composed of QD, RNA aptamer, and doxorubicin (Dox). QD-aptamer (Dox) system is initially ‘off’ as the fluorescence of QD is transferred to Dox and the fluorescence of Dox is quenched by the dsRNA aptamer due to energy transfer. (B) Once placed inside cancer cells, Dox is gradually released from QD-aptamer (Dox) system and fluorescence of the QD is recovered.
Lippard, Farokhzad, Langer, and coworkers demonstrated aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles for targeted delivery of cisplatin to prostate cancer cells [176]. Prostate-specific membrane antigen (PSMA) targeting aptamer functionalized controlled release block-copolymer nanoparticles could target and be taken up by human prostate PSMA overexpressing LNCaP epithelial cells with high efficiency, but not to PC3 cancer cells. Furthermore, the aptamer-Pt(IV) encapsulated NPs were much more effective than the free cisplatin or nontargeted nanoparticles against the LNCaP cells for drug delivery. In addition, Langer and coworkers were the first to demonstrate aptamer based drug delivery in vivo [50]. They showed that docetaxel (Dtxl) containing di-block copolymer nanoparticles functionalized with RNA aptamers could significantly decrease tumor with reduced toxicity compared to Dtxl alone.
Tan and coworkers demonstrated an advantage of using multiple aptamers conjugated nanorods as a multivalent probe for targeted cancer cell imaging [177]. Many targeting molecules, such as antibodies or aptamers, have moderate binding affinities which prevent efficient targeted cell imaging. By using Au-Ag nanorods conjugated to up to 80 fluorophore labeled aptamers, 300 fold increased fluorescence signal and 26 times higher binding affinity was obtained compared to individual aptamer strand. This methodology enables to target and image cells with low density binding sites or targeting molecules with relatively low binding affinities.
Targeted delivery of DNA or RNA molecules into cells is among one of potent therapeutic strategies for genetic diseases such as cancer. However, practical application of DNA or RNA delivery in cellular environment is difficult as those molecules can be generally degraded or digested by nucleases or enzymes. It is reported that SWNTs can be used as a protective cargo for delivery of ssDNA into cells [178]. The work from Tan and coworkers demonstrated the stability of delivered DNA against nuclease and enzymes in vitro. In addition, it has been shown that specific mRNA targeting DNA could be delivered efficiently into cells in DNA/SWNT-complexed form and the delivered DNA was active. This work demonstrated the possibility of using SWNTs as DNA/RNA delivery molecules for diagnostic and therapeutic applications. Recently, it has been reported that viral caspid carrier can also be functionalized to DNA aptamer and these can potentially be used for targeted drug delivery [179]. Capsids functionalized with 41 mer DNA aptamer specific for tyrosine kinase receptor could successfully target and be uptaken by Jurkat T cells.
We, in collaboration with Wong and Cheng groups, recently reported cancer cell specific drug delivery based on aptamer functionalized liposomes[180]. Liposome is, by far, the most widespread system used for drug delivery. By conjugating aptamers specific to nucleolin, which is overexpressed on a number of cancer cell plasma membrane [181], along with cisplatin encapsulated inside liposomes, we demonstrated targeted delivery of cisplatin onto nucleolin overexpressing breast cancer MCF-7 cells but not to control prostate cancer cells (LNCaP cell). Furthermore, the extent of drug release can be controlled by using a complementary DNA of the aptamer as an antidote so that the drug effects can be readily controlled.
5. Conclusion
By summarizing recent progress in developing methods of integrating aptamers with a diverse number of nanomaterials, we have shown that these novel bionanomaterials can be used as highly sensitive and selective diagnostic agents and targeted drug delivery agents. As each nanomaterial has different optical, electrical, magnetic, and mechanical properties, medical diagnostic and drug delivery agents with diverse characteristics can be used for different biomedical applications, making them an excellent alternative to antibody-based medical applications.
Most of the agents introduced here, however, used available aptamers specific for molecules such as adenosine, cocaine and thrombin as a proof of concept and most of tests have been carried out in vitro. To realize the full potentials of such bionanomaterials systems, one needs to carry out more selections for aptamers that are specific for more clinically relevant targets such as cancer markers and viruses, and demonstrate the effectiveness of the systems in vivo. Given progresses made in the field in such a short period of time, applications of these aptamer-nanomaterial based sensors in clinical applications will be realized in the near future.
Acknowledgments
We thank the US Department of Energy (DE-FG02-01-ER63179), the National Institute of Health (ES016865), and the National Science Foundation (DMR-0117792, CTS-0120978 and DMI-0328162) for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lieber CM. Nanoscale Science and Technology: Building a Big Future from Small Things. MRS Bull. 2003;28:486. [Google Scholar]
- 2.Daniel MC, Astruc D. Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-Size-Related Properties, and Applications toward Biology, Catalysis, and Nanotechnology. Chem Rev. 2004;104:293. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Lu Y. A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J Am Chem Soc. 2000;122:10466. [Google Scholar]
- 4.Jhaveri SD, Kirby R, Conrad R, Maglott EJ, Bowser M, Kennedy RT, Glick G, Ellington AD. Designed Signaling Aptamers That Transduce Molecular Recognition to Changes in Fluorescence Intensity. J Am Chem Soc. 2000;122:2469. [Google Scholar]
- 5.Lu Y. New Transition Metal-Dependent DNAzymes as Efficient Endonucleases and as Selective Metal Biosensors. Chem Eur J. 2002;8:4588. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Navani NK, Li Y. Nucleic Acid Aptamers and Enzymes as Sensors. Curr Opin Chem Biol. 2006;10:272. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Lu Y, Liu J. Functional DNA Nanotechnology: Emerging Applications of DNAzymes and Aptamers. Curr Opin Biotechnol. 2006;17:580. doi: 10.1016/j.copbio.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Lu Y, Liu J. Smart Nanomaterials Inspired by Biology: Dynamic Assembly of Error-Free Nanomaterials in Response to Multiple Chemical and Biological Stimuli. Acc Chem Res. 2007;40:315. doi: 10.1021/ar600053g. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Cao Z, Lu Y. Functional Nucleic Acid Sensors. Chem Rev. 2009;109:1948. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Lu Y. Functional DNA Directed Assembly of Nanomaterials for Biosensing. J Mater Chem. 2009;19:1788. doi: 10.1039/B813939C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Lu Y. Functional Nucleic Acids for Sensing and Other Analytical Applications. Springer; New York, NY: 2009. [Google Scholar]
- 12.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 13.Hazarika P, Ceyhan B, Niemeyer CM. Reversible Switching of DNA-Gold Nanoparticle Aggregation. Angew Chem Int Ed. 2004;43:6469. doi: 10.1002/anie.200461887. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Rothberg L. Colorimetric Detection of DNA Sequences Based on Electrostatic Interactions with Unmodified Gold Nanoparticles. Proc Natl Acad Sci USA. 2004;101:14036. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Rothberg L. Detection of Specific Sequences in RNA Using Differential Adsorption of Single-Stranded Oligonucleotides on Gold Nanoparticles. Anal Chem. 2005;77:6229. doi: 10.1021/ac050921y. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Lu Y. A Colorimetric Lead Biosensor Using DNAzyme-Directed Assembly of Gold Nanoparticles. J Am Chem Soc. 2003;125:6642. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Lu Y. Stimuli-Responsive Disassembly of Nanoparticle Aggregates for Light-up Colorimetric Sensing. J Am Chem Soc. 2005;127:12677. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- 18.Lee J-S, Han MS, Mirkin CA. Colorimetric Detection of Mercuric Ion (Hg2+) in Aqueous Media by DNA-Functionalized Gold Nanoparticles. Angew Chem Int Ed. 2007;46:4093. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Liu X, Hu X, Song S, Fan C. Unmodified Gold Nanoparticles as a Colorimetric Probe for Potassium DNA Aptamers. Chem Commun. 2006:3780. doi: 10.1039/b607448k. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. A Catalytic Beacon Sensor for Uranium with Parts-Per-Trillion Sensitivity and Millionfold Selectivity. Proc Natl Acad Sci USA. 2007;104:2056. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Lu Y. Rational Design Of “Turn-On” Allosteric Dnazyme Catalytic Beacons for Aqueous Mercury Ions with Ultrahigh Sensitivity and Selectivity. Angew Chem Int Ed. 2007;46:7587. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Lu Y. Colorimetric Cu2+ Detection with a Ligation DNAzyme and Nanoparticles. Chem Commun. 2007:4872. doi: 10.1039/b712421j. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Lu Y. A DNAzyme Catalytic Beacon Sensor for Paramagnetic Cu2+ Ions in Aqueous Solution with High Sensitivity and Selectivity. J Am Chem Soc. 2007;129:9838. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]
- 24.Liu CW, Hsieh YT, Huang CC, Lin ZH, Chang HT. Detection of Mercury(II) Based on Hg2+ -DNA Complexes Inducing the Aggregation of Gold Nanoparticles. Chem Commun. 2008;19:2242. doi: 10.1039/b719856f. [DOI] [PubMed] [Google Scholar]
- 25.Wernette DP, Liu J, Bohn PW, Lu Y. Functional-DNA-Based Nanoscale Materials and Devices for Sensing Trace Contaminants in Water. MRS Bull. 2008;33:34. [Google Scholar]
- 26.Li D, Wieckowska A, Willner I. Optical Analysis of Hg2+ Ions by Oligonucleotide-Gold-Nanoparticle Hybrids and DNA-Based Machines. Angew Chem Int Ed. 2008;47:3927. doi: 10.1002/anie.200705991. [DOI] [PubMed] [Google Scholar]
- 27.Dalavoy TS, Wernette DP, Gong M, Sweedler JV, Lu Y, Flachsbart BR, Shannon MA, Bohn PW, Cropek DM. Immobilization of DNAzyme Catalytic Beacons on Pmma for Pb2+ Detection. Lab on a Chip. 2008;8:786. doi: 10.1039/b718624j. [DOI] [PubMed] [Google Scholar]
- 28.Wang Z, Lee JH, Lu Y. Label Free Colorimetric Detection of Metal Ions Using Gold Nanoparticles and DNAzyme with 3 nM Detection Limit and Tunable Dynamic Range. Adv Mater. 2008;20:3263. [Google Scholar]
- 29.Wei H, Li B, Li J, Dong S, Wang E. DNAzyme-Based Colorimetric Sensing of Lead (Pb2+) Using Unmodified Gold Nanoparticle Probes. Nanotechnology. 2008;19:095501/1. doi: 10.1088/0957-4484/19/9/095501. [DOI] [PubMed] [Google Scholar]
- 30.Brown AK, Liu J, He Y, Lu Y. Biochemical Characterization of a Uranyl Ion-Specific Dnazyme. ChemBioChem. 2009;10:486. doi: 10.1002/cbic.200800632. [DOI] [PubMed] [Google Scholar]
- 31.Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: An Emerging Class of Therapeutics. Annu Rev Med. 2005;56:555. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 32.Shamah SM, Healy JM, Cload ST. Complex Target Selex. Acc Chem Res. 2008;41:130. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 33.Stojanovic MN, de Prada P, Landry DW. Fluorescent Sensors Based on Aptamer Self-Assembly. J Am Chem Soc. 2000;122:11547. doi: 10.1021/ja0022223. [DOI] [PubMed] [Google Scholar]
- 34.Saitoh H, Nakamura A, Kuwahara M, Ozaki H, Sawai H. Modified DNA Aptamers against Sweet Agent Aspartame. Nucleic Acids Research Supplement. 2002;2:215. doi: 10.1093/nass/2.1.215. [DOI] [PubMed] [Google Scholar]
- 35.Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Vargeese C, Kirschenheuter G, McGee DPC, Abesinghe P, et al. Potent 2′-Amino-2′-Deoxypyrimidine RNA Inhibitors of Basic Fibroblast Growth Factor. Biochemistry. 1995;34:11363. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 36.Pagratis NC, Bell C, Chang Y-F, Jennings S, Fitzwater T, Jellinek D, Dang C. Potent 2′-Amino-and 2′-Fluoro-2′-Deoxyribonucleotide Rna Inhibitors of Keratinocyte Growth Factor. Nat Biotechnol. 1997;15:68. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- 37.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 2′-Fluoropyrimidine RNA-Based Aptamers to the 165-Amino Acid Form of Vascular Endothelial Growth Factor (VEGF165). Inhibition of Receptor Binding and Vegf-Induced Vascular Permeability through Interactions Requiring the Exon 7-Encoded Domain. J Biol Chem. 1998;273:20556. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 38.Green LS, Jellinek D, Jenison R, Oestman A, Heldin C-H, Janjic N. Inhibitory DNA Ligands to Platelet-Derived Growth Factor B-Chain. Biochemistry. 1996;35:14413. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa A, Tomita N, Kikuchi N, Sando S, Aoyama Y. Aptamer Selection for the Inhibition of Cell Adhesion with Fibronectin as Target. Bioorg Med Chem Lett. 2004;14:4001. doi: 10.1016/j.bmcl.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 40.Mendonsa SD, Bowser MT. In Vitro Selection of Aptamers with Affinity for Neuropeptide Y Using Capillary Electrophoresis. J Am Chem Soc. 2005;127:9382. doi: 10.1021/ja052406n. [DOI] [PubMed] [Google Scholar]
- 41.Tang J, Xie J, Shao N, Yan Y. The DNA Aptamers That Specifically Recognize Ricin Toxin Are Selected by Two in Vitro Selection Methods. Electrophoresis. 2006;27:1303. doi: 10.1002/elps.200500489. [DOI] [PubMed] [Google Scholar]
- 42.Tang J, Yu T, Guo L, Xie J, Shao N, He Z. In Vitro Selection of DNA Aptamer against Abrin Toxin and Aptamer-Based Abrin Direct Detection. Biosens Bioelectron. 2007;22:2456. doi: 10.1016/j.bios.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 43.Jeon SH, Kayhan B, Ben-Yedidia T, Arnon R. A DNA Aptamer Prevents Influenza Infection by Blocking the Receptor Binding Region of the Viral Hemagglutinin. J Biol Chem. 2004;279:48410. doi: 10.1074/jbc.M409059200. [DOI] [PubMed] [Google Scholar]
- 44.Koch TH, Smith D, Tabacman E, Zichi DA. Kinetic Analysis of Site-Specific Photoaptamer-Protein Cross-Linking. J Mol Biol. 2004;336:1159. doi: 10.1016/j.jmb.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 45.Bruno JG, Kiel JL. In Vitro Selection of DNA Aptamers to Anthrax Spores with Electrochemiluminescence Detection. Biosens Bioelectron. 1999;14:457. doi: 10.1016/s0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 46.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic Evolution of a DNA Aptamer Binding to Rat Brain Tumor Microvessels. Selective Targeting of Endothelial Regulatory Protein Pigpen. J Biol Chem. 2001;276:16464. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 47.Wang C, Zhang M, Yang G, Zhang D, Ding H, Wang H, Fan M, Shen B, Shao N. Single-Stranded DNA Aptamers That Bind Differentiated but Not Parental Cells: Subtractive Systematic Evolution of Ligands by Exponential Enrichment. J Biotechnol. 2003;102:15. doi: 10.1016/s0168-1656(02)00360-7. [DOI] [PubMed] [Google Scholar]
- 48.Gragoudas ES, Adamis AP, Cunningham ET, Jr, Feinsod M, Guyer DR. VEGF Inhibition Study in Ocular Neovascularization Clinical Trial Group. Pegaptanib for Neovascular Age-Related Macular Degeneration. N Engl J Med. 2004;351:2805. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 49.Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a Targeted Anti-Vegf Aptamer for Ocular Vascular Disease. Nat Rev Drug Discov. 2006;5:123. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 50.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Targeted Nanoparticle-Aptamer Bioconjugates for Cancer Chemotherapy in Vivo. Proc Natl Acad Sci USA. 2006;103:6315. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Proske D, Blank M, Buhmann R, Resch A. Aptamers-Basic Research, Drug Development, and Clinical Applications. Appl Microbiol Biotechnol. 2005;69:367. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- 52.Fichou Y, Ferec C. The Potential of Oligonucleotides for Therapeutic Applications. Trends Biotechnol. 2006;24:563. doi: 10.1016/j.tibtech.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Ulrich H, Trujillo CA, Nery AA, Alves JM, Majumder P, Resende RR, Martins AH. DNA and RNA Aptamers: From Tools for Basic Research Towards Therapeutic Applications. Comb Chem High T Scr. 2006;9:619. doi: 10.2174/138620706778249695. [DOI] [PubMed] [Google Scholar]
- 54.Missailidis S, Perkins A. Update: Aptamers as Novel Radiopharmaceuticals: Their Applications and Future Prospects in Diagnosis and Therapy. Cancer Biother Radiopharm. 2007;22:453. doi: 10.1089/cbr.2007.357. [DOI] [PubMed] [Google Scholar]
- 55.Tombelli S, Minunni M, Mascini M. Aptamers-Based Assays for Diagnostics, Environmental and Food Analysis. Biomol Eng. 2007;24:191. doi: 10.1016/j.bioeng.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 56.Famulok M, Hartig JS, Mayer G. Functional Aptamers and Aptazymes in Biotechnology, Diagnostics, and Therapy. Chem Rev. 2007;107:3715. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 57.Balamurugan S, Obubuafo A, Soper SA, Spivak DA. Surface Immobilization Methods for Aptamer Diagnostic Applications. Anal Bioanal Chem. 2008;390:1009. doi: 10.1007/s00216-007-1587-2. [DOI] [PubMed] [Google Scholar]
- 58.Ellington AD, Szostak JW. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature. 1990;346:818. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 59.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 60.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study. Proc Natl Acad Sci USA. 2006;103:11838. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of Single-Stranded DNA Molecules That Bind and Inhibit Human Thrombin. Nature. 1992;355:564. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 62.Herr JK, Smith JE, Medley CD, Shangguan D, Tan W. Aptamer-Conjugated Nanoparticles for Selective Collection and Detection of Cancer Cells. Anal Chem. 2006;78:2918. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 63.Sassanfar M, Szostak JW. An RNA Motif That Binds ATP. Nature. 1993;364:550. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 64.Meli M, Vergne J, Decout J-L, Maurel M-C. Adenine-Aptamer Complexes. A Bipartite RNA Site That Binds the Adenine Nucleic Base. J Biol Chem. 2002;277:2104. doi: 10.1074/jbc.M107130200. [DOI] [PubMed] [Google Scholar]
- 65.Ellington AD, Szostak JW. Selection in Vitro of Single-Stranded DNA Molecules That Fold into Specific Ligand-Binding Structures. Nature. 1992;355:850. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 66.Famulok M, Szostak JW. Stereospecific Recognition of Tryptophan Agarose by In Vitro Selected Rna. J Am Chem Soc. 1992;114:3990. [Google Scholar]
- 67.Ciesiolka J, Gorski J, Yarus M. Selection of an RNA Domain That Binds Zn2+ RNA. 1995;1:538. [PMC free article] [PubMed] [Google Scholar]
- 68.Ciesiolka J, Yarus M. Small Rna-Divalent Domains. RNA. 1996;2:785. [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JH, Wernette DP, Yigit MV, Liu J, Wang Z, Lu Y. Site-Specific Control of Distances between Gold Nanoparticles Using Phosphorothioate Anchors on DNA and a Short Bifunctional Molecular Fastener. Angew Chem Int Ed. 2007;46:9006. doi: 10.1002/anie.200702569. [DOI] [PubMed] [Google Scholar]
- 70.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and Self-Assembly of Two-Dimensional DNA Crystals. Nature. 1998;394:539. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 71.Goodman RP, Schaap IA, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication. Science. 2005;310:1661. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 72.Rothemund PWK. Folding DNA to Create Nanoscale Shapes and Patterns. Nature. 2006;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 73.He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Hierarchical Self-Assembly of DNA into Symmetric Supramolecular Polyhedra. Nature. 2008;452:198. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 74.Seeman NC. DNA in a Material World. Nature. 2003;421:427. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 75.Seeman NC. The Design and Engineering of Nucleic Acid Nanoscale Assemblies. Curr Opin Struct Biol. 1996;6:519. doi: 10.1016/s0959-440x(96)80118-7. [DOI] [PubMed] [Google Scholar]
- 76.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Self-Assembled Water-Soluble Nucleic Acid Probe Tiles for Label-Free Rna Hybridization Assays. Science. 2008;319:180. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 77.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-Assembled DNA Nanostructures for Distance-Dependent Multivalent Ligand-Protein Binding. Nat Nanotechnol. 2008;3:418. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clark J, Smith SS. Application of Nanoscale Bioassemblies to Clinical Laboratory Diagnostics. Adv Clin Chem. 2006;41:23. doi: 10.1016/S0065-2423(05)41002-1. [DOI] [PubMed] [Google Scholar]
- 79.Willner I, Zayats M. Electronic Aptamer-Based Sensors. Angew Chem Int Ed. 2007;46:6408. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 80.Langer R. Drug Delivery and Targeting. Nature. 1998;392:5. [PubMed] [Google Scholar]
- 81.Wilson C, Szostak JW. Isolation of a Fluorophore-Specific DNA Aptamer with Weak Redox Activity. Chem Biol. 1998;5:609. doi: 10.1016/s1074-5521(98)90289-7. [DOI] [PubMed] [Google Scholar]
- 82.Liss M, Petersen B, Wolf H, Prohaska E. An Aptamer-Based Quartz Crystal Protein Biosensor. Anal Chem. 2002;74:4488. doi: 10.1021/ac011294p. [DOI] [PubMed] [Google Scholar]
- 83.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of Oligonucleotide Functions. Annu Rev Biochem. 1995;64:763. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 84.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD. Direct in Vitro Selection of a 2′-O-Methyl Aptamer to Vegf. Chem Biol. 2005;12:25. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 85.Park J-U, Lee JH, Paik U, Lu Y, Rogers JA. Nanoscale Patterns of Oligonucleotides Formed by Electrohydrodynamic Jet Printing with Applications in Biosensing and Nanomaterials Assembly. Nano Lett. 2008;8:4210. doi: 10.1021/nl801832v. [DOI] [PubMed] [Google Scholar]
- 86.Murray CB, Norris DJ, Bawendi MG. Synthesis and Characterization of Nearly Monodisperse CdE (E = Sulfur, Selenium, Tellurium) Semiconductor Nanocrystallites. J Am Chem Soc. 1993;115:8706. [Google Scholar]
- 87.Alivisatos AP. Semiconductor Clusters, Nanocrystals, and Quantum Dots. Science. 1996;271:933. [Google Scholar]
- 88.Peng X, Manna L, Yang W, Wickham J, Scher E, Kadavanich A, Alivisatos AP. Shape Control of CdSe Nanocrystals. Nature. 2000;404:59. doi: 10.1038/35003535. [DOI] [PubMed] [Google Scholar]
- 89.Mitchell GP, Mirkin CA, Letsinger RL. Programmed Assembly of DNA Functionalized Quantum Dots. J Am Chem Soc. 1999;121:8122. [Google Scholar]
- 90.Liang RQ, Li W, Li Y, Tan CY, Li JX, Jin YX, Ruan KC. An Oligonucleotide Microarray for Microrna Expression Analysis Based on Labeling Rna with Quantum Dot and Nanogold Probe. Nucleic Acids Res. 2005;33:e17. doi: 10.1093/nar/gni019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin CA, Sperling RA, Li JK, Yang TY, Li PY, Zanella M, Chang WH, Parak WJ. Design of an Amphiphilic Polymer for Nanoparticle Coating and Functionalization. Small. 2008;4:334. doi: 10.1002/smll.200700654. [DOI] [PubMed] [Google Scholar]
- 92.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single-Quantum-Dot-Based DNA Nanosensor. Nat Mater. 2005;4:826. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 93.Gill R, Willner I, Shweky I, Banin U. Fluorescence Resonance Energy Transfer in CdSe/ZnS-DNA Conjugates: Probing Hybridization and DNA Cleavage. J Phys Chem B. 2005;109:23715. doi: 10.1021/jp054874p. [DOI] [PubMed] [Google Scholar]
- 94.Levy M, Cater SF, Ellington AD. Quantum-Dot Aptamer Beacons for the Detection of Proteins. ChemBioChem. 2005;6:2163. doi: 10.1002/cbic.200500218. [DOI] [PubMed] [Google Scholar]
- 95.Choi JH, Chen KH, Strano MS. Aptamer-Capped Nanocrystal Quantum Dots: A New Method for Label-Free Protein Detection. J Am Chem Soc. 2006;128:15584. doi: 10.1021/ja066506k. [DOI] [PubMed] [Google Scholar]
- 96.Liu J, Lee JH, Lu Y. Quantum Dot Encoding of Aptamer-Linked Nanostructures for One Pot Simultaneous Detection of Multiple Analytes. Anal Chem. 2007;79:4120. doi: 10.1021/ac070055k. [DOI] [PubMed] [Google Scholar]
- 97.Cherukuri P, Bachilo SM, Litovsky SH, Weisman RB. Near-Infrared Fluorescence Microscopy of Single-Walled Carbon Nanotubes in Phagocytic Cells. J Am Chem Soc. 2004;126:15638. doi: 10.1021/ja0466311. [DOI] [PubMed] [Google Scholar]
- 98.Zhu Z, Tang Z, Phillips JA, Yang R, Wang H, Tan W. Regulation of Singlet Oxygen Generation Using Single-Walled Carbon Nanotubes. J Am Chem Soc. 2008;130:10856. doi: 10.1021/ja802913f. [DOI] [PubMed] [Google Scholar]
- 99.Yang R, Tang Z, Yan J, Kang H, Kim Y, Zhu Z, Tan W. Noncovalent Assembly of Carbon Nanotubes and Single-Stranded DNA: An Effective Sensing Platform for Probing Biomolecular Interactions. Anal Chem. 2008;80:7408. doi: 10.1021/ac801118p. [DOI] [PubMed] [Google Scholar]
- 100.Swearingen CB, Wernette DP, Cropek DM, Lu Y, Sweedler JV, Bohn PW. Immobilization of a Catalytic DNA Molecular Beacon on Au for Pb(II) Detection. Anal Chem. 2005;77:442. doi: 10.1021/ac0401016. [DOI] [PubMed] [Google Scholar]
- 101.Wernette DP, Swearingen CB, Cropek DM, Lu Y, Sweedler JV, Bohn PW. Incorporation of a DNAzyme into Au-Coated Nanocapillary Array Membranes with an Internal Standard for Pb(II) Sensing. Analyst. 2006;131:41. doi: 10.1039/b510071b. [DOI] [PubMed] [Google Scholar]
- 102.Wernette DP, Mead C, Bohn PW, Lu Y. Surface Immobilization of Catalytic Beacons Based on Ratiometric Fluorescent DNAzyme Sensors: A Systematic Study. Langmuir. 2007;23:9513. doi: 10.1021/la701303k. [DOI] [PubMed] [Google Scholar]
- 103.Kuo TC, Cannon DM, Shannon MA, Bohn PW, Sweedler JV. Hybrid Three-Dimensional Nanofluidic/Microfluidic Devices Using Molecular Gates, Sens. Actuators, A. 2003;102:223. [Google Scholar]
- 104.Kuo TC, Cannon DM, Chen YN, Tulock JJ, Shannon MA, Sweedler JV, Bohn PW. Gateable Nanofluidic Interconnects for Multilayered Microfluidic Separation Systems. Anal Chem. 2003;75:1861. doi: 10.1021/ac025958m. [DOI] [PubMed] [Google Scholar]
- 105.Chang I-H, Tulock JJ, Liu J, Kim W-S, Cannon DM, Jr, Lu Y, Bohn PW, Sweedler JV, Cropek DM. Miniaturized Lead Sensor Based on Lead-Specific DNAzyme in a Nanocapillary Interconnected Microfluidic Device. Environ Sci Technol. 2005;39:3756. doi: 10.1021/es040505f. [DOI] [PubMed] [Google Scholar]
- 106.Shaikh KA, Ryu KS, Goluch ED, Nam J-M, Liu J, Thaxton CS, Chiesl TN, Barron AE, Lu Y, Mirkin CA, Liu C. A Modular Microfluidic Architecture for Integrated Biochemical Analysis. Proc Natl Acad Sci USA. 2005;102:9745. doi: 10.1073/pnas.0504082102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stojanovic MN, Landry DW. Aptamer-Based Colorimetric Probe for Cocaine. J Am Chem Soc. 2002;124:9678. doi: 10.1021/ja0259483. [DOI] [PubMed] [Google Scholar]
- 108.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. Aptamer-Functionalized Au Nanoparticles for the Amplified Optical Detection of Thrombin. J Am Chem Soc. 2004;126:11768. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 109.Ho H-A, Leclerc M. Optical Sensors Based on Hybrid Aptamer/Conjugated Polymer Complexes. J Am Chem Soc. 2004;126:1384. doi: 10.1021/ja037289f. [DOI] [PubMed] [Google Scholar]
- 110.Huang C-C, Huang Y-F, Cao Z, Tan W, Chang H-T. Aptamer-Modified Gold Nanoparticles for Colorimetric Determination of Platelet-Derived Growth Factors and Their Receptors. Anal Chem. 2005;77:5735. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 111.Liu J, Lu Y. Fast Colorimetric Sensing of Adenosine and Cocaine Based on a General Sensor Design Involving Aptamers and Nanoparticles. Angew Chem Int Ed. 2006;45:90. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 112.Chen J, Saeki F, Wiley BJ, Cang H, Cobb MJ, Li Z-Y, Au L, Zhang H, Kimmey MB, Li X, Xia Y. Gold Nanocages: Bioconjugation and Their Potential Use as Optical Imaging Contrast Agents. Nano Lett. 2005;5:473. doi: 10.1021/nl047950t. [DOI] [PubMed] [Google Scholar]
- 113.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 114.Li H, Rothberg LJ. Label-Free Colorimetric Detection of Specific Sequences in Genomic DNA Amplified by the Polymerase Chain Reaction. J Am Chem Soc. 2004;126:10958. doi: 10.1021/ja048749n. [DOI] [PubMed] [Google Scholar]
- 115.Lee JH, Wang Z, Liu J, Lu Y. Highly Sensitive and Selective Colorimetric Sensors for Uranyl (UO22+): Development and Comparison of Labeled and Label-Free Dnazyme-Gold Nanoparticle Systems. J Am Chem Soc. 2008;130:14217. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Y, Li D, Ren W, Liu Z, Dong S, Wang E. Ultrasensitive Colorimetric Detection of Protein by Aptamer-Au Nanoparticles Conjugates Based on a Dot-Blot Assay. Chem Commun. 2008;(22):2520. doi: 10.1039/b801055b. [DOI] [PubMed] [Google Scholar]
- 117.Liu J, Lu Y. Smart Nanomaterials Responsive to Multiple Chemical Stimuli with Controllable Cooperativity. Adv Mater. 2006;18:1667. [Google Scholar]
- 118.Wang J, Wang L, Liu X, Liang Z, Song S, Li W, Li G, Fan C. A Gold Nanoparticle-Based Aptamer Target Binding Readout for ATP Assay. Adv Mater. 2007;19:3943. [Google Scholar]
- 119.Wei H, Li B, Li J, Wang E, Dong S. Simple and Sensitive Aptamer-Based Colorimetric Sensing of Protein Using Unmodified Gold Nanoparticle Probes. Chem Commun. 2007;36:3735. doi: 10.1039/b707642h. [DOI] [PubMed] [Google Scholar]
- 120.Zhao W, Chiuman W, Brook MA, Li Y. Simple and Rapid Colorimetric Biosensors Based on DNA Aptamer and Noncrosslinking Gold Nanoparticle Aggregation. ChemBioChem. 2007;8:727. doi: 10.1002/cbic.200700014. [DOI] [PubMed] [Google Scholar]
- 121.Zhao W, Chiuman W, Lam JCF, McManus SA, Chen W, Cui Y, Pelton R, Brook MA, Li Y. DNA Aptamer Folding on Gold Nanoparticles: From Colloid Chemistry to Biosensors. J Am Chem Soc. 2008;130:3610. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]
- 122.Chan CP, Cheung YC, Renneberg R, Seydack M. New Trends in Immunoassays. Adv Biochem Eng Biotechnol. 2008;109:123. doi: 10.1007/10_2007_075. [DOI] [PubMed] [Google Scholar]
- 123.Glynou K, Ioannou PC, Christopoulos TK, Syriopoulou V. Oligonucleotide-Functionalized Gold Nanoparticles as Probes in a Dry-Reagent Strip Biosensor for DNA Analysis by Hybridization. Anal Chem. 2003;75:4155. doi: 10.1021/ac034256+. [DOI] [PubMed] [Google Scholar]
- 124.Liu J, Mazumdar D, Lu Y. A Simple and Sensitive “Dipstick” Test in Serum Based on Lateral Flow Separation of Aptamer-Linked Nanostructures. Angew Chem Int Ed. 2006;45:7955. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 125.Su S, Nutiu R, Filipe CDM, Li Y, Pelton R. Adsorption and Covalent Coupling of Atp-Binding DNA Aptamers onto Cellulose. Langmuir. 2007;23:1300. doi: 10.1021/la060961c. [DOI] [PubMed] [Google Scholar]
- 126.Nie S, Emory SR. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science. 1997;275:1102. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 127.Fleischmann M, Hendra PJ, McQuillan AJ. Raman Spectra of Pyridine Adsorbed at a Silver Electrode. Chem Phys Lett. 1974;26:163. [Google Scholar]
- 128.Willets KA, Van Duyne RP. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu Rev Phys Chem. 2007;58:267. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 129.Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Single Molecule Detection using Surface-Enhanced Raman Scattering. Phys Rev Lett. 1997;78:1667. [Google Scholar]
- 130.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science. 2002;297:1536. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 131.Barhoumi A, Zhang D, Tam F, Halas NJ. Surface-Enhanced Raman Spectroscopy of DNA. J Am Chem Soc. 2008;130:5523. doi: 10.1021/ja800023j. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y, Wei H, Li B, Ren W, Guo S, Dong S, Wang E. SERS Opens a New Way in Aptasensor for Protein Recognition with High Sensitivity and Selectivity. Chem Commun. 2007;48:5220. doi: 10.1039/b709492b. [DOI] [PubMed] [Google Scholar]
- 133.Tasset DM, Kubik MF, Steiner W. Oligonucleotide Inhibitors of Human Thrombin That Bind Distinct Epitopes. J Mol Biol. 1997;272:688. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 134.Chen JW, Liu XP, Feng KJ, Liang Y, Jiang JH, Shen GL, Yu RQ. Detection of Adenosine Using Surface-Enhanced Raman Scattering Based on Structure-Switching Signaling Aptamer. Biosens Bioelectron. 2008;24:66. doi: 10.1016/j.bios.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 135.Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. In Vivo Tumor Targeting and Spectroscopic Detection with Surface-Enhanced Raman Nanoparticle Tags. Nat Biotechnol. 2008;26:83. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 136.Keren S, Zavaleta C, Cheng Z, de la Zerda A, Gheysens O, Gambhir SS. Noninvasive Molecular Imaging of Small Living Subjects Using Raman Spectroscopy. Proc Natl Acad Sci USA. 2008;105:5844. doi: 10.1073/pnas.0710575105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Josephson L, Perez JM, Weissleder R. Magnetic Nanosensors for the Detection of Oligonucleotide Sequences. Angew Chem Int Ed. 2001;40:3204. doi: 10.1002/1521-3773(20010903)40:17<3204::AID-ANIE3204>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 138.Perez JM, Josephson L, O’Loughlin T, Hoegemann D, Weissleder R. Magnetic Relaxation Switches Capable of Sensing Molecular Interactions. Nat Biotechnol. 2002;20:816. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 139.Zhao M, Josephson L, Tang Y, Weissleder R. Magnetic Sensors for Protease Assays. Angew Chem Int Ed. 2003;42:1375. doi: 10.1002/anie.200390352. [DOI] [PubMed] [Google Scholar]
- 140.Tsourkas A, Hofstetter O, Hofstetter H, Weissleder R, Josephson L. Magnetic Relaxation Switch Immunosensors Detect Enantiomeric Impurities. Angew Chem Int Ed. 2004;43:2395. doi: 10.1002/anie.200352998. [DOI] [PubMed] [Google Scholar]
- 141.Perez JM, Josephson L, Weissleder R. Use of Magnetic Nanoparticles as Nanosensors to Probe for Molecular Interactions. ChemBioChem. 2004;5:261. doi: 10.1002/cbic.200300730. [DOI] [PubMed] [Google Scholar]
- 142.Yigit MV, Mazumdar D, Kim HK, Lee JH, Odintsov B, Lu Y. Smart “Turn-On” Magnetic Resonance Contrast Agents Based on Aptamer-Functionalized Superparamagnetic Iron Oxide Nanoparticles. ChemBioChem. 2007;8:1675. doi: 10.1002/cbic.200700323. [DOI] [PubMed] [Google Scholar]
- 143.Yigit MV, Mazumdar D, Lu Y. Mri Detection of Thrombin with Aptamer Functionalized Superparamagnetic Iron Oxide Nanoparticles. Bioconjug Chem. 2008;19:412. doi: 10.1021/bc7003928. [DOI] [PubMed] [Google Scholar]
- 144.Katz E, Willner I. Biomolecule-Functionalized Carbon Nanotubes: Applications in Nanobioelectronics. ChemPhysChem. 2004;5:1084. doi: 10.1002/cphc.200400193. [DOI] [PubMed] [Google Scholar]
- 145.Patolsky F, Katz E, Willner I. Amplified DNA Detection by Electrogenerated Biochemiluminescence and by the Catalyzed Precipitation of an Insoluble Product on Electrodes in the Presence of the Doxorubicin Intercalator. Angew Chem Int Ed. 2002;41:3398. doi: 10.1002/1521-3773(20020916)41:18<3398::AID-ANIE3398>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 146.Patolsky F, Lichtenstein A, Willner I. Highly Sensitive Amplified Electronic Detection of DNA by Biocatalyzed Precipitation of an Insoluble Product onto Electrodes. Chem Eur J. 2003;9:1137. doi: 10.1002/chem.200390131. [DOI] [PubMed] [Google Scholar]
- 147.Patolsky F, Lichtenstein A, Willner I. Detection of Single-Base DNA Mutations by Enzyme-Amplified Electronic Transduction. Nat Biotechnol. 2001;19:253. doi: 10.1038/85704. [DOI] [PubMed] [Google Scholar]
- 148.Polsky R, Gill R, Kaganovsky L, Willner I. Nucleic Acid-Functionalized Pt Nanoparticles: Catalytic Labels for the Amplified Electrochemical Detection of Biomolecules. Anal Chem. 2006;78:2268. doi: 10.1021/ac0519864. [DOI] [PubMed] [Google Scholar]
- 149.Mir M, Vreeke M, Katakis I. Different Strategies to Develop an Electrochemical Thrombin Aptasensor. Electrochem Commun. 2006;8:505. [Google Scholar]
- 150.Ikebukuro K, Kiyohara C, Sode K. Novel Electrochemical Sensor System for Protein Using the Aptamers in Sandwich Manner. Biosens Bioelectron. 2005;20:2168. doi: 10.1016/j.bios.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 151.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. An Electronic, Aptamer-Based Small-Molecule Sensor for the Rapid, Label-Free Detection of Cocaine in Adulterated Samples and Biological Fluids. J Am Chem Soc. 2006;128:3138. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 152.Wu Z-S, Guo M-M, Zhang S-B, Chen C-R, Jiang J-H, Shen G-L, Yu R-Q. Reusable Electrochemical Sensing Platform for Highly Sensitive Detection of Small Molecules Based on Structure-Switching Signaling Aptamers. Anal Chem. 2007;79:2933. doi: 10.1021/ac0622936. [DOI] [PubMed] [Google Scholar]
- 153.Zuo X, Song S, Zhang J, Pan D, Wang L, Fan C. A Target-Responsive Electrochemical Aptamer Switch (TREAS) for Reagentless Detection of Nanomolar ATP. J Am Chem Soc. 2007;129:1042. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 154.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Label-Free Electronic Detection of Thrombin in Blood Serum by Using an Aptamer-Based Sensor. Angew Chem Int Ed. 2005;44:5456. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 155.Radi A-E, Acero Sanchez JL, Baldrich E, O’Sullivan CK. Reagentless, Reusable, Ultrasensitive Electrochemical Molecular Beacon Aptasensor. J Am Chem Soc. 2006;128:117. doi: 10.1021/ja053121d. [DOI] [PubMed] [Google Scholar]
- 156.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. A Reagentless Signal-on Architecture for Electronic, Aptamer-Based Sensors Via Target-Induced Strand Displacement. J Am Chem Soc. 2005;127:17990. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 157.Du Y, Li B, Wei H, Wang Y, Wang E. Multifunctional Label-Free Electrochemical Biosensor Based on an Integrated Aptamer. Anal Chem. 2008;80:5110. doi: 10.1021/ac800303c. [DOI] [PubMed] [Google Scholar]
- 158.Hansen JA, Wang J, Kawde A-N, Xiang Y, Gothelf KV, Collins G. Quantum-Dot/Aptamer-Based Ultrasensitive Multi-Analyte Electrochemical Biosensor. J Am Chem Soc. 2006;128:2228. doi: 10.1021/ja060005h. [DOI] [PubMed] [Google Scholar]
- 159.Zayats M, Huang Y, Gill R, Ma C-a, Willner I. Label-Free and Reagentless Aptamer-Based Sensors for Small Molecules. J Am Chem Soc. 2006;128:13666. doi: 10.1021/ja0651456. [DOI] [PubMed] [Google Scholar]
- 160.Iijima S. Helical Microtubules of Graphitic Carbon. Nature. 1991;354:56. [Google Scholar]
- 161.Ajayan PM. Nanotubes from Carbon. Chem Rev. 1999;99:1787. doi: 10.1021/cr970102g. [DOI] [PubMed] [Google Scholar]
- 162.Dai H. Carbon Nanotubes: Synthesis, Integration, and Properties. Acc Chem Res. 2002;35:1035. doi: 10.1021/ar0101640. [DOI] [PubMed] [Google Scholar]
- 163.Kam NW, Liu Z, Dai H. Carbon Nanotubes as Intracellular Transporters for Proteins and DNA: An Investigation of the Uptake Mechanism and Pathway. Angew Chem Int Ed. 2006;45:577. doi: 10.1002/anie.200503389. [DOI] [PubMed] [Google Scholar]
- 164.Nguyen CV, Delzeit L, Cassell AM, Li J, Han J, Meyyappan M. Preparation of Nucleic Acid Functionalized Carbon Nanotube Arrays. Nano Lett. 2002;2:1079. [Google Scholar]
- 165.Hazani M, Naaman R, Hennrich F, Kappes MM. Confocal Fluorescence Imaging of DNA-Functionalized Carbon Nanotubes. Nano Lett. 2003;3:153. [Google Scholar]
- 166.Dwyer C, Guthold M, Falvo M, Washburn S, Superfine R, Erie D. DNA-Functionalized Single-Walled Carbon Nanotubes. Nanotechnology. 2002;13:601. [Google Scholar]
- 167.Baker SE, Cai W, Lasseter TL, Weidkamp KP, Hamers RJ. Covalently Bonded Adducts of Deoxyribonucleic Acid (DNA) Oligonucleotides with Single-Wall Carbon Nanotubes: Synthesis and Hybridization. Nano Lett. 2002;2:1413. [Google Scholar]
- 168.Chen RJ, Zhang Y, Wang D, Dai H. Noncovalent Sidewall Functionalization of Single-Walled Carbon Nanotubes for Protein Immobilization. J Am Chem Soc. 2001;123:3838. doi: 10.1021/ja010172b. [DOI] [PubMed] [Google Scholar]
- 169.Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NWS, Shim M, Li Y, Kim W, Utz PJ, Dai H. Noncovalent Functionalization of Carbon Nanotubes for Highly Specific Electronic Biosensors. Proc Natl Acad Sci USA. 2003;100:4984. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.So H-M, Won K, Kim YH, Kim B-K, Ryu BH, Na PS, Kim H, Lee J-O. Single-Walled Carbon Nanotube Biosensors Using Aptamers as Molecular Recognition Elements. J Am Chem Soc. 2005;127:11906. doi: 10.1021/ja053094r. [DOI] [PubMed] [Google Scholar]
- 171.Maehashi K, Katsura T, Kerman K, Takamura Y, Matsumoto K, Tamiya E. Label-Free Protein Biosensor Based on Aptamer-Modified Carbon Nanotube Field-Effect Transistors. Anal Chem. 2007;79:782. doi: 10.1021/ac060830g. [DOI] [PubMed] [Google Scholar]
- 172.So HM, Park DW, Jeon EK, Kim YH, Kim BS, Lee CK, Choi SY, Kim SC, Chang H, Lee JO. Detection and Titer Estimation of Escherichia Coli Using Aptamer-Functionalized Single-Walled Carbon-Nanotube Field-Effect Transistors. Small. 2008;4:197. doi: 10.1002/smll.200700664. [DOI] [PubMed] [Google Scholar]
- 173.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Quantum Dot-Aptamer Conjugates for Synchronous Cancer Imaging, Therapy, and Sensing of Drug Delivery Based on Bi-Fluorescence Resonance Energy Transfer. Nano Lett. 2007;7:3065. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 174.Bagalkot V, Farokhzad OC, Langer R, Jon S. An Aptamer-Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform. Angew Chem Int Ed. 2006;45:8149. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 175.Wang AZ, Bagalkot V, Vasilliou CC, Gu F, Alexis F, Zhang L, Shaikh M, Yuet K, Cima MJ, Langer R, Kantoff PW, Bander NH, Jon S, Farokhzad OC. Superparamagnetic Iron Oxide Nanoparticle-Aptamer Bioconjugates for Combined Prostate Cancer Imaging and Therapy. ChemMedChem. 2008;3:1311. doi: 10.1002/cmdc.200800091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Targeted Delivery of Cisplatin to Prostate Cancer Cells by Aptamer Functionalized Pt(IV) Prodrug-Plga-Peg Nanoparticles. Proc Natl Acad Sci USA. 2008;105:17356. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang Y-F, Chang H-T, Tan W. Cancer Cell Targeting Using Multiple Aptamers Conjugated on Nanorods. Anal Chem. 2008;80:567. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 178.Wu Y, Phillips JA, Liu H, Yang R, Tan W. Carbon Nanotubes Protect DNA Strands During Cellular Delivery. ACS Nano. 2008;2:2023. doi: 10.1021/nn800325a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tong GJ, Hsiao SC, Carrico ZM, Francis MB. Viral Capsid DNA Aptamer Conjugates as Multivalent Cell-Targeting Vehicles. J Am Chem Soc. 2009;131:11174. doi: 10.1021/ja903857f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cao Z, Tong R, Mishra A, Xu W, Wong GCL, Cheng J, Lu Y. Reversible Cell-Specific Drug Delivery with Aptamer-Functionalized Liposomes. Angew Chem Int Ed. 2009;48:6494. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 181.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. Antiproliferative Activity of G-rich Oligonucleotides Correlates with Protein Binding. J Biol Chem. 1999;274:26369. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]